Abstract
Human immunodeficiency virus type 1 (HIV1) vectors poorly transduce rhesus hematopoietic cells due to species-specific restriction factors, including the tripartite motif-containing 5 isoformα (TRIM5α) which targets the HIV1 capsid. We previously developed a chimeric HIV1 (χHIV) vector system wherein the vector genome is packaged with the simian immunodeficiency virus (SIV) capsid for efficient transduction of both rhesus and human CD34+ cells. To evaluate whether χHIV vectors could efficiently transduce rhesus hematopoietic repopulating cells, we performed a competitive repopulation assay in rhesus macaques, in which half of the CD34+ cells were transduced with standard SIV vectors and the other half with χHIV vectors. As compared with SIV vectors, χHIV vectors achieved higher vector integration, and the transgene expression rates were two- to threefold higher in granulocytes and red blood cells and equivalent in lymphocytes and platelets for 2 years. A recipient of χHIV vector-only transduced cells reached up to 40% of transgene expression rates in granulocytes and lymphocytes and 20% in red blood cells. Similar to HIV1 and SIV vectors, χHIV vector frequently integrated into gene regions, especially into introns. In summary, our χHIV vector demonstrated efficient transduction for rhesus long-term repopulating cells, comparable with SIV vectors. This χHIV vector should allow preclinical testing of HIV1-based therapeutic vectors in large animal models.
Introduction
Hematopoietic stem cell (HSC)-targeted gene therapy is potentially curative for a number of congenital and acquired disorders and indisputable efficacy has now been demonstrated in several clinical trials involving mostly subjects with immunodeficiencies using autologous HSC transplantation after ex vivo γ-retroviral gene transfer.1,2,3,4,5 However, four out of nine patients in an X-linked severe combined immunodeficiency gene therapy trial developed T cell type acute lymphoblastic leukemia, which was caused by insertional mutagenesis of the proto-oncogenes LMO2 and CCND2.4 In another trial, correction of X-linked chronic granulomatous disease was associated with an in vivo clonal expansion of transduced cells (no leukemia development) with viral insertion into known γ-retroviral vector common integration sites, such as MDS1-EVI1, PRDM16, and SETBP1.6 Evidence now confirms that γ-retroviral vectors tend to integrate near transcription start sites,7,8,9 which implies an inherent risk for integration into promoter regions that might induce insertional mutagenesis. In contrast, lentiviral vectors based on human immunodeficiency virus type 1 (HIV1) have a tendency toward equal integration into all gene regions.8,9 Therefore, HIV1 vectors are thought to possess a lower likelihood of insertional mutagenesis as compared with that of γ-retroviral vectors. In addition, current HIV1 vectors have been further modified to prevent activation of genes around viral integration sites through self-inactivation (SIN) of the long terminal repeats (LTR).10
HIV1 vectors have additional features which render them more suitable for gene therapy approaches to treat hemoglobin disorders. Gene transfer has long been envisioned as a tenable approach to correct hemoglobin disorders through the use of ex vivo transduction of autologous HSCs. The first demonstration of success in rodents was through the use of an HIV1-based lentiviral vector, TNS9, having erythroid specific β-globin expression sufficient to correct a murine model of thalassemia.11Confirmatory studies followed in both β-thalassemia and sickle cell disease models.12,13,14,15,16,17As a preclinical step, we initiated studies to evaluate whether HIV1 vectors have an ability to drive therapeutically relevant levels of β-globin production in large animals. In rhesus macaques, HIV1 transduction is partially blocked by species-specific retroviral restriction.18,19 In order to circumvent this restriction, we modified the TNS9 vector by exchange of the cyclophilin A binding region in