Abstract
Although recombinant adeno-associated viral (rAAV) vectors are promising tools for gene therapy of genetic disorders, they remain mostly episomal and hence are lost during cell replication. For this reason, rAAV vectors capable of chromosomal integration would be desirable. Ribosomal DNA (rDNA) repeat sequences are overrepresented during random integration of rAAV. We therefore sought to enhance AAV integration frequency by including 28S rDNA homology arms into our vector design. A vector containing ~1 kb of homology on each side of a cDNA expression cassette for human fumarylacetoacetate hydrolase (FAH) was constructed. rAAV of serotypes 2 and 8 were injected into Fah-deficient mice, a model for human tyrosinemia type 1. Integrated FAH transgenes are positively selected in this model and rDNA-containing AAV vectors had a ~30× higher integration frequency than controls. Integration by homologous recombination (HR) into the 28S rDNA locus was seen in multiple tissues. Furthermore, rDNA-containing AAV vectors for human factor IX (hFIX) demonstrated increased transgene persistence after liver regeneration. We conclude that rDNA containing AAV vectors may be superior to conventional vector design for the treatment of genetic diseases, especially those associated with increased hepatocyte replication.
Introduction
Recombinant adeno-associated viral (rAAV) vector currently is one of the most promising gene therapy vectors. It has been used with success in many animal models, but recently also in human clinical trials.1 Treatment of Leber's congenital amaurosis by intraocular injection of serotype 2 rAAV expressing the RPE65 gene resulted in clinical improvement in multiple studies.2,3 More recently, hepatic gene transfer with AAV serotype 8 was successfully used in hemophilia B (factor IX deficiency).4 Generally, the safety profile of these vectors has been excellent.5
For genetic liver diseases, long-term AAV-mediated transgene expression at therapeutic levels would be highly desirable. However, in animal models the vast majority of rAAV genomes in liver remain episomal and <5% of the expression persists after partial hepatectomy.6 In healthy humans, hepatocytes are known to divide every 200–300 days7 and this rate is predicted to be significantly higher with liver injury such as viral hepatitis. Many common infections are associated with mild elevations in serum transaminases8 suggesting that the therapeutic benefit of non-integrated rAAV in the liver will be transient.
AAV2 vectors can be pseudotyped with capsids from diverse AAV types and many different serotypes have been developed.3,9,10 Much effort has been invested into the modification and evolution of AAV capsids to increase transduction efficiency of different tissues.11,12,13 To date, however, limited progress has been made in terms of developing rAAV vectors capable of more efficient chromosomal integration.
Large-scale analyses of AAV integration sites in hepatocytes have been performed to understand the mechanisms and patterns of integration.14,15 Studies in the mouse model of HT1 showed that about 60% of rAAV vector integrations occurred in active genes.16 In addition, integration “hot spots” were found and one of the surprising findings was that 3–8% of integrated vectors resided in ribosomal DNA (rDNA) repeats. Even though this sequence has ~200 copies/haploid genome, this frequency corresponds to a strong overrepresentation in terms of integration specificity. Given the preference of unmodified rAAV vectors for integration into rDNA and knowing the uncoating of the virus occurs in the nucleolus,14,15,17 we wished to ask whether introduction of rDNA homology into rAAV vectors would increase integration frequency, especially into rDNA repeats, by homologous recombination (HR) or nonhomologous end joining.
Here, we report the application of a rDNA homology-containing vector for in vivo gene therapy focusing on a model system, which permits selection of hepatic transgenes. The accompanying paper by Lisowski et al. explores this system with a non-selective transgene for the clinically important disease hemophilia B.
Results
AAV-rDNA-hFAH results in significantly higher integration frequency
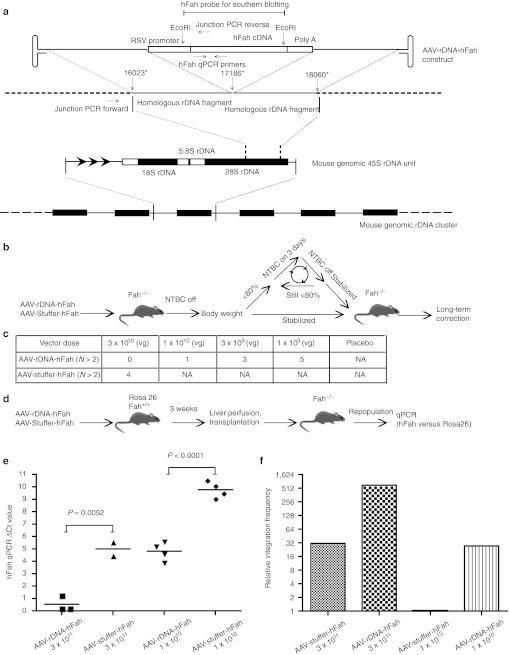
In order to increase the integration frequency of rAAV into chromosomal DNA, a novel vector genome containing DNA from the 28S ribosomal repeat DNA gene was designed with the purpose of achieving site-specific HR. This specific sequence was chosen because rAAV has a propensity to integrate into rDNA,14,16 a sequence which is repeated in 100s of very similar copies throughout the genome. A fumarylacetoacetate hydrolase (FAH) expression cassette was cloned into the middle of a ~2.2 kb fragment of 28S rDNA generating homology arms of ~1 kb on either side (Figure 1a) and then inserted into an AAV2 genome.
Figure 1.
AAV-rDNA-hFah yields about 30 times higher integration frequency. (a) AAV-rDNA-hFah structure and homologous recombination into 28S rDNA. The overall structure of the mouse 45S rDNA unit and the location of the region used in the rDNA vector are shown. (b) Schematic of Fah−/− liver function rescue experiment. (c) Comparison of the dose responses of AAV8-rDNA-hFah and AAV-stuffer-hFah. The NTBC cycle number needed to rescue the Fah−/− mice is given. At each virus genome (vg) dose, AAV-rDNA-hFah required fewer NTBC cycles to rescue liver function. (d) Experimental design for the comparison of AAV-rDNA-hFah and AAV-stuffer-hFah without transgene selection. (e) Copy number determination by qPCR. The δCt for hFAH versus Rosa26, (donor hepatocyte marker) is shown for AAV-rDNA-hFah and AAV-stuffer-hFah. All hFAH and Rosa26 PCR Ct values were normalized to the single copy gene Maai. (f) Relative total integration frequencies at the indicated dose. All integration frequencies were normalized to that of AAV-stuffer-hFAH at 1 × 1010 (3rd bar), which was arbitrarily set to 1. AAV, adeno-associated viral vector; NA, not applicable; NTBC, 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclohexanedione; qPCR, quantitative PCR; RSV, Rous sarcoma virus; rDNA, ribosomal DNA.
