Abstract
The Sleeping Beauty (SB) transposon is a nonviral, integrating vector system with proven efficacy in preclinical animal models, and thus holds promise for future clinical applications. However, SB has a close-to-random insertion profile that could lead to genotoxic effects, thereby presenting a potential safety issue. We evaluated zinc finger (ZF) DNA-binding domains (DBDs) for their abilities to introduce a bias into SB's insertion profile. E2C, that binds a unique site in the erbB-2 gene, mediated locus-specific transposon insertions at low frequencies. A novel ZF targeting LINE1 repeats, ZF-B, showed specific binding to an 18-bp site represented by ~12,000 copies in the human genome. We mapped SB insertions using linear-amplification (LAM)-PCR and Illumina sequencing. Targeted insertions with ZF-B peaked at approximately fourfold enrichment of transposition around ZF-B binding sites yielding ~45% overall frequency of insertion into LINE1. A decrease in the ZF-B dataset with respect to transposon insertions in genes was found, suggesting that LINE1 repeats act as a sponge that “soak up” a fraction of SB insertions and thereby redirect them away from genes. Improvements in ZF technology and a careful choice of targeted genomic regions may improve the safety profile of SB for future clinical applications.
Introduction
Transposons have recently been developed as potent, nonviral gene delivery tools.1 Transposon-based gene vectors typically consist of two components: a DNA component comprised of two terminal inverted repeats that flank a transgene cargo of interest, and a protein component, the transposase, that binds to the terminal inverted repeats and catalyzes transposition. During transposition, the transposase excises the cargo from a plasmid and inserts it at a genomic target site (Figure 1a). One thoroughly studied transposon is the Sleeping Beauty (SB) system2 that, owing to stable genomic insertion of expression cassettes, can lead to both long-term and efficient transgene expression in preclinical animal models.3,4 Thus, the establishment of nonviral, integrating vectors generated considerable interest in developing efficient and safe vectors for human gene therapy.3,4 Indeed, the first clinical application of the SB system has been launched using autologous T cells genetically modified to redirect specificity for B-lineage malignancies.5
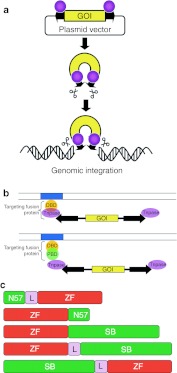
Figure 1.
Sleeping Beauty transposition for gene delivery. (a) The transposon is used as a bi-component vector system for delivering transgenes that are maintained in plasmids. One component contains a gene of interest (GOI) between the transposon terminal inverted repeats (black arrows), the other component is the transposase protein (pink spheres). The transposon is excised from the donor plasmid and is integrated at a chromosomal site by the transposase. (b) Experimental strategies for target-selected transgene integration by transposable element gene vectors. A DNA-binding protein domain (DBD, yellow sphere) recognizes a specific sequence (blue box) in the target DNA (parallel lines). Targeting can be achieved by fusing a DBD either directly to the transposase or by fusing a DBD to a protein binding domain (PBD, green sphere) that interacts with the transposase. (c) Engineered SB transposase constructs used in this study. The ZF domains (red) were fused either N- or C-terminally to the SB transposase (green) or to the N57 protein interaction domain of the SB transposase. In some of the constructs, the ZF and transposase domains were separated by peptide linkers (L).
Similar to most other transposons, target site selection of SB is non-random at the primary DNA sequence level, as it always integrates into TA dinucleotide sites.6 At the genomic scale, SB's insertion profile is close-to-random, as it shows no pronounced preference for inserting into transcription units or transcriptional regulatory regions of genes.6,7,8 Even though chromosomal integration of SB transposons is precise, and no SB-associated adverse effects have been reported,4 random genomic insertion carries the risk of inadvertent mutagenesis of endogenous cellular genes. Thus, target site selection of SB and its experimental manipulation represent an important field of research, because guiding SB transposon insertions to “safe harbors” in the human genome would make the use of the SB transposon system safer for human applications.3,9
For targeted transposon insertion at least one component of the transposon system, either the transposon vector DNA or the transposase (or factors interacting with either of these components) need to be engineered to be physically linked or interact with a heterologous DNA-binding domain (DBD), which is to tether the transposase/transposon complex to defined sites in the human genome, and to facilitate integration of the transposon into adjacent DNA (Figure 1b). Fusions of the SB transposase with the GAL4 DBD showed an enrichment of transposon insertions in ~400-bp window around the targeted sites in plasmid-based assays in cultured human cells.10 Potential SB targeting was also assessed by engineering a LexA operator into a benign site within an SB transposon vector. Targeted transposition events into endogenous chromosomal MAR sequences as well as a chromosomally integrated tetracycline response element were recovered by employing targeting fusion proteins containing LexA and either the SAF-box, a protein domain that binds to chromosomal MARs, or the tetracycline repressor (TetR).11 Finally, a molecular strategy was successfully adapted for targeted SB transposition by taking advantage of the N-terminal helix-turn-helix domain of the SB transposase spanning 57 amino acids (N57), previously shown to mediate protein–protein interactions between transposase subunits.12 Coexpression of the SB transposase with a targeting fusion protein consisting of TetR and N57 promoted targeted SB transposition within a 2.5-kb window around a chromosomally located tetracycline response element at a frequency of >10%.11 The most significant advantage of such molecular design is that the transposase itself does not need to be engineered, thereby eliminating negative effects on transposition activity of direct transposase fusions.
In order to target several, physiologically relevant sites in the human genome at will, a repertoire of DBDs that can be designed to bind essentially any sequence would be highly beneficial. Zinc finger (ZF) proteins of the C2H2-type with their modular structure could provide such DBDs, because stitching together several individual fingers is expected to result in proteins with substantial specificities in target DNA binding.13 In the present work, two approaches were taken to target SB transposon insertions in the human genome, both dependent on engineered fusions of ZFs with the SB transposase and/or its N57 protein interaction domain (Figure 1c). First, we evaluated an already established artificial six-finger ZF called E2C,14 which binds a unique, 18-bp site in the promoter region of the human erbB-2 gene. In a second approach, novel six-finger ZFs, which bind 18-bp sequences that exist in multiple copies in the human genome, were designed. One of the novel ZF proteins was tested in fusions to SB transposase for targeting SB transposon insertions in the human genome. We provide proof-of-concept for biased SB insertion profiles in human cells by ZF proteins, thereby establishing an experimental platform for further developments and refinement towards manipulating target site selection properties of mobile genetic elements.
Results
DNA binding and transpositional activities of E2C/SB fusions
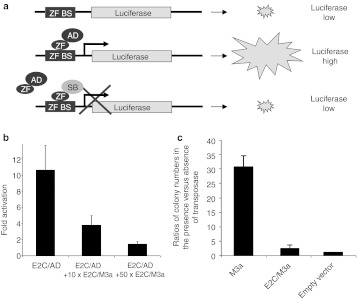
The efficiency of E2C to its binding site was assessed by a cell-based one-hybrid assay that measures transcriptional activation of a luciferase reporter by engineered transcription factors consisting of the ZFs and the VP16 transactivation domain (AD) upon ZF-dependent binding to binding sites situated upstream of the promoter driving luciferase expression (Figure 2a). The E2C ZF protein induced reporter gene expression tenfold as compared to luciferase values obtained in the presence of AD only (Figure 2b). The DNA-binding capacity of E2C in the context of a transposase fusion was examined in competitive luciferase assays that measure reporter gene expression in the presence of ZF/transposase fusions (Figure 1c) that compete with ZF/AD fusions for binding to the ZF binding sites (Figure 2a). As fusion partner, a hyperactive version of the SB transposase, M3a,15 was used. Cotransfection of E2C/M3a reduced reporter gene expression to as low as ~10% (Figure 2b). Thus, E2C retains its ability to bind to its binding site in the context of a fusion protein with the SB transposase.
