Abstract
We have constructed a transformation-defective polyoma virus mutant (Py 1387-T) that directs the synthesis of a normal small tumor antigen, a functional large tumor antigen, and a truncated (51,000-dalton) middle-sized tumor (mT) antigen that lacks 37 amino acids at its COOH terminus. The shortened mT polypeptide is missing the hydrophobic "tail" thought to be responsible for the anchorage of this protein into the plasma membrane and is in fact in cytosol fractions. This truncated mT polypeptide is inactive in an in vitro protein kinase assay and is altered in its phosphorylation in vivo. Mutant 1387-T differs from wild-type virus in having a T.A base pair instead of a C.G base at nucleotide position 1387. This change was introduced into viral DNA by using a synthetic undecanucleotide as a specific mutagen. Wild-type polyoma DNA was rendered single stranded by molecular cloning into coliphage M13. The oligonucleotide, which hybridizes with a mismatch at the site to be altered, was used to prime the synthesis of double-stranded closed circular DNA. Progeny recombinant phage were screened by DNA sequence analysis for the desired base change. The polyoma mutant was reconstructed from recombinant phage replicative form DNA molecules containing the mutation.
Full text
PDF

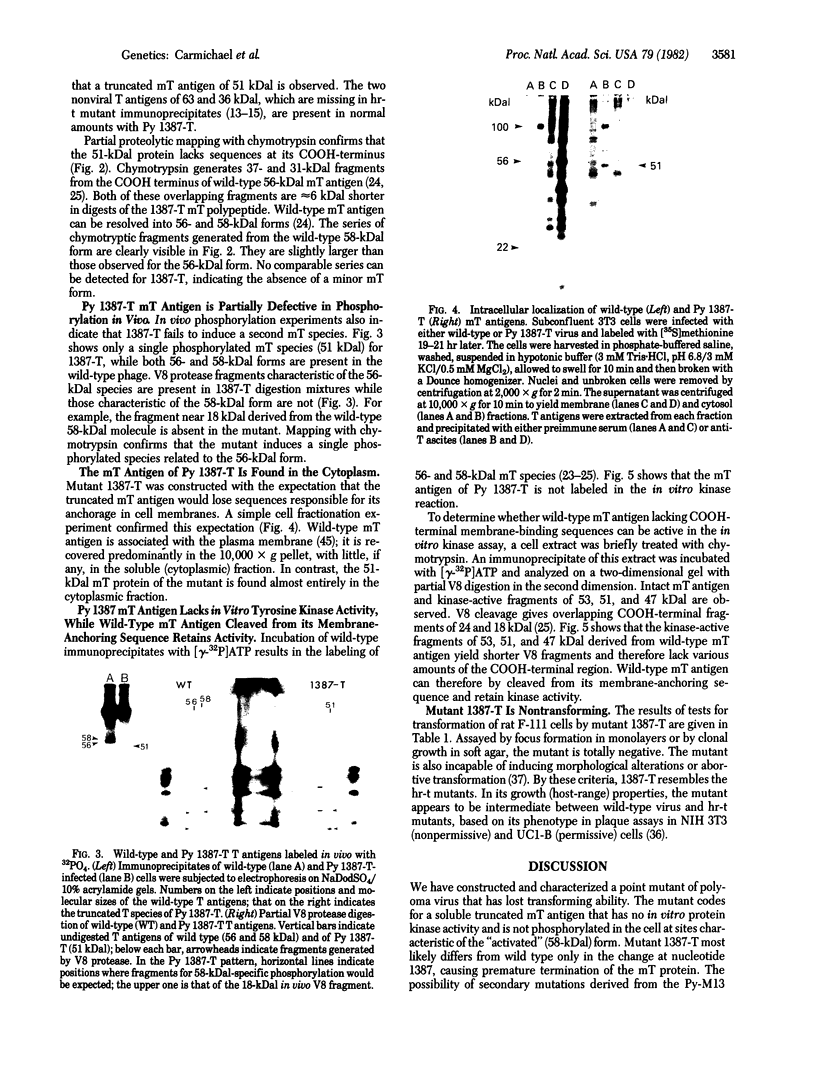


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin T. L., Schaffhausen B. S., Silver J. E. Polyoma T (tumor) antigen species in abortively and stably transformed cells. J Supramol Struct. 1979;12(1):127–137. doi: 10.1002/jss.400120110. [DOI] [PubMed] [Google Scholar]
- Di Mayorca G., Callender J., Marin G., Giordano R. Temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):126–133. doi: 10.1016/0042-6822(69)90134-2. [DOI] [PubMed] [Google Scholar]
- Eckhart W. Complementation and transformation by temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):120–125. doi: 10.1016/0042-6822(69)90133-0. [DOI] [PubMed] [Google Scholar]
- Eckhart W., Hutchinson M. A., Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979 Dec;18(4):925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- FRIED M. CELL-TRANSFORMING ABILITY OF A TEMPERATURE-SENSITIVE MUTANT OF POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1965 Mar;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam S., Jahnke P., Astell C., Phillips S., Hutchison C. A., 3rd, Smith M. Defined transversion mutations at a specific position in DNA using synthetic oligodeoxyribonucleotides as mutagens. Nucleic Acids Res. 1979 Jul 11;6(9):2973–2985. doi: 10.1093/nar/6.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillam S., Smith M. Site-specific mutagenesis using synthetic oligodeoxyribonucleotide primers: I. Optimum conditions and minimum ologodeoxyribonucleotide length. Gene. 1979 Dec;8(1):81–97. doi: 10.1016/0378-1119(79)90009-x. [DOI] [PubMed] [Google Scholar]
- Gillam S., Smith M. Site-specific mutagenesis using synthetic oligodeoxyribonucleotide primers: II. In vitro selection of mutant DNA. Gene. 1979 Dec;8(1):99–106. doi: 10.1016/0378-1119(79)90010-6. [DOI] [PubMed] [Google Scholar]
- Goldman E., Benjamin T. L. Analysis of host range of nontransforming polyoma virus mutants. Virology. 1975 Aug;66(2):372–384. doi: 10.1016/0042-6822(75)90210-x. [DOI] [PubMed] [Google Scholar]
- Hassell J. A., Topp W. C., Rifkin D. B., Moreau P. E. Transformation of rat embryo fibroblasts by cloned polyoma virus DNA fragments containing only part of the early region. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3978–3982. doi: 10.1073/pnas.77.7.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Phillips S., Edgell M. H., Gillam S., Jahnke P., Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978 Sep 25;253(18):6551–6560. [PubMed] [Google Scholar]