This vector (rDNA-hFAH AAV) was packaged into AAV8 and AAV2 capsids and injected into Fah−/− mice of different ages. As in our previous experiments using conventional vector designs,16 we used Fah knockout mice, because this model permits in vivo selection of Fah-expressing hepatocytes. Episomal transgenes are lost because of injury-induced hepatocyte turnover and only cells with integrated transgenes can survive.6,16 In order to exclude simple size differences as the reason for different integration frequencies, a control vector was generated in which the identical FAH-expression cassette was flanked by single copy intronic sequence of the same size. Thus both the rDNA-hFAH and the control stuffer-hFAH vectors were about 4.3 kb in total length.
To determine whether the rDNA-hFAH vector yielded a higher integration frequency, we first established the minimal vector dose required to rescue the lethal liver failure phenotype of Fah knockout mice. One of the important features of this model is that near-complete liver repopulation and phenotypic rescue of treated animals can be achieved even if the frequency of Fah+ hepatocytes is very low.18 To obtain clinical correction, multiple cycles of 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclohexanedione (NTBC) withdrawal and readministration are used. The animals initially loose weight but recover from liver failure while back on NTBC. In each withdrawal cycle, the Fah+ nodules grow in size until a sufficient mass of healthy hepatocytes is reached and liver function is fully restored. The number of NTBC cycles required for animals to achieve normal liver function can be used to estimate the frequency of functional hepatocyte nodules. AAV8-rDNA-hFah and AAV8-stuffer-hFah were injected into five 2-month-old Fah−/− mice each at four different doses (20 mice total for each vector), ranging from 3 × 1010 down to 1 × 109 vector genome (vg)/mouse. After injection, NTBC was stopped to induce liver failure and to permit selection of Fah+-hepatocytes. If drug withdrawal produced weight loss exceeding 20% of the starting weight, NTBC therapy was reinstituted to restore health as schematically shown in Figure 1b. Interestingly, the rDNA-hFAH and stuffer-hFAH cohorts displayed markedly different clinical dose responses. As shown in Figure 1c, AAV-rDNA-hFah directly rescued Fah−/− mice at a dose of 3 × 1010 vg in the very first cycle. None of the treated animals lost more than 20% of body weight and NTBC did not have to be restarted. In stark contrast, animals injected with the same dose of AAV-stuffer-hFAH vector required four cycles of NTBC withdrawal before they stabilized. At lower doses (1 × 1010 or lower), the stuffer vector was not able to rescue Fah−/− mice at all, whereas the AAV-rDNA-hFah vector produced permanently corrected mice at all doses tested. Based on the number of NTBC cycles required, it could be estimated that the clinical rescue dose for the rDNA vector was about 10–30 times lower than for the control vector, lacking rDNA homology. Similar data were obtained with AAV2 (data not shown).
Next, we tested whether the more effective dose-response of the rDNA vector was due to a higher integration frequency. Since the improved functional rescue obtained with the rDNA vector could also be the result of better Fah expression rather than higher integration, we sought to uncouple the measurement of integration frequency from the levels of Fah expression. This was achieved by injecting the AAV vectors into Rosa26 mice, which have normal Fah-activity, followed by transplantation into Fah−/− mice (Figure 1d). In this experiment, every hepatocyte from the AAV-injected primary animal expresses Fah and can therefore be selected in the recipient regardless of whether AAV has integrated. During the expansion of the donor hepatocytes episomal AAV vectors are lost, but integrated AAV genomes are retained, regardless of whether they result in Fah expression or not. Two doses (3 × 1011 and 1 × 1010 vg) of rAAV8-rDNA-hFah and rAAV8-stuffer-hFah were injected into 6-week-old Rosa26 mice (n = 4/each dose). Injected mice were kept for 3 weeks to allow for AAV vector integration and then their hepatocytes were isolated and transplanted into secondary Fah−/− recipients (n = 4/each dose). After repopulation of the secondary recipients by donor hepatocytes, mice were killed and liver genomic DNA was extracted. Quantitative real-time PCR was performed to measure the copy numbers of human FAH cDNA (AAV vector), the Rosa26 gene (marker for donor hepatocytes), and mouse Maai gene (a single copy gene, common to both donor and recipient as internal control). If the AAV-rDNA-hFah had a higher integration frequency than AAV-stuffer-hFah, the copy number of human FAH cDNA would be expected to be higher when normalized to the donor input (Rosa26 gene). Indeed, as seen in Figure 1e, the copy number of hFah was ~21× higher for AAV-rDNA-hFah than the control vector at the lower dose and ~31× higher at the higher dose. These figures fit the clinical dose-response very well and suggested that the superior performance of AAV-rDNA-hFAH was due to a higher integration frequency.
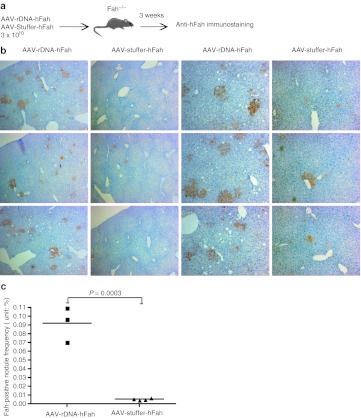
To further confirm this finding, we also measured integration by determining the frequency of Fah+ nodules, similar to experiments done previously for sleeping beauty transposons.19,20 AAV8-rDNA-hFah and control AAV8-stuffer-hFah were injected into 2-month-old Fah−/− mice at a dose of 3 × 1010 vg/mouse (Figure 2a). NTBC was stopped to permit positive selection of Fah+-hepatocyte nodules and mice were killed 3 weeks after injection. Fah immunohistochemistry was done on multiple liver sections and nodule size and number were ascertained. At this low dose, the average nodule frequency for AAV-rDNA-hFah was 1/1,080 (0.092%) and 1/20,000 (0.005%) for the control (Figure 2c), once again confirming the significantly higher integration frequency of the rDNA vector.