the HIV1 capsid with that of a macrophage-tropic HIV1 strain. This vector was previously shown to allow efficient transduction of simian (baboon) cells.20 Shortly following transplantation of the TNS9 transduced rhesus CD34+ cells, human β-globin expression rates of 5% or higher were detectable by flow cytometry. However, long-term gene marking levels decreased to ~0.001% at 2 years, potentially due to additional species-specific HIV1 restriction factors such as the tripartite motif-containing 5 isoform α (TRIM5α).18,19 The use of simian immunodeficiency virus (SIV)-based lentiviral vectors can circumvent this restriction.21 However, the SIV-based vector system is not able to use HIV1-based vector plasmids for preparing functional viral particles,22 a major limitation given that the majority of therapeutic vector constructs have been developed using HIV1-based vector systems.11,12,16,23,24,25,26
To address this issue, we developed a chimeric HIV1-based lentiviral vector system (χHIV vector), in which the HIV1 vector genome is packaged in the context of SIV capsid sequences, by passing the species-specific restriction. Using this χHIV vector, we demonstrated efficient transduction of both human and rhesus CD34+ cells and superior short-term multi-lineage hematopoietic marking to a conventional HIV1 vector in the rhesus transduced CD34+ cell competitive repopulation transplantation model for 3–6 months.22 This χHIV vector system has the advantage that it can use HIV1-based therapeutic vector constructs to prepare viral particles. In this study, we sought to evaluate whether χHIV vectors efficiently transduce long-term repopulating rhesus hematopoietic cells, by comparison with standard SIV vectors in the rhesus HSC competitive repopulation model.
Results
Rhesus hematopoietic stem cell transplantation with lentiviral transduction
We evaluated whether χHIV vectors could transduce rhesus blood cells as efficiently as SIV vectors by performing a competitive repopulation assay in two rhesus macaques (RQ7307 and RQ7280) in which half of the CD34+ cells were transduced with the standard SIN-SIV vector and the other half with the SIN-χHIV vector under otherwise identical conditions (Figure 1a). In a third animal (RQ7387), we used the SIN-χHIV vector alone to evaluate both the transduction efficiency and integration sites of our χHIV vector in rhesus hematopoietic repopulating cells.
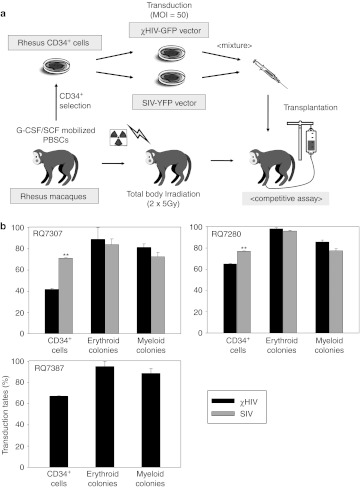
Figure 1.
Competitive repopulation assay for transplantation of rhesus CD34+ cells transduced with the chimeric human immunodeficiency virus type 1 (χHIV) vector and simian immunodeficiency virus (SIV) vector. (a) We evaluated whether the χHIV vector can transduce rhesus hematopoietic cells as efficiently as SIV vectors by performing a competitive repopulation assay in two rhesus macaques (RQ7307 and RQ7280), in which half of the rhesus mobilized CD34+ cells were transduced with the χHIV vector and the other half with the SIV vector under otherwise identical conditions. The third animal (RQ7387) received transplantation of CD34+ cells transduced with the χHIV-green fluorescent protein vector alone to simply evaluate transduction efficiency and integration sites. The transduced CD34+ cells were infused into rhesus macaques following myeloablative total body irradiation (total 10 Gy). Transgene expression in rhesus peripheral blood was followed for over two years after the transduced hematopoietic stem cell transplantation. (b) A small aliquot of transduced CD34+ cells were cultured for an additional 2 days in liquid culture and for an additional 6 days in CFU assay, to evaluate in vitro transduction efficiency. Lower transduction rates in cultured bulk CD34+ cells were found for the χHIV vector (P < 0.01), whereas no significant differences were noted for transduction rates in CFUs between the two vectors. G-CSF, granulocyte colony-stimulating factor; MOI, multiplicity of infection; PBSCs: peripheral blood stem cells;SCF, stem cell factor. *P < 0.01 (two-tailed t-test).