- Israel M. A., Chan H. W., Hourihan S. L., Rowe W. P., Martin M. A. Biological activity of polyoma viral DNA in mice and hamsters. J Virol. 1979 Mar;29(3):990–996. doi: 10.1128/jvi.29.3.990-996.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Griffin B. E. Middle T antigen as primary inducer of full expression of the phenotype of transformation by polyoma virus. J Virol. 1980 Jul;35(1):219–232. doi: 10.1128/jvi.35.1.219-232.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- MACPHERSON I., MONTAGNIER L. AGAR SUSPENSION CULTURE FOR THE SELECTIVE ASSAY OF CELLS TRANSFORMED BY POLYOMA VIRUS. Virology. 1964 Jun;23:291–294. doi: 10.1016/0042-6822(64)90301-0. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- Novak U., Dilworth S. M., Griffin B. E. Coding capacity of a 35 percent fragment of the polyoma virus genome is sufficient to initiate and maintain cellular transformation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3278–3282. doi: 10.1073/pnas.77.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U., Griffin B. E. Requirement for the C-terminal region of middle T-antigen in cellular transformation by polyoma virus. Nucleic Acids Res. 1981 May 11;9(9):2055–2073. doi: 10.1093/nar/9.9.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Hirose T., Itakura K., Riggs A. D. Efficient correction of a mutation by use of chemically synthesized DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4268–4270. doi: 10.1073/pnas.75.9.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D. D., Kreibich G., Morimoto T., Adesnik M. Mechanisms for the incorporation of proteins in membranes and organelles. J Cell Biol. 1982 Jan;92(1):1–22. doi: 10.1083/jcb.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Phosphorylation of polyoma T antigens. Cell. 1979 Dec;18(4):935–946. doi: 10.1016/0092-8674(79)90206-x. [DOI] [PubMed] [Google Scholar]
- Schaffhausen B. S., Silver J. E., Benjamin T. L. Tumor antigen(s) in cell productively infected by wild-type polyoma virus and mutant NG-18. Proc Natl Acad Sci U S A. 1978 Jan;75(1):79–83. doi: 10.1073/pnas.75.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B., Benjamin T. L. Comparison of phosphorylation of two polyoma virus middle T antigens in vivo and in vitro. J Virol. 1981 Oct;40(1):184–196. doi: 10.1128/jvi.40.1.184-196.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel R., Benjamin T. L. Cellular alterations dependent upon the polyoma virus Hr-t function: separation of mitogenic from transforming capacities. Cell. 1978 Jul;14(3):587–599. doi: 10.1016/0092-8674(78)90244-1. [DOI] [PubMed] [Google Scholar]
- Silver J., Schaffhausen B., Benjamin T. Tumor antigens induced by nontransforming mutants of polyoma virus. Cell. 1978 Oct;15(2):485–496. doi: 10.1016/0092-8674(78)90018-1. [DOI] [PubMed] [Google Scholar]
- Smith A. E., Smith R., Griffin B., Fried M. Protein kinase activity associated with polyoma virus middle T antigen in vitro. Cell. 1979 Dec;18(4):915–924. doi: 10.1016/0092-8674(79)90204-6. [DOI] [PubMed] [Google Scholar]
- Staneloni R. J., Fluck M. M., Benjamin T. L. Host range selection of transformation-defective hr-t mutants of polyoma virus. Virology. 1977 Apr;77(2):598–609. doi: 10.1016/0042-6822(77)90485-8. [DOI] [PubMed] [Google Scholar]
- Templeton D., Eckhart W. Mutation causing premature termination of the polyoma virus medium T antigen blocks cell transformation. J Virol. 1982 Mar;41(3):1014–1024. doi: 10.1128/jvi.41.3.1014-1024.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Novak U., Favaloro J., Kamen R. Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature. 1981 Aug 13;292(5824):595–600. doi: 10.1038/292595a0. [DOI] [PubMed] [Google Scholar]
- WINOCOUR E. Purification of polyoma virus. Virology. 1963 Feb;19:158–168. doi: 10.1016/0042-6822(63)90005-9. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson P. F., Tanaka S., Schöld M., Itakura K., Abelson J. Directed deletion of a yeast transfer RNA intervening sequence. Science. 1980 Sep 19;209(4463):1396–1400. doi: 10.1126/science.6997991. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Derbyshire R., Guy A., Molko D., Roget A., Téoule R., Chambon P. Specific in vitro transcription of conalbumin gene is drastically decreased by single-point mutation in T-A-T-A box homology sequence. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7024–7028. doi: 10.1073/pnas.77.12.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]