Figure 2.
Assessment of integration frequency by nodule analysis. (a) Experimental design. (b) Representative FAH staining (brown color) of liver specimens after AAV infusion after 25 days of selection. Left two panels: ×50 magnification. Right two panels: ×200 magnification. Sections from three separate animals are shown in each column. The vector used is indicated above. The increased nodule frequency obtained with the rDNA vector is clear. (c) Quantitative analysis of nodule frequency. AAV, adeno-associated viral vector; FAH, fumarylacetoacetate hydrolase; rDNA, ribosomal DNA.
AAV-rDNA-hFAH can integrate specifically into rDNA
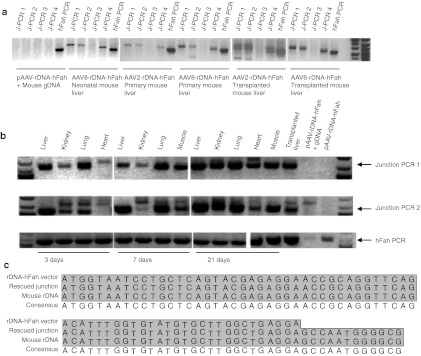
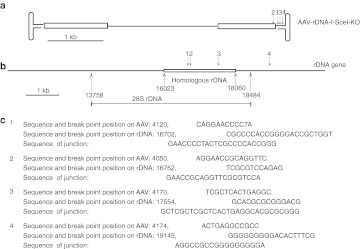
It has been shown previously that rDNA is one of the hot spots for all rAAV vector integration.14,15 We therefore wanted to determine whether the inclusion of rDNA homology into the vector genome would enhance integration into this region by site-specific HR. The presence of 28S rDNA HR events was ascertained by performing PCR with primers specifically designed to amplify junction fragments unique to such events. Four independent primer sets were developed in order to ensure maximum sensitivity and specificity of the assay. As a control for template switching during PCR, we mixed circular vector plasmid with mouse genomic DNA and demonstrated that no junction-specific PCR products could be amplified from such samples (Supplementary Figure S1); 1 × 1010 vg of AAV8-rDNA-hFAH were injected into four neonatal Fah−/− mice by facial vein injection and NTBC was stopped immediately. Interestingly, all injected mice showed complete phenotypic rescue and became NTBC independent. Given the high turnover of liver cells in neonates and the associated loss of episomes, this result again supports a high frequency of integration by the rDNA vector. Multiple junction PCRs were positive in the liver of all injected animals (Figure 3a and Supplementary Figure S1), indicating that site-specific HR indeed occurred. To better understand the kinetics of rDNA-mediated integration, two 2-month-old mice injected with different doses of AAV-rDNA-hFAH vector were harvested at 3, 7, and 21 days after injection and multiple tissues were analyzed by a junction PCR assay. Site-specific rDNA locus integration was detected as early as 3 days in liver, kidney, and lung, but not in heart (Figure 3b). At 7 days, HR was seen in skeletal muscle and by 21 days the heart became positive. These results demonstrate that rDNA vectors can integrate into the genome by HR in multiple different tissues, but have different temporal kinetics with muscle tissues being delayed in comparison to solid organs. To confirm the specificity of the PCR reaction, junction PCR products were cloned and sequenced (Figure 3c), showing perfect alignment with genomic 28S rDNA sequences.
Figure 3.
Site-specific integration of AAV-rDNA-hFah. (a) AAV-rDNA-hFah–mediated site-specific rDNA integration was detected by different sets of junction PCRs (J-PCR1, J-PCR2, J-PCR3, J-PCR4) in primary infected and secondary transplanted liver as well as neonatal mouse liver. One primer was located within the Fah transgene (junction PCR reverse in Figure 1a) and one within rDNA adjacent to the homology (junction PCR forward, Figure 1a). A PCR for the human Fah transgene (hFah PCR) served as positive control for presence of the vector. J-PCRs 1, 2, a nd 4 were positive in multiple samples of AAV-rDNA-hFah–injected liver. The far left panel shows that junction PCRs were negative when genomic DNA was mixed with unintegrated pAAV-rDNA-hFah plasmid. (b) Site-specific integration was detected in liver, kidney, lung, heart, and muscle, as early as 3 days postinjection. (c) Alignment of rescued junction fragment sequences with genomic rDNA sequence. AAV, adeno-associated viral vector; gDNA, genomic DNA; rDNA, ribosomal DNA.
AAV-rDNA-hFAH copy number and site-specific integration ratio
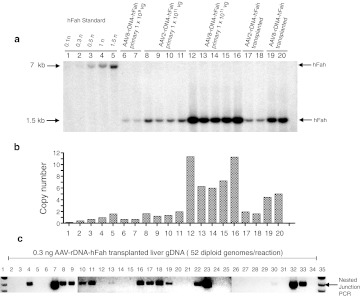
The data described above clearly indicated that site-specific 28S rDNA integration did indeed occur reproducibly, but the precise frequency of such events remained unknown. Given the fact, that the AAV-rDNA-hFAH vector integrates about 30× more frequently than the stuffer control, one possibility was that HR accounted for the majority of this increase. To address this possibility, we performed Southern blot analysis of liver DNA from Fah−/− mice which had been rescued by AAV-rDNA-hFAH injection, i.e., undergone selection for vector integration. DNA was digested with an enzyme that cuts once within the vector genome and we reasoned that a junction-specific band would be visible, if at least 10% of the integration events had occurred by HR in the 28S rDNA locus. While the integrated AAV genomes could be readily detected by using a human FAH probe, no junction fragments were seen (Figure 4a and Supplementary Figure S2). The band sizes seen with single cutters were indicative of concatemeric genomes (Supplementary Figure S2). This result indicated that the frequency of integration by HR was below the detection limit of a Southern blot and hence represented <10% of all integration events. The Southern blot was informative in regards to the number of integrated AAV copies per hepatocyte (Figure 4a). With serotype 2, each selected hepatocyte had 1–3 copies of hFAH per diploid genome equivalent. AAV8 produced higher copy numbers of ~6–13 vg/cell. Thus, the majority of AAV integration events found were concatemeric with multiple copies.
Figure 4.