Figure 2.
DNA binding and transpositional activity of a Sleeping Beauty transposase fusion with the E2C zinc finger DNA-binding domain. (a) Schematic of a luciferase reporter assay. The binding site of a zinc finger DNA-binding domain (ZF BS) is cloned in front of a minimal promoter driving expression of a luciferase reporter. Binding of an artificial transcription factor composed of a ZF and the VP16 transcriptional transactivation domain (ZF/AD) to the ZF BS activates luciferase expression. Coexpression of a ZF/SB fusion protein interferes with transcriptional activation of the reporter, because it competes with ZF/AD for binding at the ZF BS. (b) Luciferase transactivation by the E2C ZF in the absence and presence of a tenfold and a 50-fold molar excess of E2C/M3a as a competitor. Bars represent normalized relative luminescence units (RLUs) for activator proteins (ZF/AD) divided by RLUs for a negative control (AD only). (c) Transposition activity of the E2C/M3a fusion protein. Transposition efficiency is measured in a cell-based colony formation assay based on cotransfection of an antibiotic resistance-marked transposon donor plasmid and a helper plasmid that expresses the transposase into HeLa cells. The ratio of colony numbers obtained in the presence versus the absence of the transposase provides a measure of transpositional activity. The graph represents such ratios obtained by E2C/M3a relative to unfused M3a transposase. Error bars represent the SEM.
The SB transposase is believed to undergo several conformational changes during the transposition process. Addition of a tag or foreign protein domain may constrain the activities of the transposase. Indeed, fusions to the C-terminus of the SB transposase were invariably found to completely abolish transposition activity, whereas fusion of tags or foreign domains to the N-terminus were shown to be tolarated to some extent, although at the expense of diminished transposition activity.10,11,16 Transposition activity of the E2C/M3a fusion protein was examined using a cell culture transposition assay, that measures the formation of antibiotic-resistant colonies in the presence or absence of transposase, as previously described.2 The E2C/M3a transposase showed only twofold increase in colony numbers compared to negative controls (Figure 2c), and inclusion of linkers of various amino acid composition had no effect on its transposition efficiency (data not shown). Thus, in contrast to E2C fusions with the SB10 transposase (the originally reconstructed, first-generation SB transposase)2 that lacked transposition activity,11 use of a hyperactive transposase was able to rescue some fraction of transpositional activity, thereby enabling tests for target-selected transposition.
Biased transposition by the E2C/SB transposase fusion protein into plasmid targets
The targeting ability of the E2C/M3a fusion was first examined in a plasmid context, because shifts in transposition patterns might be easier to observe on a 5.5-kb plasmid than in the context of 3 billion base pairs in the human genome. Three plasmids were transfected into HeLa cells: a transposase-expressing helper plasmid, a donor plasmid carrying a kanamycin-marked transposon, and an ampicillin-resistant target plasmid. It should be noted that about half of the target plasmid are off-limits for recovery of integration products, because insertions into the ampicillin resistance gene and the origin of replication compromise bacterial growth. Plasmid DNA was purified from the transfected cells 48 hours post-transfection and transformed into Escherichia coli, which were plated on amp/kan plates to select for transposition events of the kanamycin-marked transposon into the ampicillin-resistant target plasmid. The target plasmid contained nucleotides −758 to −1 relative to the ATG initiation codon of the human erbB-2 gene including the E2C binding site (Figure 3a).14 In addition to transfections with E2C/M3a and unfused M3a transposases alone, coexpression of E2C/M3a together with limited amounts of unfused M3a transposase was also tested.
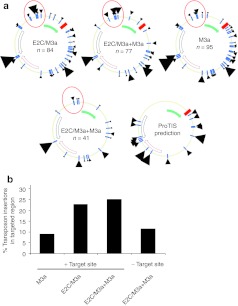
Figure 3.
Target site usage of Sleeping Beauty transposition by E2C/SB fusions in target plasmids carrying an E2C binding site. A donor plasmid containing an SB transposon carrying a kanamycin resistance gene, an ampicillin-resistant target plasmid containing the E2C binding site and a helper plasmid expressing SB transposase were cotransfected into HeLa cells. Plasmids were recovered from the cells 48 hours post-transfection and electroporated into Escherichia coli. Using kan/amp double selection only target plasmids containing a transposon insertion were able to grow. Positions of transposon insertion sites were identified by sequencing. (a) The interplasmid transposition assay was done to compare transposon integration profiles for E2C/M3a (84 insertions), E2C/M3a mixed with unfused M3a (77 insertions), and unfused M3a transposase (95 insertions). Transposon integration sites are represented by black arrowheads. Transposon insertions into the same TA or close to each other were combined into one arrowhead. Larger arrowheads represent multiple insertions at a particular region. Target plasmids contained the E2C recognition site (red square) and an adjacent 758-bp fragment from the erbB2 gene (filled green arrow). No insertions into the antibiotic resistance gene (black unfilled arrow) and only few integrations into the ori (red unfilled arrow) were recovered due to the experimental set up. As control, transposon integrations into an identical target plasmid but lacking the E2C binding site were also mapped (41 insertions). A prediction of SB transposon insertion preferences using the ProTIS software is depicted. Predicted hotspots for transposon insertions are depicted with black arrowheads, large arrowheads indicate preferred TA dinucleotides, small arrowheads indicate semi-preferred TA dinucleotides. (b) Quantification of targeted SB insertion by E2C into a region upstream of the E2C binding site.
Altogether 297 transposon insertions were recovered from kanr/ampr bacteria, sequenced, and mapped onto the target plasmids. In these experiments, targeting is defined as a bias in the overall insertion patterns that is dependent on both the E2C protein as well as on the binding site of E2C. As expected, insertions into the ampicillin resistance gene and the origin of replication were not recovered. Clear insertion hotspots were visible, some of which were predicted by the ProTIS software that was developed to predict preferred target sites of SB (Figure 3a).17 Importantly, in a region about 1 kb upstream of the E2C binding site just outside the cloned erbB-2 promoter sequence an enrichment of transposon insertions was observed using E2C/M3a and E2C/M3a + M3a (23 and 25%, respectively) compared to 9.4% of the insertions mapping to this region with unfused M3a transposase (Figure 3a,b). Transposon insertions within this region dropped back to 12% with a control target plasmid lacking the E2C binding site (Figure 3a,b). Intriguingly, these insertions occurred in a region of the target plasmid not predicted to have preferred sites by ProTIS (Figure 3a), suggesting that target choice in this region was imposed by the ZF component of the transposase fusion protein. Even though these insertion frequencies, likely due to the relatively low numbers of insertions analyzed per dataset, were statistically not significant, they were nevertheless dependent on both the E2C protein as well as on the binding site of E2C, and thus prompted us to investigate biased insertion patterns at the genome level.