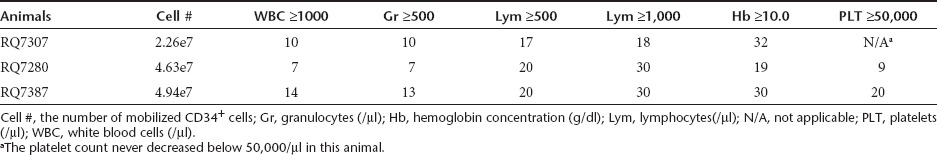
After transplantation, bone marrow suppression followed by rapid reconstitution of peripheral blood cells was observed in all three animals (Table 1). The nadirs of peripheral blood counts were total white blood cells: 290 ± 120/µl (day 6.3 ± 0.3), granulocytes: 120 ± 67/µl (day 6.3 ± 0.3), lymphocytes: 56 ± 17/µl (day 7.3 ± 0.3), hemoglobin concentration 7.6 ± 0.34g/dl (day 12.3 ± 4.8), and platelets 30,000 ± 8,200/µl (day 9.8 ± 1.7). The durations of myelosuppression were 5.0 ± 2.1 days for white blood cells <1,000/µl, and 4.7 ± 1.9 days for granulocytes <500/µl. The recovery dates of each blood cell lineage were day 10.3 ± 2.0 for white blood cells ≥1,000/µl, day 10.0 ± 1.7 for granulocytes ≥500/µl and day 14.5 for platelets ≥50,000/µl (the baseline platelets count for animal RQ7307 was never <50,000/µl) (Table 1). No animal required red blood cell (RBC) transfusion, and 60-ml platelet transfusion was needed for animal RQ7387 on day 13 after transplantation. No complications were observed in the three transplanted animals. After peripheral blood reconstitution, similar blood counts continued for 2 years (Supplementary Figure S1 online).
Table 1. Number of days to recovery of peripheral blood counts after transplantation.
Competitive repopulation assay between the χHIV and SIV vectors in rhesus macaques
After an additional 2 days of liquid culture following transduction, lower transduction rates were found for the χHIV vector than for the SIV vector (Figure 1b, P < 0.01) among bulk cultured CD34+ cells. A small aliquot of cells from the end of transduction was also cultured in methylcellulose media to evaluate their in vitro transduction. Transduction rates among colony forming units were, in contrast, not significantly different between the χHIV and SIV vectors (Figure 1b, not significant ).