Analysis of integrated AAV-rDNA-hFah. (a) hFAH southern blot of liver DNA after selection for AAV-rDNA-hFah integration and full repopulation (>90%). Each lane is from one mouse from either primary repopulation after virus injection (lanes 6–16) or from serial transplant recipients of virally rescued livers (lanes 17–20). The Fah+ hepatocytes from serial transplantation have undergone at least 16 rounds of replication. Both AAV2 and AAV8 were used. A dilution series of linear 7.0 kb plasmid pAAV-rDNA-hFah (0.1–1.5 copies per diploid genome) was used for quantitation (left lanes).Location of probe and EcoR1 restriction sites shown in Figure 1a. (b) Quantification of integrated vector copy numbers by phosphorimager readings. (c) Nested junction PCR on 0.3 ng of genomic DNA/sample from liver fully repopulated with AAV-rDNA-hFah–transduced hepatocytes. Lanes 2–33: individual 0.3 ng samples. Lane 34: water control. Lanes 1 and 35: size markers. AAV, adeno-associated viral vector; FAH, fumarylacetoacetate hydrolase; gDNA, genomic DNA; rDNA, ribosomal DNA; vg, vector genome.
Since the frequency of 28S integration by HR was below the detection limit for Southern blot analysis, we devised a PCR-based method to estimate the rate. The DNA from hepatocytes, which had undergone selection for hFAH vector integration was diluted such that each PCR tube contained the equivalent of 1, 5, 20, 50, and 100 hepatocytes. Next, 30–50 samples of each DNA amount was subjected to a highly sensitive nested junction PCR (Figure 4c). The number of samples positive for the junction PCR in each group was determined and the Poisson distribution was used to infer the number of cells that had at least one HR event (Figure 4c). When 0.3 ng of AAV8-injected adult hepatocyte repopulated liver genomic DNA per tube was used (= ~52 diploid genomes), 16/32 samples were PCR positive. Using Poisson distribution mathematics (see Materials and Methods) this indicated that 1.32% of hepatocytes had a HR event. This number represents a minimal estimate, as some events could have been missed due to limitations of the sensitivity of the PCR. In addition, not all liver DNA comes from hepatocytes and the majority of hepatocytes are polyploid.21 Taken these factors into account, we estimated that ~2–3% of all selected hepatocytes had a 28S rDNA site-specific integration event. This number is compatible with the lack of a detectable junction fragment on Southern blot.
Since site-specific HR could not explain the clearly documented increase in integration frequency, we used a plasmid rescue strategy14,15,16 (Supplementary Figure S3) and were able to isolate 12 genomic integration sites. Of these 4 (33%) were located within the rDNA locus (Supplementary Table S1), ~100× more frequent than expected since rDNA represents only 0.3% of the genome, and about 10 times higher than previously reported frequency of integration into rDNA loci for traditional AAVs.14 Junctions were not perfect and contained deletions, indicating that integration did not occur by homologous recombination (Figure 5).
Figure 5.
Analysis of AAV-rDNA integration sites in the rDNA region. (a) Schematic of four integration breakpoints (from Supplementary Table S1) on the AAV-rDNA vector. The AAV-rDNA vector genome is 4,354 nucleotides in length. The terminal 145 nucleotides (positions 1–145 and 4110–4254) represent inverted terminal repeats (ITRs). (b) Location of integration breakpoints on rDNA relative to the homologous sequence (position on rDNA: 13412–15451) present within the AAV-rDNA vector. Position is based on rDNA sequence GenBank: X82564.1. (c) Junction sites sequence alignments with the AAV-rDNA vector and rDNA locus. AAV, adeno-associated viral vector; rDNA, ribosomal DNA.
Improved persistence of factor IX levels after gene therapy with an rDNA AAV vector
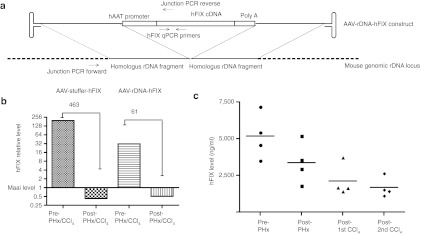
As a preliminary test, whether the rDNA vector design could also work for another disease model lacking the feature of transgene selection, we generated an AAV-rDNA-hFIX as shown in Figure 6a. This vector contained rDNA homology arms similar to those of the hFAH vector. Four 8-week-old wild-type mice were injected with AAV serotype 8 at a dose of 3 × 1011 vg/mouse. Fourteen days after injection the baseline levels of human factor IX (hFIX) were measured and then, the mice were sequentially subjected to partial hepatectomy and two sublethal CCl4 treatments. These manipulations induce high levels of hepatocyte turnover and hence eliminate most episomal AAV genomes. The serum levels of hFIX were measured after each manipulation. Importantly, about 40% of the initial hFIX level was still present despite massive hepatocyte replication in mice injected with the rDNA vector (Figure 6c). In contrast, as previously shown, only 5–10% of the hFIX levels were present in those mice injected with the control stuffer-hFIX vector.6 In order to determine whether the improved persistence of FIX levels was due to a higher frequency of transgene integration, quantitative PCR was used to measure hFIX DNA copy number before and after liver replication. This was done by comparing the DNA of the liver lobe removed by partial hepatectomy to liver DNA obtained at final harvest. On average, the copy number of hFIX had dropped 61-fold in AAV-rDNA-hFIX–injected mice while the difference was 480-fold when the stuffer was used (Figure 6b). Most of this drop of course reflects loss of episomes, but the data show that a higher percentage of the vector genomes are resistant to cell turnover when a rDNA vector was used.
Figure 6.
AAV-rDNA-hFIX demonstrates improved transgene persistence. (a) Structure of the AAV-rDNA-hFIX vector. (b) Copy number changes of the hFIX transgene after liver regeneration by qPCR. The Maai copy number served as an internal control. (c) hFIX serum levels after partial hepatectomy and two sublethal CCl4 treatments. About 30% of the expression persisted. AAV, adeno-associated viral vector; hFIX, human factor IX; qPCR, quantitative PCR; rDNA, ribosomal DNA.