Targeted SB transposon insertions into the human genome by the E2C ZF domain
Encouraged by detecting an insertion bias in target plasmids, we next addressed the question whether the E2C ZF can direct SB transposon insertions close to the endogenous erbB-2 locus in the human genome. HeLa cells were transfected with E2C/M3a or the M3a transposase helper plasmid and a transposon donor plasmid carrying a neomycin-marked SB transposon. Cells were selected for transposon insertions by G418 selection and 48 cell clones per transposase were individually picked. Semi-nested, erbB-2 locus-specific PCR was performed using one primer annealing in the erbB-2 locus (711 bp upstream of the E2C binding site) and two nested primers annealing in the transposon terminal inverted repeats. A PCR product would thus only be generated if a transposon insertion occurred near the E2C binding site. No PCR product was seen in any of the 48 samples tested for unfused M3a transposase (Figure 4a). However, for the E2C/M3a transposase fusion protein one out of the 48 clones produced a band (Figure 4a). This PCR product was sequenced, and found to represent a bona fide SB transposon insertion into a TA dinucleotide target site 179 bp upstream the E2C binding site (Figure 4b).
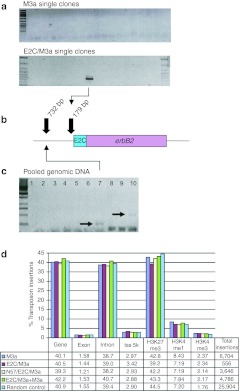
Figure 4.
Insertion into the erbB-2 locus by E2C/SB transposase fusion proteins. (a) Locus-specific PCR with primers specific to the human genomic erbB-2 locus and to the TIRs of SB transposons on genomic DNA samples from individual transgenic HeLa cell clones obtained by the E2C/M3a fusion and by unfused M3a transposase. (b) Relative positions of targeted SB transposon insertions in the human erbB-2 locus upstream of the E2C binding site. (c) Locus-specific PCR with primers specific to the human genomic erbB-2 locus and to the TIRs of SB transposons on genomic DNA samples from pooled HeLa cell clones. Lanes: 1) water control; 2) transposon only; 3) E2C/SB10; 4) E2C/SB10 + SB10; 5) 10× E2C/SB10 + SB10; 6) SB10/E2C + SB10; 7) 10× SB10/E2C + SB10; 8) SB10; 9) N57/E2C + SB10; 10) 10× N57/E2C + SB10. Ten times transposase fusion protein + SB indicates transfections with fusion transposase plasmids and unfused SB transposase in a ratio of 10:1. (d) Distribution of transposon insertions in the human genome catalyzed by E2C/SB. The graphs show relative distributions of transposon insertions with respect to genes, exons, introns, 5-kb regions around transcription start sites (tss_5k) and different histone methylations indicating intergenic and silent-coding regions (H3K27me3), active genes (H3K4me1), and transcriptional start sites of expressed genes (H3K4me3). Color-coded bars represent the percentage of transposon insertions within a genomic region catalyzed by the indicated transposase. Random control represents bioinformatically calculated random distribution of any TA dinucleotide in the respective genomic region. TIR, terminal inverted repeat.
In a different experiment, HeLa cells were transfected with a neomycin-marked transposon donor plasmid and either unfused SB10 transposase, or E2C fusions to N57 or full-length SB10 transposase that were produced previously.11 N57 was fused N-terminally to the E2C ZF, whereas full-length SB10 transposase was connected either C- or N-terminally to the E2C ZF (Figure 1c); all of these fusions contained a 10x glycine linker connecting the transposase and the ZF domains (Figure 1c).11 Because these SB10-based fusion proteins have previously been shown to lack transposition activity,11 the question addressed in this experiment was whether a mixture of transpositionally active transposase and transpositionally inactive transposase/ZF fusion proteins might target SB transposition through heterodimerization. Cells containing transposon insertions were selected by G418 and pooled. A diagnostic PCR specific for the erbB-2 locus was performed on genomic DNA samples from pooled cells (representing ~500 individual cell clones), as described above. No products were observed for control transfections with unfused SB10 transposase (Figure 4c, lane 8), with the transposon donor plasmid only (Figure 4c, lane 2), and with the E2C/SB10 fusion either without or with cotransfected SB10 transposase (Figure 4c, lanes 3–5). However, a PCR product was detected in the SB10/E2C sample (Figure 4c, lane 7), and another in the N57/E2C + unfused SB10 transposase sample (Figure 4c, lane 10), both from transfections where the plasmids expressing the catalytically inactive targeting fusions were represented at 10-times higher concentrations than that of unfused SB10 transposase. The PCR product shown in lane 7 of Figure 4c was sequenced, and found to represent a bona fide SB transposon insertion into a TA dinucleotide target site 732 bp upstream the E2C binding site in the erbB-2 promoter region (Figure 4b). In sum, the data suggest that the E2C ZF protein, both in the context of fusions with full-length SB transposase as well as with N57, is able to direct SB transposon insertions near the E2C binding site in the promoter region of the erbB-2 promoter in the human genome, at detectable frequencies.
Assessment of the effect of the E2C protein on target site selection by genome-wide analysis of SB insertion sites
Linear-amplification (LAM)-PCR offers a more unbiased method to map transposon insertions than locus-specific PCR.18 To examine the potential of E2C ZF fusion proteins to target SB transposon integrations, the unfused M3a transposase, the E2C/M3a transposase, the E2C/M3a transposase mixed with unfused M3a transposase, and N57/E2C mixed with unfused M3a transposase were tested in transfected HeLa cells. After selecting for transposon insertions with G418, cells were pooled and genomic DNA extracted. LAM-PCR was then applied to generate complex insertion site libraries that were analyzed by Illumina sequencing. A computer-generated TA dinucleotide set randomly picked from the human genome was used as reference (random control in Figure 4d). After mapping the integration sites onto the human genome, they were annotated with respect to insertions in genes, transcription start sites, and histone marks including H3K27me3 (a heterochromatic mark), and H3K4me1 and H3K4me3 (euchromatic marks) (Figure 4d).19,20 No profound difference in integration patterns between the transposition samples was observed (Figure 4d), with one notable exception: for E2C/M3a+M3a a slight, but significant (P value ≤0.05) increase in transposon integrations into genes (42.2 versus 40.1% for the unfused M3a transposase) was found; these intragenic insertions localized in introns (40.7 versus 38.7% for the unfused M3a transposase) (Figure 4d). The frequencies of transposon insertions into chromatin regions characterized by the different H3 methylation marks were not different across the samples and from the random control dataset (Figure 4d). Surprisingly, not a single insertion into a 20-kb window around the E2C binding site was detected for any of the transposition samples, and relaxing stringency of the analysis by allowing up to two mismatches in the E2C binding site did not improve recovery of targeted events (Supplementary Table S1). A flexing model for targeted transposition was previously proposed.10 The model posits that a protein that remains too tightly bound to DNA, such as E2C/M3a to the canonical, 18-bp E2C binding site, may not be able to efficiently catalyze transposition due to physical constraints. However, binding to a mutant E2C site by interactions mediated by only three fingers of E2C could improve the flexibility of the transposase domain, which might enhance the acquisition of neighboring target sites. Indeed, insertion site mapping in 20-kb windows around genomic sequences matching the 5′ E2C half site (consisting of 9 bp as opposed to the full, 18-bp E2C binding site) revealed a slight enrichment of transposon insertions for E2C/M3a when compared to unfused M3a (4.5 versus 3.7%) (Supplementary Table S1).