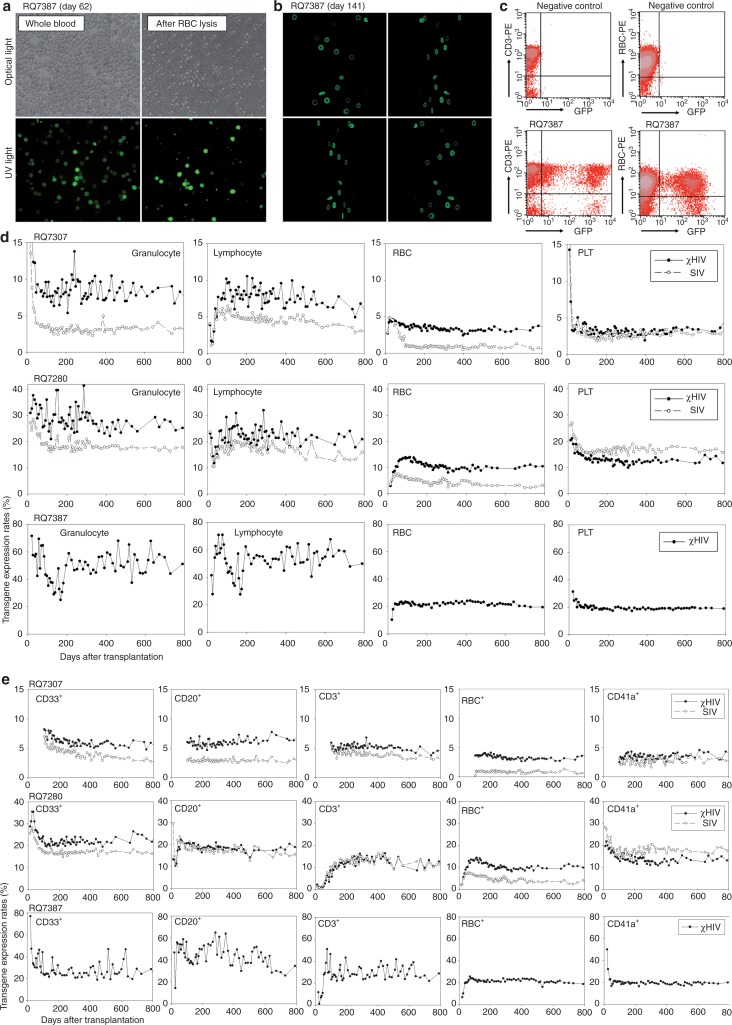
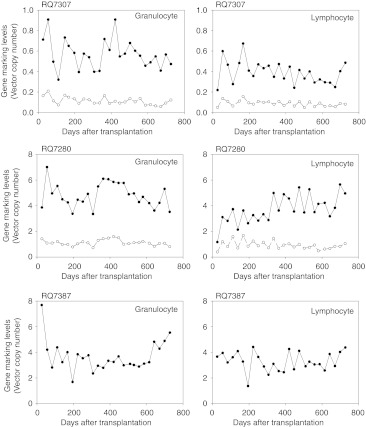
After reconstitution of peripheral blood cells, enhanced green fluorescent protein (GFP) and enhanced yellow fluorescent protein (YFP) expression was determined by fluorescence microscopy (Figure 2a) and by flow cytometry (Figure 2c). We detected the biconcave shape of RBCs by contrast of GFP (and YFP) fluorescence under UV light by a phase-contrast microscope (Figure 2a) and confocal microscope (Figure 2b), suggesting high expression of the transgene from viral constructs in erythroid cells. One month following transplantation, transgene expression rates had not yet reached plateau levels by flow cytometry, but higher GFP transgene expression rates with the χHIV vector as compared with the SIV vector were seen in granulocytes (RQ7307: 12.4 versus 6.1% and RQ7280: 36.1 versus 27.2%) and equivalent transgene expression rates were seen in lymphocytes, RBCs, and platelets. Three to four months after transplantation, in vivo transgene expression rates plateaued, and two- to threefold higher transgene expression rates for the χHIV vector as compared with the SIV vector were seen in granulocytes (RQ7307: 6.3 versus 2.5% and RQ7280: 30.4 versus 17.0%) and RBCs (RQ7307: 3.5 versus 1.0% and RQ7280: 13.8 versus 5.2%), and equivalent transgene expression rates in lymphocytes (RQ7307: 6.0 versus 5.1% and RQ7280: 25.9 versus 17.3%) and platelets (RQ7307: 2.8 versus 2.3% and RQ7280: 13.3 versus 15.4%) (Figure 2d). In peripheral blood cells of the two animals, similar transgene expression rates continued in all cell lineages for at least 2 years (Figure 2d).
Figure 2.