Discussion
rAAV-mediated gene therapy is rapidly emerging as the currently best technology for the treatment of some monogenic disorders, particularly those affecting the retina2 and liver,22 but also skeletal23 and cardiac muscle24 and the central nervous system.25 Although cell turnover rates in these tissues are thought to be low or absent in the absence of injury, cell division clearly occurs in hepatocytes even during normal homeostasis.26 Mild liver damage and hepatocellular death are a very common feature of many disorders, including viral infections27as well as common nutritional28 and drug-induced29 insults. For this reason, it can be predicted that therapeutic gene expression will not be permanent after gene transfer into hepatocytes if the transgene does not integrate into the chromosome. rAAV vectors are well known to remain mostly episomal and be lost quickly even after one round of cell replication.6,30 Permanent, life-long gene expression is, of course, desirable in the treatment of inherited disorders and therefore transgene persistence is an important issue. There are multiple strategies that may overcome this problem. One way to achieve transgene persistence is to readminister another round of rAAV therapy. The immune responses to readministration22 could be overcome by switching serotypes.31 Alternatively, chromosomal integration of the rAAV vector would also achieve the desired goal. Here, two basic strategies can be considered. The first is gene repair, in which the disease-causing mutation is removed by HR.32,33,34 This approach has the advantage of reproducing the natural levels of gene regulation and expression. Indeed, AAV vectors are particularly capable of producing gene repair33,35 even in vivo.32,33,36 Furthermore, the induction of DNA double-strand breaks near the mutation, using for example zinc finger nucleases, can enhance the efficiency of this approach.36 A major disadvantage of the gene repair strategy, however, is the fact that rAAV to be used is mutation specific. In the case of larger genes, different rAAV vectors would be required for each and every different mutation, causing practical and regulatory hurdles.
The second strategy is the one used by us and the accompanying paper.37 Chromosomal integration is targeted to a safe harbor,38 which could be applicable for transgene expression in many different disorders. We targeted 28S rDNA in our experiments, but other loci could also be considered in the future. Overall, we observed a significant increase (~10–30×) of transgene integration and persistence when rAAV vectors contained 28S rDNA homology were used. It is important to note that the integration frequency of ~1/1,000 in Figure 2 was achieved at low vector doses for the purpose of retaining a countable number of integration nodules. At higher doses, the rDNA-hFAH AAV vector produced integration in 2–5% of all hepatocytes. This increased frequency of stable integration lies considerably below frequencies that have been reported for retroviral vectors.39 Nonetheless, the increased integration frequency had functional significance. The rescue dose for the treatment of tyrosinemia was ~30-fold lower than for conventional vectors. In addition, a higher percentage of FIX expression was retained after liver regeneration (see also accompanying paper).
In the case of the hFAH transgene, we found only 2–4% of all integration events to have resulted from HR in 28S rDNA. Thus, the 30-fold increase in overall integration frequency cannot be explained by homologous integration. Nonetheless, our own limited integration site analysis, as well as the more extensive linear amplification-mediated–PCR done in the accompanying paper37 suggest a strong enrichment for nonhomologous integrations into rDNA. It is interesting to note that wild-type AAV also displays the capability of integrating into a particular genomic location (AAVS1 on human chromosome 19) without using HR.38 The exact mechanism by which the presence of rDNA homology used here can change the pattern of genomic integration is unclear. Nonetheless, the increased integration frequency of such vectors is desirable for the treatment of genetic diseases. rDNA-containing AAV vectors should be considered when therapeutic transgenes are small enough to accommodate the inclusion of rDNA homology arms.
Materials and Methods
AAV vector construction, virus preparation, and injection. A 2,038 bp fragment of the 28S rDNA locus of the mouse was amplified from wild-type 129s4 strain mouse genomic DNA with the following primers: forward: 5′-GCGGCCGCAAAGGGAGTCGGGTTCAGAT-3′, reverse: 5′-GCGGCC GCTCCTCAGCCAAGCACATACA-3′. This fragment was cloned into the blunted NotI sites of a pAAV2 vector16 generating the plasmid pAAV-rDNA. A human FAH-expression cassette containing a Rous sarcoma virus (RSV) promoter, hFAH cDNA, and bovine growth hormone polyadenylation sequence A was amplified from plasmid pRSV-hFah40 (forward primer: CTTAAGGCTGCTTCGCGATGTACG, reverse: CTTAA GCCATAGAGCCCACCGC ATC) and cloned into a unique Afl II site centered in the middle of pAAV-rDNA, thus generating pAAV-rDNA-hFAH. The overall insert length was 2,250 bp. To make a control vector of comparable size, pAAV-stuffer-hFah was constructed. A 1,968 bp fragment of mouse Fah gene intron 3 was subcloned to pAAV plasmid Not I sites after amplification with the following primers, forward: 5′-TTGCGGCC GCATTCCCAC CCACTCTCCTCT-3′, reverse: GCGGCCGCGGGGTTGGATTCAATGTTTG, generating pAAV-stuffer. Next, the RSV-hFah-PolyA expression cassette was subcloned into the Afl II site of pAAV-stuffer. To construct pAAV-rDNA-hFIX, a 2,280 fragment of a hFIX expression cassette was amplified from pNEB193-SynEnh-TTR-hFIX10-90-spa (courtesy of M.A. Kay at Stanford University, Stanford, CA) and subcloned into the AflII site of pAAV-rDNA.
These plasmids were used to generate rAAV vectors of capsid serotypes 2 and 816,41 by transient transfection of HEK293 using published technology.42 In adult animals, all virus injections were done intravenously by retro-orbital administration in 200 µl of sterile saline at the doses indicated. For neonatal animals, facial vein injections were done with 30 µl.
Animal husbandry and HT1 mouse model. All animal experiments were performed according to the guidelines for animal care at Oregon Health & Science University, Portland, OR. Fah-deficient mice were of the Fahδexon5 strain on the 129s4 background as previously described.43 Fah−/− mice were maintained with drinking water containing 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclohexanedione (NTBC; http://www.yecuris.com) at a concentration of 4 mg·l−1.20 Rosa26 transgenic mice were developed originally by Dr Philippe Soriano and maintained on 129S4 background.44
Hepatocyte transplantation and in vivo selection. For in vivo selection of FAH-expressing hepatocytes with integrated rAAV2 vector genomes, we withdrew NTBC from the drinking water as previously described.20
To further select for hepatocytes containing integrated vector genomes and to dilute non-integrated vector genomes, serial transplantation was performed.45 Primary recipients of AAV injections were subjected to NTBC withdrawal and positive selection of Fah-expressing hepatocytes. After their weight and health had stabilized, the livers of primary animals were perfused and hepatocytes were isolated for serial transplantation according to a previously described protocol.45 For transplantation, 300,000 hepatocytes from AAV-infused primary liver were intrasplenically injected into secondary Fah−/− recipient mice. NTBC was discontinued 1 week after transplant to permit selection of donor hepatocytes.