DNA-binding activities of novel ZF proteins designed for endogenous human repeat elements
The above data suggest that ZFs might be suitable reagents for targeted transposition, but the E2C-based system is inefficient, likely because either one or more of the components are suboptimal. As an alternative, we selected human endogenous LINE1 (L1) retrotransposons as potentially suitable targets for SB, because (i) they are present in the human genome in high copy numbers, (ii) they are enriched in AT-rich regions of the genome thereby representing attractive targets for SB,6,7 (iii) they are enriched in gene-poor chromosomal regions,21 thereby fulfilling the criterion that safe transgene insertions should preferentially be located away from genes, and (iv) when present in genes, L1 sequences are in introns and thereby would have little insertional consequences should a transposon land in or close by. About 17% of the human genome consists of L1 elements,21 most of which are retrotranspositionally inactive due to mutations or 5′-truncation. The 3′-region of L1.3, belonging to the Ta subfamily of L1 elements,22 was chosen as target for ZF binding (Figure 5a). Even though targeting SB transpositions into L1 elements may not fully eliminate the genotoxic risk associated with transgene insertion, the features of these dispersed repeat elements listed above make them attractive to probe a ZF protein-based targeting system in a proof-of-concept experimental setup.
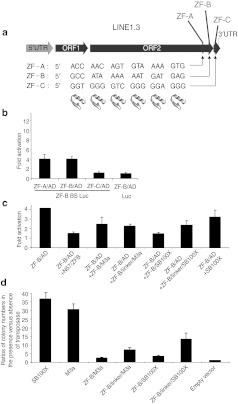
Figure 5.
DNA binding and transpositional activities of novel zinc finger domains binding to 18-bp sequences at the 3′-end of the LINE1.3 human retrotransposon. (a) The general organization of the LINE1 retrotransposon containing a 5′-UTR, coding regions for ORF1 and ORF2 and a 3′-UTR is depicted. Newly designed zinc finger (ZF) DNA-binding domains designated ZF-A, ZF-B, and ZF-C recognize 18-bp sequences localized in the 3′-end of the retrotransposon. (b) Luciferase transactivation by different ZF/AD fusions representing the three novel ZF domains in the presence or absence of a ZF-B binding site (BS). (c) Luciferase transactivation by ZF-B in the absence and presence of a 50-fold excess of different ZF-B/SB fusion proteins. (d) Transposition activity of ZF-B/SB fusion proteins as described in Figure 2c. The graphs represent transpositional activities of ZF-B/SB fusions relative to unfused M3a and SB100X transposase. Error bars represent the SEM. UTR, untranslated region.
For the selection of suitable ZF binding sites and for the design of the corresponding ZF proteins the “Zinc Finger Tools” website developed by the Barbas laboratory23 was used. Three 18-bp ZF binding sites were selected (Figure 5a), and ZF proteins predicted to bind to these sites were generated. Efficiencies of these ZFs to their binding sites were assessed by the luciferase transactivation assay as described above. Two ZF proteins out of the three tested (ZF-A and ZF-B) induced reporter gene expression at levels fourfold higher than those obtained in the presence of AD only (Figure 5b). Transcriptional activation by ZF-B was investigated further, and found to be dependent on the presence of its binding site in the reporter contruct (Figure 5b), indicating a specific interaction between this ZF protein and its 18-bp target site. The DNA-binding capacity of the ZF-B protein in the context of transposase fusions was examined in competitive luciferase assays as described above. As fusion partners, two hyperactive versions of the SB transposase, M3a and SB100X,24 as well as the N57 transposase interactor domain11,12 were used. Cotransfection of ZF-B/SB reduced reporter gene expression on average to about 50% compared to negative controls that contained ZF-B/AD either without any competitor or with unfused SB transposase as competitor (Figure 5c). The presence of an 18-aa linker between the ZF and transposase domains apparently did not affect DNA binding of the fusion proteins (Figure 5c). The data indicate that the ZF-B protein retains its ability to bind to its cognate recognition site in the context of fusion proteins with the SB transposase.
Transposition activities of ZF/transposase fusion proteins
Transposition activities of ZF-B/SB transposase fusion proteins were examined using the cell culture transposition assay described above. A direct fusion between ZF-B and M3a transposase retained <10% of transposition activity of unfused M3a, whereas including a linker between the two moieties resulted in an increase of transposition activity to about 23% of transposition activity of unfused transposase (Figure 5d). Replacing the M3a transposase with the hyperactive transposase version SB100X resulted in only a slight increase of transposition activity in a direct fusion, but yielded as high as about 40% of transposition activity of unfused SB100X when the ZF and transposase domains were separated by the linker (Figure 5d). Even though some fraction of the fusion proteins underwent processing likely by proteolysis, the extent of this was apparently independent of the different fusion partners (Supplementary Figure S1). In sum, even though tagging generally had a negative effect on the transpositional activities of ZF-B/SB fusions, a reasonable fraction of transpositional activity can be rescued by applying hyperactive versions of the transposase.
Biased insertion by ZF-B assessed by genome-wide analysis of SB insertion sites
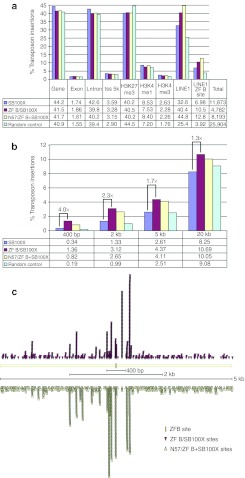
To examine the potential of ZF-B to target SB transposon integrations to L1 sequences, we next analyzed target site preferences of the unfused SB100X transposase in relation with the ZF-B/SB100X fusion protein as well as N57/ZF-B mixed with unfused SB100X transposase. A control dataset was calculated for sites randomly picked at any TA dinucleotide in the human genome (random control). Significant (P value ≤0.001) differences in transposon distribution were found for insertions into genes: 41.5% for ZF-B/SB100X and 41.7% for N57/ZF-B+SB100X versus 44.2% for unfused SB100X transposase; these intragenic insertions localized in introns (39.8% for ZF-B/SB100X, 40.2% for N57/ZF-B+SB100X, and 42.6% for SB100X) (Figure 6a). In contrast to what was seen before for the E2C datasets, transposition in all samples was significantly underrepresented in chromosomal regions characterized by H3K27me3 marks (as low as 40.2% in SB100X and N57/ZF-B+SB100X versus 44.5% for the random control) (Figure 6a).
Figure 6.
Distribution of transposon insertions in the human genome catalyzed by ZF-B/SB fusion proteins. (a) The graphs show relative distributions of transposon insertions as detailed in Figure 4d. (b) Enrichment of SB transposon insertions near ZF-B binding sites in the human genome using ZF-B/SB fusion constructs. Depicted are percentages of transposon insertions within defined windows around ZF-B binding sites. (c) Distribution of SB insertions around ZF-B binding sites. Each arrowhead marks the position of a single SB insertion site relative to a nearby ZF-B site (yellow rectangle).