High transgene expression rates with the chimeric human immunodeficiency virus type 1 (χHIV) vector in rhesus peripheral blood cells. (a,b, and c) Following engraftment in the transplanted animals (RQ7387), transgene expression in peripheral blood cells was evaluated by fluorescence microscopy (a) and flow cytometry (c). The brightness of green fluorescent protein (GFP) allowed detection of the biconcave shape of red blood cells by a phase-contrast microscope (a) and confocal microscopy (b). (d) Three to four months after transplantation, in vivo transgene expression rates reached plateau levels in all animals, and 1.5–2 fold higher marking levels, in granulocytes, two- to threefold higher marking levels in red blood cells, and equivalent marking levels in lymphocytes and platelets, were found for the χHIV vector as compared with the SIV vector, over 2 years (RQ7307 and RQ7280). In the third animal (RQ7387) who received cells transduced with the χHIV-GFP vector alone, the transgene expression rates of peripheral blood cells demonstrated high transgene expression rates of 30–40% in granulocytes and lymphocytes and ~20% in red blood cells (RBCs) and platelets. (e) Transgene expression was followed by cell surface analysis of CD33+ granulocytes, CD20+ B lymphocytes, CD3+ T lymphocytes, rhesus RBC-antibody+ red blood cells, and CD41a+ platelets over 2 years. The CD33+ and RBC+ cells showed higher transgene expression rates with the χHIV vector, whereas CD20+, CD3+, and CD41a+ cells had similar transgene expression rates with both vectors. PE, phycoerythrin; PLT, platelets; RBC, red blood cells; UV, ultraviolet.
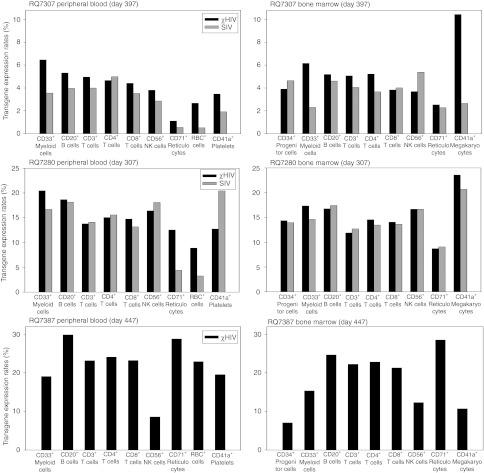
Using cell type specific surface marker analysis in peripheral blood cells of the two animals, the χHIV vector showed 1.3–7.5-fold higher transgene expression rates in CD33+cells (granulocytes), CD71+ cells (reticulocytes), and RBC+ cells, and equivalent transgene expression rates in CD3+ cells (T cells), CD4+ cells (T cells), CD8+ cells (T cells), CD20+ cells (B cells), CD41a+ cells (platelets), and CD56+ cells (natural killer cells) ~1 year after transplantation (Figure 3). However, bone marrow samples demonstrated no distinct difference in transgene expression rates between the χHIV and SIV vectors (Figure 3). When we followed transgene expression rates in CD3+ cells (T cells), CD20+ cells (B cells), CD33+ cells (granulocytes), RBC+ cells, and CD41a+ cells (platelets), similar transgene expression rates were detected in peripheral blood cells over 2 years (Figure 2e). These data demonstrated that after the transgene expression rates plateau, the χHIV vector has equivalent transgene expression rates in rhesus macaques, comparable with the SIV vector. The χHIV vector showed higher transgene expression rates in granulocytes and RBCs, as compared with the SIV vector.
Figure 3.
Multi-lineage transgene expression in peripheral blood cells and bone marrow cells. The chimeric human immunodeficiency virus type 1 (χHIV) vector showed multi-lineage marking in both peripheral blood cells and bone marrow cells that was comparable to that of simian immunodeficiency virus (SIV), with superior transduction rates observed in red blood cells (RBCs).
Efficient viral integration with the χHIV vector in rhesus macaques
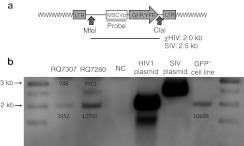
We also evaluated the integrated vector copy number of the χHIV and SIV vectors in both transplanted animals (RQ7307 and RQ7280) by Southern blot analysis (Figure 4). Each vector signal was separated by band size, and the band densities were compared with those of a control cell line that has a single copy of vector integration. We then calculated the average vector copy numbers in the peripheral blood mononuclear cells. The χHIV vector showed several fold higher average vector copy numbers as compared with the SIV vector (RQ7307: 0.37 versus 0.07 and RQ7280: 1.19 versus 0.37) (Figure 4b).
Figure 4.