Histology and immunohistology. AAV-infected or -transplanted Fah−/− mice were killed and specimens were fixed in 10% buffered formalin, sectioned, and immunostained by custom rabbit polyclonal FAH antibody diluted to 1:1,000 and detected with 10 ng/ml goat anti-rabbit secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) as previously described.20
Southern blot analysis. Genomic DNA was isolated from 350 mg of liver tissue from AAV-infected primary or secondary mice using the Qiagen genomic DNA isolation kit (Qiagen, Valencia, CA). Ten microgram of genomic DNA was digested with the restriction enzyme EcoR I (NEB, Ipswich, MA) and subjected to electrophoresis in a 0.9% agarose gel overnight. Capillary transfer and hybridizations were performed according to standard protocols.46 A 1.5 kb fragment containing hFAH cDNA from plasmid pRSV-hFah40 was digested with EcoRI and gel purified with a Qiagen gel purification kit (Qiagen). This fragment was radioactively labeled with the Random Primed DNA Labeling Kit (Roche Applied Science, Indianapolis, IN) with α-32P dCTP (PerkinElmer, Waltham, MA) and used as the probe. Hybridizations were conducted in 0.5 mol/l sodium phosphate pH 7.2, 1 mmol/l EDTA, 7% SDS containing 2 × 106 cpm probe/ml and 100 µg of herring sperm DNA/ml at 65 °C for 36 hours. Washing was performed with 40 mmol/l sodium phosphate pH 7.2, 1% SDS, once at room temperature for 15 minutes and twice at 60 °C for 40 minutes. Signal was visualized and quantitated by a PhosphorImager (GE Healthcare Life Sciences, Piscataway, NJ).
Quantitative real-time PCR. Quantitative real-time PCR was used to measure copy numbers of the human FAH and hFIX transgenes and the donor marker Rosa26. This was done with the Bio-Rad MyiQ single-color real-time PCR detection system using SYBR green dye. The single copy mouse gene Maai (maleylacetoacetate isomerase) was used for normalization. The following primer pairs were used: hFAH: 5′-CCCGTATATGGTGCCTGCAA-3′ and 5′-TCGTGCACTCCAGTCGTTCA-3′ Rosa26: 5′-CATCAGCCGCTACA GTCAACAG-3′ and 5′-CAGCCATGTGCCTTCTTCCGC-3′ Maai: 5′-GT ACCATTGAGGTGGGCCTA-3′ and 5′-GCTGGTTCGTCCACTCTTT C-3′ hFIX: 5′-AAGATGCCAAACCAGGTCAATT-3′ and 5′-GATAGAG CCTCCACAGAATGCA-3′.
The real-time PCR program parameters were: 95 °C × 5′ (95 °C × 15′′, 62 °C × 20′′, 72 °C × 20′′) × 45.
PCR for the detection of site-specific 28S rDNA integration. Junction PCR program (touch-down PCR):95 °C × 5′ (95 °C × 15′′, 65 °C > 55 °C × 20′′, 72 °C × 2′); (95 °C × 15′′, 55 °C × 20′′, 72 °C × 2′) × 35; 72 °C × 20′.
The following primer pairs were used: In each case, one primer was located within the hFAH expression cassette and one was located within 28S rDNA sequence, but outside the homology sequence contained within the rDNA vector (see Figure 1a).
For J-PCR1, forward primer: 5′-CCGCTTTTCGCCTAAACACAC-3′, reverse primer: 5′-CCCAGAAGCAGCAGGTCGTCTCTACG-3′ for J-PCR2, forward primer: 5′-GCCGTATCGTTCCGCCTG-3′, reverse primer: 5′-GTATATCTGGCCCGTACAT-3′for J-PCR3, forward primer: 5′-GTATATCTGGCCCGTACAT-3′, reverse primer: 5′-GCTCCTCAGCC AAGCACA-3′ for J-PCR4, forward primer: 5′-CACCCCGTTTCCC AAGACG-3′, reverse primer: 5′-AACACACCCTAGTCCCCT-3′.
To increase the sensitivity of detecting junction fragments, a nested PCR assay was developed.
The first round of the nested PCR was done with primers located outside of the J-PCR4 primer pair. These were, forward primer: 5′-ccgcttTTCGCCTAAACACAC-3′, reverse primer: 5′-cccagaagcagcaggtcg tctctacg-3′.
One microliter from the first 50 µl PCR reaction was then used as a template for the second round PCR using the same conditions as described for the junction PCR 4 described above.
hFIX analysis. Mouse blood was collected by retro-orbital bleeding into heparin tubes and an ELISA assay was employed to analyze for total hFIX antigen as described previously.6 The specific activity of hFIX in a mouse plasma background may be slightly varied because of unknown interactions of the human protein with mouse plasma proteins.
Quantitation of HR integration by serial dilution junction PCR. Liver genomic DNA from serially transplanted Fah−/− mice was diluted sequentially from 1 µg to 0.01 ng. Thirty separate nested junction PCRs were performed for each DNA amount to identify a concentration at which only a percentage of all reactions was positive. To calculate the integration frequency, we used a Poisson distribution analysis.47,48 P0 = e−x, where P0 is the percentage of samples (PCR reactions) which were positive at a given DNA concentration and x represents the frequency of the event analyzed (integration by HR).
This formula rearranges to: x = ln (1/P0). In our study, the percentage of positive site-specific PCRs was 16 out of 32 using 0.3 ng genomic liver DNA as template per reaction. x = ln (1/(16/32)) = ln2 = 0.693. Therefore, on average 0.693 integrations were present in 0.3 ng genomic DNA. Each diploid mouse genome contains ~5.7 pg genomic DNA, which means 0.3 ng genomic DNA has 52.6 diploid genomes. Hence, the site-specific integration frequency in our sample can be estimated at: 0.693/52.6 = 1.32%.