To evaluate the targeting capacity of the ZF-B/transposase fusions, the frequencies of SB transposon insertions into L1 elements were determined. A significant increase of transposon insertions into L1 elements (up to 44.8% of all insertions) was detected for ZF-B containing transposases (Figure 6a and Supplementary Table S3), and this enrichment was especially pronounced for a subset of L1 elements that contains the 18-bp binding site of ZF-B (12.8% for N57/ZF-B+SB100X and 6.98% for SB100X) (Figure 6a). Next, genomic DNA sequences around transposon insertions were searched for the presence of predicted ZF-B binding sites. Transposon insertions obtained with the ZF-B/SB100X fusion protein and with N57/ZF-B+SB100X were enriched approximately fourfold and ~2.4-fold, respectively, in a 400-bp window around the ZF-B binding sites, as compared to the unfused SB100X control (Figure 6b). Widening the search window around the ZF-B binding sites up to 20 kb revealed a less pronounced enrichment of transposon insertions around the ZF-B binding sites (Figure 6b), suggesting that the observed bias in transposon insertions in close vicinity of the targeted sites is indeed due to physical interaction between the ZF-B DBD and the targeted 18-bp sites. Indeed, mapping of transposon insertions with respect to ZF-B binding sites and their flanking chromosomal regions revealed that the highest peaks of transposon insertions are proximal to the ZF-B binding sites (Figure 6c). Allowing up to two mismatches in the ZF-B binding site as well as repeating the analyses with 9-bp half sites did not reveal further enrichment in targeted transposition (Supplementary Table S2). The data are consistent with targeted SB integration near ZF-B binding sites at the 3′ end of human L1 retrotransposons.
Discussion
Random, uncontrolled insertion of transgene vectors can lead to transgene silencing or can have mutagenic effects, both of which are highly undesired for gene therapy approaches. For example, lentiviral vectors derived from HIV-1 have a tendency to insert into transcription units,25 whereas γ-retroviral vectors preferentially target transcription start sites,26 bearing the risk of transactivating endogenous genes.27,28,29 Indeed, a major setback for gene therapy struck when some patients who otherwise had been successfully treated for severe combined immunodeficiency (SCID-X) by γ-retroviral vector-based gene therapy developed leukemias.27,29,30 In these patients the vector inserted close to or inside an oncogene,27 whose promoter got upregulated by the retrovirus-borne enhancer, thereby leading to deregulated cell proliferation. Compared with retroviral systems, the SB vectors lack a preference for insertion into genes,7,8 have an inherently low enhancer/promoter activity,31 and were shown to have attenuated trans-activation of promoters of neighboring genes by flanking the transcription units of their cargo with insulator sequences.31 Nevertheless, introducing a bias into SB's target site selection profile in order to avoid or limit potentially hazardous insertions would represent a significant improvement of SB's safety profile.3
Introducing a bias into target selection profiles of integrating genetic elements has been a challenging task. Fusions of the ASV IN to the LexA protein32 or HIV IN to the Λ-repressor,33 the LexA protein,34 the Zif268 ZF,35 and E2C36 have been shown to introduce a bias into viral integration patterns in vitro. Transposable elements, including SB, have also been engineered to allow targeted insertion by heterologous DBDs into plasmid targets,10,37,38 and we previously reported that the SB transposon can be targeted into genomically located tetracycline operator sites.11 There are numerous differences between the approaches that were previously used for transposon targeting, which collectively make a direct comparison difficult. First, different transposases react differently to protein fusions. Second, different transposases have different sequence preferences for insertion, and this fundamentally affects any attempt to introduce a bias into their natural insertion profile. Third, strategies based on protein–protein interactions were only explored for the SB system, where the N57 domain has been characterized. Functionally equivalent domains or interacting proteins that could be exploited for transposon targeting have not been described for other transposon systems. Finally, most previous attempts of transposon targeting have established insertional biases in target plasmids or engineered chromosomal sites containing idealized target sequences. To our knowledge, our present work establishing reasonable efficiencies of redirecting SB insertions by applying ZF DBDs provides the first proof-of-concept for biased transposition into endogenous sites in the human genome. Our results reinforce the concept that, beyond direct fusions of a DBD to the transposase, protein domains interacting with a component of the integration machinery are capable of tethering the integration complex to a defined chromosomal region or site. Importantly, several, naturally occurring transposable elements evolved strategies for targeted insertion into defined chromosomal sites or regions, and the mechanisms of targeted insertions often rely on protein–protein interactions between a transposon-encoded factor and a cellular, DNA-binding host factor. For example, based upon observations for a role of LEDGF/p75 in directing HIV integration into expressed transcription units, in vitro studies have shown increased integration near Λ-repressor binding sites by fusing either the full-length LEDGF/p75 or the LEDGF/p75 IN-binding domain to the DBD of phage Λ-repressor protein.39 This approach was successfully applied for targeting lentiviral vectors into chromosomal regions bound by CBX1, a cellular factor interacting with histone H3 di- or trimethylated on K9 associated with pericentric heterochromatin and intergenic regions by fusing the LEDGF/p75 IN-binding domain to CBX1.40 In an analogous fashion, Sir4p (which mediates targeted insertion of the yeast Ty5 retrotransposon into heterochromatin) fused to the Escherichia coli LexA DBD was shown to result in integration hot spots for Ty5 near LexA operators.41
We designed and characterized a novel ZF protein binding an 18-bp site within the 3′-UTR of a subset of L1 retrotransposons that exist in multiple copies scattered throughout the human genome. The total number of L1 elements in the genome is >800,000, the total number of the 18-bp ZF-B binding sites (GCCATAAAAAATGATGAG) is 12,766, and the number of L1 elements that contain the 18-bp ZF-B binding site is 12,703. Thus, almost all of the 18-bp binding site of ZF-B are associated with L1 elements. We demonstrate that ZF-B, in fusion with either the SB transposase or N57, results in a fourfold enrichment of transposon insertions towards the targeted sites as compared to unfused SB transposase. We consider it likely that this is an underestimate of the true efficiency of transposon targeting, because our LAM-PCR procedure followed by deep sequencing and bioinformatic analysis cannot detect independent targeting events at the same TA dinucleotide in the human genome, because multiple independent transposon insertions at the same TA dinucleotide will be scored as one single insertion. Even though these results provide proof-of-concept for the use of artificial ZF proteins for target-selected gene insertion, it is also evident that a large fraction of SB insertions remained untargeted. This is because the SB transposase in ZF/transposase fusion proteins retain their intrinsic ability to bind target DNA independent from the ZF. Thus, the targeted DNA sequences, even if they exists in multiple copies, are easily outnumbered by ~108 TA dinucleotides in the human genome. This situation is markedly different from the biochemistry of ZF nucleases, in which sequence-specific DNA cleavage is dependent on heterodimerization of FokI endonuclease domains, thereby providing considerable specificity to the reaction.42
We consider three factors whose choice and design could contribute to further enhancement of the efficiency of targeted transposition. The first is the transposase itself. It was shown before that fusions to the SB transposase have an inhibitory effect on transposition; however, we show here that by using hyperactive transposases a significant fraction of transposition activity can be rescued. Future work will need to be dedicated to the generation of transposase mutants that have an attenuated affinity to target DNA in the hope to suppress off-target transposition events. The second factor that likely has a profound effect on targeted transposition is the DBD responsible for guiding the transpositional complex to the targeted DNA sequence. We applied here ZF proteins designed by modular assembly, based on a simple base triplet-amino acid code. However, recent developments indicate that selection-based approaches, such as the OPEN method43 might be more appropriate for the generation of novel ZF proteins with superior binding characteristics, due to context-dependent binding of ZF modules to DNA sequence motifs. Novel ZF proteins generated by applying these principles might yield enhanced frequencies of targeted insertion due to their higher specificity to their defined target sites. In this context, the recently discovered and developed TALE domains might hold considerable promise in providing highly efficient heterologous DBDs as well as straightforward design.44
Finally, selection of an appropriate target site appears to be of paramount importance. The minimal requirements for such sites are that they have to be accessible to the transpositional complex and provide DNA flanking the DBD binding site that is suitable for transgene insertion by the respective vector system. Specifically, the targeted sites should be flanked by TA dinucleotides to support SB transposition; in fact, SB was reported to prefer insertion into TA-rich DNA in general.45 It is noteworthy that the human erbB-2 locus around the E2C binding site is rather GC-rich: GC-content of a region covering 1,500 bp upstream of the E2C binding site is 51%, whereas GC-content of a region covering 1,500 bp downstream the E2C binding site is 60%. Analysis of the erbB-2 promoter region with ProTIS,17 a software that analyzes DNA for potential SB integration hot spots, showed only a handful of TA dinucleotides predicted to be preferred by SB within a 4-kb window around the E2C binding site (Figure 7). In fact, the total number of TAs (also including unpreferred sites) is only 8 within a 800-bp window around the E2C binding site, consistent with the relatively poor targeting of this locus by SB, and in agreement with earlier work from Yant et al.10 In contrast, as we demonstrate in this work, L1 elements can be more efficiently targeted by SB. The LINE1.3 element has an overall TA-content of 58%, and 63 TA dinucleotides offering potential SB insertion sites map within 1 kb of its 3′-end containing the ZF-B binding site. Analysis of the 3′-UTR of the LINE1.3 element with the ProTIS program indeed predicted several TA dinucleotides with the highest ranking for preferred SB transposon insertion (Figure 7). The importance of DNA composition in the vicinity of targeted sites was highlighted in a recent report on targeted piggyBac transposition in human cells.46 In that study, biased transposition was only observed with engineered loci that contained numerous TTAA sites (the target site of piggyBac transposons) in the flanking regions of a DNA sequence bound by a ZF protein. Furthermore, high-resolution probing of targeted transposition of the ISY100 transposon (which, like SB, is a member of the Tc1/mariner transposon superfamily) by the Zif268 ZF fused to the C-terminus of the ISY100 transposase has shown highly specific integration into TA dinucleotides positioned 6–17 bp to one side of a Zif268 binding site in Escherichia coli.47 This suggests a considerable constrain in target site usage once the transposase is docked on the target DNA by the DBD. These observations indicates that future design of ZFs and TALEs should take into account that targeted sequences should ideally be flanked by suitable TA sites in the direct vicinity. Collectively, these considerations should assist in the design of target-selected gene insertion systems with enhanced efficiency and specificity.
Figure 7.
Analysis of genomic DNA around ZF target sites for potential Sleeping Beauty insertion site hot spots using ProTIS. Two kb up- and downstream of the E2C BS in the erbB-2 genomic region and the 3′-terminal 2,814 bp of the LINE1.3 element are depicted. Stars indicate TA dinucleotides that are ranked as the highest preferred for SB transposon integration by ProTIS analysis.
Materials and Methods
Plasmid constructs
E2C/SB transposase fusion constructs. The vector pFV4aE2C was created by removing the sequence coding for 10xGly/SB by NheI and NotI digest from pFV4aE2C/10xGly/SB.11 The coding region of M3a was PCR-amplified using primers fw_NheI/SalI-SB and SB-Not-rv (all primer sequences can be found in Supplementary Materials and Methods), the PCR fragment was digested with NheI and NotI and cloned into pFV4aE2C resulting in pFV4aE2C/M3a. The ampicillin resistance gene in pFV4aE2C/M3a was replaced with the chloramphenicol resistance gene. To clone pFV4aM3a, the M3a coding region was cloned as a blunted XhoI/BamHI fragment into the SmaI site of pFV4a. The plasmid pFV4aN57/E2C was generated by inserting an N57 fragment in place of SB in FV4aSB/E2C.11 The E2C/VP64 plasmid was kindly provided by Carlos Barbas.
LINE1-specific ZF constructs. Three ZF binding sites and ZF proteins potentially binding these sites were selected in the 3′-terminal region of the LINE1.3 element using the “Zinc Finger Tools” website (http://www.scripps.edu/mb/barbas/zfdesign/zfdesignhome.php).23 DNA sequences of the binding sites and the amino acid sequences of the ZF proteins are detailed in Supplementary Materials and Methods. Human codon-optimized synthetic genes encoding the designed ZF proteins were synthesized by GENEART (Regensburg, Germany). In order to clone the ZF domains into vectors containing the VP16 transactivation domain, the vector pcDNA.Rep.TZ.AD48 was cut using EcoRI and NotI resulting in pcDNA_AD, followed by ligation of the ZF-A, ZF-B, and ZF-C coding regions resulting in pcDNA_ZFA/AD, pcDNA_ZFB/AD, and pcDNA_ZFC/AD, respectively.
The E2C ZF domain was replaced in pFV4aE2C/M3acam with the ZF-B coding region following a digest with SacII and NheI, resulting in pFV4aZFB/M3a. A peptide linker (KLGGGAPAVGGGPKAADK)49 was introduced as a double-stranded oligonucleotide between ZF-B and the M3a transposase following NheI and partial SalI digest. M3a was cut out of pFV4aZF4/M3acam with NheI and NotI digest. The SB100X coding sequence was PCR-amplified from pCMV(CAT)T7-SB100X24 using primers fw_NheI/SalI-SB and SB-NotI-rv, digested with NheI and NotI and cloned into the prepared vector backbone pFV4aZFB yielding pFV4aZFB/SB100Xcam. To create pFV4aZFB/linker/SB100Xcam, the same procedure was performed as detailed above. To create pFV4aSB100Xcam, pFV4acam was digested with ApaI and SpeI, followed by insertion of the SB100X coding region. The E2C ZF coding sequence was removed from pFVN57/E2C by ApaI digest, blunt ended, and XhoI digested. ZF-B was PCR-amplified using primers XhoI ohne ATG ZF4 fw and ZF4 STOP ApaI rv, digested with XhoI, followed by insertion into pFVN57.
Luciferase reporter plasmids. The luciferase reporter plasmid erbB214 was PCR-amplified excluding the E2C binding site using primers erbB2ΔE2C_fw and erbB2ΔE2C_rv and ligated together yielding the plasmid erbB2ohneE2C. Double-stranded oligonucleotides encoding ZF-A, ZF-B, and ZF-C were cut with NheI and AgeI and cloned into the luciferase reporter plasmid pGLtk.11.Luc.50
Cell culture, transfection, and transposition assay. One day prior transfection 3 × 105 or 1.5 × 105 HeLa cells were seeded per 6-well or 12-well, respectively. Transfections were done with QIAGEN-purified endotoxin-free plasmid DNA using jetPEI (Polyplus-transfection) according to the manufacturer's protocol. Transposition assays were done as described.11 Typically, 3 × 105 cells were transfected with 100 ng of transposase expression plasmid and 20 ng of pTneo for E2C experiments and 200 ng of transposase expression plasmid and 200 ng of pTneo for ZF-B experiments. Forty-eight hours after transfection, a fraction (1/2–1/5) of transfected cells was replated on 10 cm dishes and selected for transposon integration using 1.4 mg/ml G418 (Biochrom, Berlin, Germany). After 3 weeks of selection, cell colonies were fixed with 10% vol/vol formaldehyde in phosphate-buffered saline (PBS), stained with methylene blue in PBS, and counted.