Measurement of absolute integration frequency. The integration frequency of Fah-expressing AAV vectors was determined by counting FAH+ nodules stained by immunohistochemistry.32 NTBC was withdrawn for only 25 days in order to prevent repopulation nodules from becoming too large and confluent for accurate counting. The nodule size seen in 2D sections was used to calculate a correction factor for the estimation of the number of clonal nodules as previously described.49 The total number of hepatocytes in a section was determined by precisely measuring the surface area of each section; 1 mm2 of a mouse liver section contains about 1,860 hepatocytes.49 Surface area was measured by scanning the slides along with a size standard using a Canon scanner at a resolution of 300 dpi. The pixels of each liver section were counted using Adobe PhotoShop7 software (http://www.adobe.com) and converted into mm2 based on the size standard.
Integration site rescue. The shuttle vector AAV-rDNA-I-SceI-KO was constructed based on plasmids pAAV-rDNA and pNKan (Supplementary Figure S3). AAV8-rDNA-I-SceI-KO was produced and purified as described before.16 We injected 12 adult male wild-type mice via the retro-orbital route with AAV8-rDNA-I-SceI-KO at a dose of 3.0 × 1011 vg/mouse. After 1 month, these mice were killed and livers were immediately frozen in liquid nitrogen and genomic DNA was made from these frozen livers. AAV8-rDNA-I-SceI-KO integration library was constructed as described before.16 Library screening was based on the strategy shown in Supplementary Figure S3. The screened clones were sequenced and the bioinformatics process was done as described.14
SUPPLEMENTARY MATERIAL Figure S1. Homologous recombination junction PCR test and junction enzyme digestion. Figure S2. Southern blotting on AAV-rDNA-hFah–infected liver concatemeric integrations. Figure S3. Schematic of shuttle vector AAV-rDNA-I-Sce-I-KO and PCR screening strategy. Table S1. rAAV integration sites isolated by plasmid rescue.
Acknowledgments
This work was supported by grants from the National Institute of Diabetes & Digestive & Kidney Diseases to M.A.K. (HL064274), H.N. (DK 78388), and M.G. (RO1-DK48252). We thank Angela Major (Texas Children's Hospital) for histology support. We thank Annelise Haft and Jessica Coleman (Oregon Health & Science University) for assistance in animal husbandry. M.G. is a shareholder of Yecuris, Inc. Portland, which sells NTBC and has licensed Fah−/− mice. The other authors declared no conflict of interest.
Supplementary Material
Homologous recombination junction PCR test and junction enzyme digestion.
Southern blotting on AAV-rDNA-hFah–infected liver concatemeric integrations.
Schematic of shuttle vector AAV-rDNA-I-Sce-I-KO and PCR screening strategy.
rAAV integration sites isolated by plasmid rescue.
REFERENCES
- Mingozzi F., and, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J.et al. (2008Safety and efficacy of gene transfer for Leber's congenital amaurosis N Engl J Med 3582240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonelli F, Maguire AM, Testa F, Pierce EA, Mingozzi F, Bennicelli JL.et al. (2010Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration Mol Ther 18643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC.et al. (2011Adenovirus-associated virus vector-mediated gene transfer in hemophilia B N Engl J Med 3652357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High KA., and, Aubourg P. rAAV human trial experience. Methods Mol Biol. 2011;807:429–457. doi: 10.1007/978-1-61779-370-7_18. [DOI] [PubMed] [Google Scholar]
- Nakai H, Yant SR, Storm TA, Fuess S, Meuse L., and, Kay MA. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald RA. “Lifespan” of liver cells. Autoradio-graphic study using tritiated thymidine in normal, cirrhotic, and partially hepatectomized rats. Arch Intern Med. 1961;107:335–343. doi: 10.1001/archinte.1961.03620030023003. [DOI] [PubMed] [Google Scholar]
- Giboney PT. Mildly elevated liver transaminase levels in the asymptomatic patient. Am Fam Physician. 2005;71:1105–1110. [PubMed] [Google Scholar]
- Vandenberghe LH, Wilson JM., and, Gao G. Tailoring the AAV vector capsid for gene therapy. Gene Ther. 2009;16:311–319. doi: 10.1038/gt.2008.170. [DOI] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH., and, Wilson JM. New recombinant serotypes of AAV vectors. Curr Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- Wu P, Xiao W, Conlon T, Hughes J, Agbandje-McKenna M, Ferkol T.et al. (2000Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism J Virol 748635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Asokan A, Grieger JC, Govindasamy L, Agbandje-McKenna M., and, Samulski RJ. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J Virol. 2006;80:11393–11397. doi: 10.1128/JVI.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod A, Ried M, Wobus C, Lahm H, Leike K, Kleinschmidt J.et al. (1999Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2 Nat Med 51438. [DOI] [PubMed] [Google Scholar]
- Nakai H, Wu X, Fuess S, Storm TA, Munroe D, Montini E.et al. (2005Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver J Virol 793606–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Trobridge GD, Petek LM, Jacobs MA, Kaul R., and, Russell DW. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J Virol. 2005;79:11434–11442. doi: 10.1128/JVI.79.17.11434-11442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai H, Montini E, Fuess S, Storm TA, Grompe M., and, Kay MA. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat Genet. 2003;34:297–302. doi: 10.1038/ng1179. [DOI] [PubMed] [Google Scholar]
- Johnson JS., and, Samulski RJ. Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J Virol. 2009;83:2632–2644. doi: 10.1128/JVI.02309-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M.et al. (1996Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I Nat Genet 12266–273. [DOI] [PubMed] [Google Scholar]
- Montini E, Held PK, Noll M, Morcinek N, Al-Dhalimy M, Finegold M.et al. (2002In vivo correction of murine tyrosinemia type I by DNA-mediated transposition Mol Ther 6759–769. [DOI] [PubMed] [Google Scholar]
- Held PK, Olivares EC, Aguilar CP, Finegold M, Calos MP., and, Grompe M. In vivo correction of murine hereditary tyrosinemia type I by phiC31 integrase-mediated gene delivery. Mol Ther. 2005;11:399–408. doi: 10.1016/j.ymthe.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Hickey RD, Paulk NK, Culberson AJ, Olson SB, Finegold MJ.et al. (2009Ploidy reductions in murine fusion-derived hepatocytes PLoS Genet 5e1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ.et al. (2006Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response Nat Med 12342–347. [DOI] [PubMed] [Google Scholar]
- Fisher KJ, Jooss K, Alston J, Yang Y, Haecker SE, High K.et al. (1997Recombinant adeno-associated virus for muscle directed gene therapy Nat Med 3306–312. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhu T, Qiao C, Zhou L, Wang B, Zhang J.et al. (2005Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart Nat Biotechnol 23321–328. [DOI] [PubMed] [Google Scholar]
- Eberling JL, Jagust WJ, Christine CW, Starr P, Larson P, Bankiewicz KS.et al. (2008Results from a phase I safety trial of hAADC gene therapy for Parkinson disease Neurology 701980–1983. [DOI] [PubMed] [Google Scholar]
- Bucher NLR., and, Malt RA. Little Brown: Boston; 1971. Regeneration of Liver and Kidney. [Google Scholar]
- McIntyre N., and, Heathcote J. The laboratory in the diagnosis and management of viral hepatitis. Clin Gastroenterol. 1974;3:317–336. [PubMed] [Google Scholar]
- Ghouri N, Preiss D., and, Sattar N. Liver enzymes, nonalcoholic fatty liver disease, and incident cardiovascular disease: a narrative review and clinical perspective of prospective data. Hepatology. 2010;52:1156–1161. doi: 10.1002/hep.23789. [DOI] [PubMed] [Google Scholar]
- Larsen LC., and, Fuller SH. Management of acetaminophen toxicity. Am Fam Physician. 1996;53:185–190. [PubMed] [Google Scholar]
- Nakai H, Storm TA., and, Kay MA. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J Virol. 2000;74:9451–9463. doi: 10.1128/jvi.74.20.9451-9463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangeli A, Harvey BG, Yao J, Wolff G, Kovesdi I, Crystal RG.et al. (1996“Sero-switch” adenovirus-mediated in vivo gene transfer: circumvention of anti-adenovirus humoral immune defenses against repeat adenovirus vector administration by changing the adenovirus serotype Hum Gene Ther 779–87. [DOI] [PubMed] [Google Scholar]
- Paulk NK, Wursthorn K, Wang Z, Finegold MJ, Kay MA., and, Grompe M. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology. 2010;51:1200–1208. doi: 10.1002/hep.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Wang PR, Petek LM, Hirata RK, Sands MS., and, Russell DW. Gene targeting in vivo by adeno-associated virus vectors. Nat Biotechnol. 2006;24:1022–1026. doi: 10.1038/nbt1231. [DOI] [PubMed] [Google Scholar]
- Paulk NK, Loza LM, Finegold MJ., and, Grompe M. AAV-mediated gene targeting is significantly enhanced by transient inhibition of NHEJ or the proteasome in vivo. Hum Gene Ther. 2012;23:658–665. doi: 10.1089/hum.2012.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Petek LM., and, Russell DW. Human gene targeting by adeno-associated virus vectors is enhanced by DNA double-strand breaks. Mol Cell Biol. 2003;23:3550–3557. doi: 10.1128/MCB.23.10.3550-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS.et al. (2011In vivo genome editing restores haemostasis in a mouse model of haemophilia Nature 475217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowski L, Lau A, Wang Z, Zhang F, Grompe M., and, Kay MA.2012Non-random integrating rAAV-rDNA vectors allow for stable transgene expression from rDNA lociIn this issue). [DOI] [PMC free article] [PubMed]
- McCarty DM, Young SM., Jr, and, Samulski RJ. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu Rev Genet. 2004;38:819–845. doi: 10.1146/annurev.genet.37.110801.143717. [DOI] [PubMed] [Google Scholar]
- Kalpana GV. Retroviral vectors for liver-directed gene therapy. Semin Liver Dis. 1999;19:27–37. doi: 10.1055/s-2007-1007095. [DOI] [PubMed] [Google Scholar]
- Overturf K, al-Dhalimy M, Ou CN, Finegold M, Tanguay R, Lieber A.et al. (1997Adenovirus-mediated gene therapy in a mouse model of hereditary tyrosinemia type I Hum Gene Ther 8513–521. [DOI] [PubMed] [Google Scholar]
- Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J., and, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi VW, Asokan A, Haberman RA., and, Samulski RJ. Production of recombinant adeno-associated viral vectors for in vitro and in vivo use. Curr Protoc Mol Biol. 2007;Chapter 16:Unit 16.25. doi: 10.1002/0471142727.mb1625s78. [DOI] [PubMed] [Google Scholar]
- Grompe M, al-Dhalimy M, Finegold M, Ou CN, Burlingame T, Kennaway NG.et al. (1993Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice Genes Dev 712A2298–2307. [DOI] [PubMed] [Google Scholar]
- Friedrich G., and, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Overturf K, al-Dhalimy M, Ou CN, Finegold M., and, Grompe M. Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol. 1997;151:1273–1280. [PMC free article] [PubMed] [Google Scholar]
- Grompe M, Caskey CT., and, Fenwick RG. Improved molecular diagnostics for ornithine transcarbamylase deficiency. Am J Hum Genet. 1991;48:212–222. [PMC free article] [PubMed] [Google Scholar]
- Fuller SA, Takahashi M., and, Hurrell JG. Cloning of hybridoma cell lines by limiting dilution. Curr Protoc Mol Biol. 2001;Chapter 11:Unit11.8. doi: 10.1002/0471142727.mb1108s01. [DOI] [PubMed] [Google Scholar]
- Brisco MJ, Latham S, Bartley PA., and, Morley AA. Incorporation of measurement of DNA integrity into qPCR assays. BioTechniques. 2010;49:893–897. doi: 10.2144/000113567. [DOI] [PubMed] [Google Scholar]
- Wang X, Montini E, Al-Dhalimy M, Lagasse E, Finegold M., and, Grompe M. Kinetics of liver repopulation after bone marrow transplantation. Am J Pathol. 2002;161:565–574. doi: 10.1016/S0002-9440(10)64212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Homologous recombination junction PCR test and junction enzyme digestion.
Southern blotting on AAV-rDNA-hFah–infected liver concatemeric integrations.
Schematic of shuttle vector AAV-rDNA-I-Sce-I-KO and PCR screening strategy.
rAAV integration sites isolated by plasmid rescue.