Luciferase assays. HeLa cells were seeded in 12-well plates and transfected in duplicates with 250 ng luciferase reporter plasmid carrying a ZF binding site upstream of the luciferase gene, 125 ng of a plasmid expressing a ZF/AD fusion protein, and 50 ng of a β-galactosidase expression plasmid using jetPEI (Polyplus-transfection) following the manufacturer's protocol. Forty-eight hours post-transfection cells were lysed in 200 µl CCLR buffer (125 mmol/l Tris-phosphate pH 7.8 H3PO4, 10 mmol/l DTT, 10 mmol/l 1,2-Diaminocyclohexan-N,N,N′,N′-tetraacetic acid, 50% vol/vol glycerol, 5% vol/vol Triton X-100) per well, vortexed for 15 seceonds, centrifuged, and the supernatant was kept. For normalization of transfection efficiency, a β-galactosidase assay was performed as described in Supplementary Materials and Methods. For determination of luciferase activity, 15 µl sample was added to 50 µl luciferase assay reagent (20 mmol/l Tricine pH 7.8 NaOH, 1.07 mmol/l (MgCO3)4Mg(OH)2 × 5 H2O, 2.67 mmol/l MgSO4, 0.1 mmol/l EDTA, 270 µmol/l Coenzyme A, 470 µmol/l luciferin, 530 µmol/l adenosine 5′-triphosphate), vortexed and immediately measured with a 10-second integration period in a luminometer (Lumat LB 9507; Berthold, Bad Wildbad, Germany). Raw luciferase reads are normalized by using the following formula: normalized luciferase reads = raw luciferase reads/β-gal units.
For competition assays, HeLa cells in 12-well plates were transfected in duplicates with 50 ng luciferase reporter plasmid carrying a ZF binding site upstream of the luciferase gene, 15 ng of a plasmid expressing an AD/ZF fusion protein, 935 ng of a plasmid expressing a ZF/transposase fusion protein, and 50 ng of a β-galactosidase expression plasmid complexed with jetPEI (Polyplus-transfection) following the manufacturer's protocol.
Interplasmid transposition assay. Six-well plates were triple-transfected with 200 ng transposon donor plasmid pTkan,45 200 ng target plasmid erbB2 and 50 ng of transposase helper plasmid. In case of transfections containing both E2C/M3a and M3a, 50 ng E2C/M3a and 5 ng M3a were applied. The target plasmid carried a promoter fragment encompassing nucleotides −758 to −1 relative to the ATG start codon of the erbB-2 gene cloned into pGL3basic.14 As additional negative control a target plasmid (erbB2ohneE2C) identical to erbB2 but lacking the E2C binding site was used. Forty-eight hours post-transfection cells were washed twice with PBS, and lysed in 400 µl lysis buffer (0.6% SDS, 0.01 M EDTA). Genomic DNA was precipitated by addition of 100 µl 5 mol/l NaCl. Low molecular weight DNA was subjected to phenol/chloroform and chloroform extraction and precipitated using 0.1 vol 2.5 mol/l KAc pH 8, 10 µg glycogen, and 0.8 vol isopropanol. DNA pellets were washed twice with 70% ethanol and resuspended in 10 µl ddH2O. Plasmid DNA was electroporated into ElectroMAX DH10B cells according to the manufacturer's protocol. To avoid division and consequently amplification of individual transposition events, bacteria were plated directly after electroporation on LB agar plates containing 100 µg/ml amp and 25 µg/ml kan. Plasmids from individual clones were sequenced for transposon insertions.
Semi-nested locus-specific PCR. HeLa cells were transfected and selected as described for the cell culture transposition assay. Three weeks after selection with G418, cells were either pooled or single clones were picked and further propagated in 12- or 24-well plates. Cells were washed twice in PBS and lysed by adding PBS containing 0.2 mg/ml Proteinase K at 50 °C for 2 hours. After heat inactivation of Proteinase K at 94 °C for 15 minutes, a first PCR reaction containing 500 ng of genomic DNA, 20 mmol/l Tris pH 8.4, 50 mmol/l KCl, 3 mmol/l MgCl2, 0.4 mmol/l dNTP, 0.2 pmol/l primer erbB2/5, and BalRev3 or T-Jobb1, and 2 U Taq-Polymerase was performed with a program consisting of: 94 °C for 4 minutes, 30 cycles of 94 °C for 30 seconds, ramp to 59 °C, 1 °C/second, 59 °C for 30 seconds, 72 °C for 2.5 minutes. The second PCR contained 1 µl of the first PCR, 20 mmol/l Tris pH 8.4, 50 mmol/l KCl, 2 mmol/l MgCl2, 0.2 mmol/l dNTP, 0.2 pmol/l primer erbB2/5, and BalRev or T-Jobb2, and 2 U Taq-Polymerase with a program consisting: 94 °C for 4 minutes, 30 cycles of 94 °C for 30 seconds, ramp to 59 °C, 1 °C/second, 59 °C for 30 seconds, 72 °C for 2.5 minutes.
LAM-PCR for Illumina sequencing. HeLa cells were transfected as described for a cell culture transposition assay. For transfections where E2C/M3a, N57/E2C or N57/ZF-B were supplemented with unfused transposase, 1/20 of the amount of plasmid encoding the fusion protein was transfected from the plasmid expression of the unfused transposase. At least 10,000 HeLa cell clones carrying at least one transposon insertion were pooled. Genomic DNA was extracted from cells using the DNeasy blood and tissue kit (QIAGEN, Valencia, CA) according to manufacturer's protocol. Genomic DNA was sheared with a Covaris S2 (Covaris, Woburn, MA) ultrasonicator. Sheared genomic DNA fragments were of around 300 bp after ultrasonication. Five hundred nanogram of genomic DNA was applied to a linear-amplification procedure as detailed in Supplementary Materials and Methods.
Bioinformatic analyses. We used a set of error-correcting barcodes for distinguishing different data sets pooled into a single Illumina Genome Analyzer IIx flow cell lane. The 76-bp long sequencing reads were checked for starting exactly with the barcode (4 bp) followed by the intact SB transposon ends (21 bp). The remaining parts of the read (51 bp) starting with a TA dinucleotide (indicating an SB transposase-mediated transposition event as opposed to random integration) were mapped using Bowtie to the chromosomes of the human genome (NCBI built GRCh37/hg19, February 2009) excluding the Y-chromosome, because HeLa is a female cell line. Only reads occurring without mismatches at a single genomic position (exact uniquely mapped reads) were used for the subsequent analysis, except for the analysis of repetitive regions (Supplementary Table S3), for which we also took reads with multiple exact matches into account. Multiple reads mapping to the same genomic position and strand were treated as a single integration site.
For statistical analysis, we generated a random control set containing 25,904 unique sites and 3,862 non-unique sites as follows. We randomly selected 51-mers from the reference genome starting with the TA dinucleotide sequence. Then we applied Bowtie for mapping them back to the genome using the same settings as described above.
Databases of genomic regions (RefSeq) were downloaded from UCSC (http://genome.ucsc.edu). We used the H3K27me3 domains as determined previously19 (GEO accession no. GSM325898). Regions enriched for H3K4me1 and H3K4me3 were determined as follows: the raw ChIP-Seq reads20 (http://www.bcgsc.ca/data/histone-modification) were mapped to the human genome using Bowtie, then peaks were called using MACS, and H3K4me1/3 domains are then defined as 5-kb windows around the centers of the peaks. The search for ZF binding sites was done by Python scripts.
SUPPLEMENTARY MATERIAL Figure S1. Partial proteolytic processing of SB transposase fusions. Table S1. Distribution of Sleeping Beauty transposon insertions catalyzed by E2C/SB in the human genome. Table S2. Distribution of Sleeping Beauty transposon insertions catalyzed by ZF-B/SB in the human genome. Table S3. Transposon insertions into repetitive regions of the human genome for ZF-B/SB fusion proteins and controls. Materials and Methods.
Acknowledgments
We thank Carlos Barbas for kindly providing a luciferase reporter plasmid containing an E2C binding site as well as the gene construct encoding E2C. We are grateful to Gerald Schumann for valuable discussions on the LINE1.3 element. This work was supported by EU FP7 (PERSIST, grant no. 222878), and grants from the Deutsche Forschungsgemeinschaft “Mechanisms of gene vector entry and persistence” (SPP1230, grant no. IV 21/4-2) and from the Bundesministerium für Bildung und Forschung (InTherGD, grant no. 01GU0815). The authors declared no conflict of interest.
Supplementary Material
Partial proteolytic processing of SB transposase fusions.
Distribution of Sleeping Beauty transposon insertions catalyzed by E2C/SB in the human genome.
Distribution of Sleeping Beauty transposon insertions catalyzed by ZF-B/SB in the human genome.
Transposon insertions into repetitive regions of the human genome for ZF-B/SB fusion proteins and controls.
REFERENCES
- Ivics Z, Li MA, Mátés L, Boeke JD, Nagy A, Bradley A.et al. (2009Transposon-mediated genome manipulation in vertebrates Nat Methods 6415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH., and, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Hackett PB, Cooper LJ., and, Ivics Z. Translating Sleeping Beauty transposition into cellular therapies: victories and challenges. Bioessays. 2010;32:756–767. doi: 10.1002/bies.201000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett PB, Largaespada DA., and, Cooper LJ. A transposon and transposase system for human application. Mol Ther. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DA. Sleeping beauty vector system moves toward human trials in the United States. Mol Ther. 2008;16:1515–1516. doi: 10.1038/mt.2008.169. [DOI] [PubMed] [Google Scholar]
- Vigdal TJ, Kaufman CD, Izsvák Z, Voytas DF., and, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM., and, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt B, Miskey C, Staunstrup NH, Gogol-Döring A, Bak RO, Sharma N.et al. (2011Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells Mol Ther 191499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, Izsvák Z., and, Ivics Z. Targeted gene insertion for molecular medicine. J Mol Med. 2008;86:1205–1219. doi: 10.1007/s00109-008-0381-8. [DOI] [PubMed] [Google Scholar]
- Yant SR, Huang Y, Akache B., and, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Katzer A, Stüwe EE, Fiedler D, Knespel S., and, Izsvák Z. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Khare D, Behlke J, Heinemann U, Plasterk RH., and, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS., and, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B., and, Barbas CF. 3rdToward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albieri I, Onorati M, Calabrese G, Moiana A, Biasci D, Badaloni A.et al. (2010A DNA transposon-based approach to functional screening in neural stem cells J Biotechnol 15011–21. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Kaminski JM., and, George AL., Jr Functional zinc finger/sleeping beauty transposase chimeras exhibit attenuated overproduction inhibition. FEBS Lett. 2005;579:6205–6209. doi: 10.1016/j.febslet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Hackett CS, Bell JB, Bergemann TL, Collier LS, Carlson CM.et al. (2006Structure-based prediction of insertion-site preferences of transposons into chromosomes Nucleic Acids Res 342803–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I.et al. (2007High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods 41051–1057. [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K., and, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T.et al. (2007Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing Nat Methods 4651–657. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J.et al. (2001Initial sequencing and analysis of the human genome Nature 409860–921. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P., and, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Mandell JG., and, Barbas CF. 3rdZinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34 Web Server issue:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A.et al. (2009Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates Nat Genet 41753–761. [DOI] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P.et al. (2003LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 Science 302415–419. [DOI] [PubMed] [Google Scholar]
- Baum C, von Kalle C, Staal FJ, Li Z, Fehse B, Schmidt M.et al. (2004Chance or necessity? Insertional mutagenesis in gene therapy and its consequences Mol Ther 95–13. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E.et al. (2008Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1 J Clin Invest 1183132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrasher AJ., and, Gaspar HB. ASGT press release: Milwaukee, Wisconsin; 2007. Severe Adverse Event in Clinical Trial of Gene Therapy for X-SCID. [Google Scholar]
- Walisko O, Schorn A, Rolfs F, Devaraj A, Miskey C, Izsvák Z.et al. (2008Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators Mol Ther 16359–369. [DOI] [PubMed] [Google Scholar]
- Katz RA, Merkel G., and, Skalka AM. Targeting of retroviral integrase by fusion to a heterologous DNA binding domain: in vitro activities and incorporation of a fusion protein into viral particles. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- Bushman FD. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc Natl Acad Sci USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulaouic H., and, Chow SA. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD., and, Miller MD. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Zhu K, Segal DJ, Barbas CF., 3rd, and, Chow SA. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J Virol. 2004;78:1301–1313. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragathavally KJ, Kaminski JM., and, Coates CJ. Chimeric Mos1 and piggyBac transposases result in site-directed integration. FASEB J. 2006;20:1880–1882. doi: 10.1096/fj.05-5485fje. [DOI] [PubMed] [Google Scholar]
- Szabó M, Müller F, Kiss J, Balduf C, Strähle U., and, Olasz F. Transposition and targeting of the prokaryotic mobile element IS30 in zebrafish. FEBS Lett. 2003;550:46–50. doi: 10.1016/s0014-5793(03)00814-7. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Diamond TL, Hwang Y, Marshall HM., and, Bushman FD. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum Gene Ther. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M.et al. (2010LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin Mol Ther 18552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Dai J, Fuerst PG., and, Voytas DF. Controlling integration specificity of a yeast retrotransposon. Proc Natl Acad Sci USA. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ., and, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF., and, Joung JK. Oligomerized pool engineering (OPEN): an 'open-source' protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF.et al. (2011A TALE nuclease architecture for efficient genome editing Nat Biotechnol 29143–148. [DOI] [PubMed] [Google Scholar]
- Liu G, Geurts AM, Yae K, Srinivasan AR, Fahrenkrug SC, Largaespada DA.et al. (2005Target-site preferences of Sleeping Beauty transposons J Mol Biol 346161–173. [DOI] [PubMed] [Google Scholar]
- Kettlun C, Galvan DL, George AL, Jr, Kaja A., and, Wilson MH. Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol Ther. 2011;19:1636–1644. doi: 10.1038/mt.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Bednarz AL., and, Colloms SD. Precise targeted integration by a chimaeric transposase zinc-finger fusion protein. Nucleic Acids Res. 2010;38:1204–1216. doi: 10.1093/nar/gkp1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathomen T, Collete D., and, Weitzman MD. A chimeric protein containing the N terminus of the adeno-associated virus Rep protein recognizes its target site in an in vivo assay. J Virol. 2000;74:2372–2382. doi: 10.1128/jvi.74.5.2372-2382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szüts D., and, Bienz M. LexA chimeras reveal the function of Drosophila Fos as a context-dependent transcriptional activator. Proc Natl Acad Sci USA. 2000;97:5351–5356. doi: 10.1073/pnas.97.10.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF., 3rd, , Segal DJ.et al. (2005Custom zinc-finger nucleases for use in human cells Mol Ther 12610–617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Partial proteolytic processing of SB transposase fusions.
Distribution of Sleeping Beauty transposon insertions catalyzed by E2C/SB in the human genome.
Distribution of Sleeping Beauty transposon insertions catalyzed by ZF-B/SB in the human genome.
Transposon insertions into repetitive regions of the human genome for ZF-B/SB fusion proteins and controls.