Integrated copy numbers of chimeric human immunodeficiency virus type 1 (χHIV) and simian immunodeficiency virus (SIV) vectors in vivo. (a) We evaluated the integrated vector copy number of the χHIV vector and SIV vector in transplanted animals (RQ7307 in 11 months after transplantation and RQ7280 in 8 months) by Southern blot analysis. As control, we used a monoclonal GFP+ cell line in which one copy of HIV1 vector was integrated into single cells. The vector sequences were cut out by MfeI and ClaI restriction enzymes, and these signals were detected by the MSCV promoter probe as 2.0 k bands for the χHIV vector and 2.5 k bands for the SIV vector. (b) The densities of vector signals are described next to the appropriate bands. The vector signals were divided by the control signal to calculate the average vector copy numbers. The χHIV vector showed three- to fivefold higher average vector copy numbers as compared with the SIV vector (RQ7307: 0.37 versus 0.07 and RQ7280: 1.19 versus 0.37). LTR, long terminal repeat; MSCV, murine stem cell virus; NC, negative control.
In addition, at various time points following transplantation, we evaluated the gene marking levels of both χHIV and SIV vectors in granulocytes and lymphocytes using real time polymerase chain reaction (PCR) (Figure 5). In the all three animals, similar gene marking levels continued in the peripheral blood cells for 2 years. In the competitive assay animals (RQ7307 and RQ7280), the χHIV vector demonstrated around fourfold higher gene marking levels, as compared with the SIV vector. These integration data suggested higher transduction efficiency for rhesus hematopoietic repopulating cells for the χHIV vector than for the SIV vector.
Figure 5.
Long-term gene marking in peripheral blood cells of transplanted rhesus macaques. We evaluated gene marking levels of the chimeric human immunodeficiency virus type 1 (χHIV) vector and simian immunodeficiency virus (SIV) vector in granulocytes and lymphocytes by real time polymerase chain reaction at various time points following transplantation. In all three animals, similar gene marking levels continued in peripheral blood cells for 2 years. The χHIV vector had around fourfold higher gene marking levels, as compared with the SIV vector, in the competitive assay animals (RQ7307 and RQ7280).
Integration site analysis of the χHIV vector in hematopoietic repopulating cells
We performed transplantation using cytokine mobilized, immunoselected rhesus CD34+ cells transduced with the χHIV vector alone to evaluate integration sites. The transgene expression rates among peripheral blood cells in this animal (RQ7387) plateaued 1–3 months after transplantation, with transgene expression rates of 30–40% in granulocytes and lymphocytes and ~20% in RBCs and platelets (Figure 2d). Multi-lineage marking was observed for 2 years after transplantation by cell surface marker analysis (Figures 2e and 3).
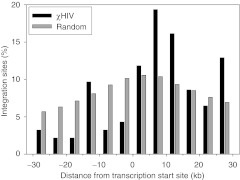
We evaluated integration sites for the χHIV vector in rhesus hematopoietic repopulating cells by linear amplification-mediated PCR, using peripheral blood cells of animal RQ7387 0.5–1.5 years after transplantation. The integration site data were compared with computer-generated random controls (Table 2 and Figure 6).27,28,29 We amplified a total of 344 integration sites for the χHIV vector, and these data demonstrated that the χHIV vector integrated into gene regions, especially introns, as compared with the integration of random controls (Table 2, P < 0.01). On the other hand, the data revealed fewer integrations of the χHIV vector into the regions 30 kb upstream of genes than would be expected at random (Table 2, P < 0.01 and Figure 6). Most of the integration sites were in gene density regions of 0–10 genes within 1 Mb upstream or downstream of integration sites (P < 0.01), as compared with that of random controls (Table 2). No specific trend was noted for the number of integration sites around CpG islands and the common integration sites with the χHIV vector (Table 2). These data suggest that the χHIV vector has integration patterns comparable with HIV1 and SIV vectors.
Table 2. Distribution of integration sites for the χHIV vector in peripheral blood cells.
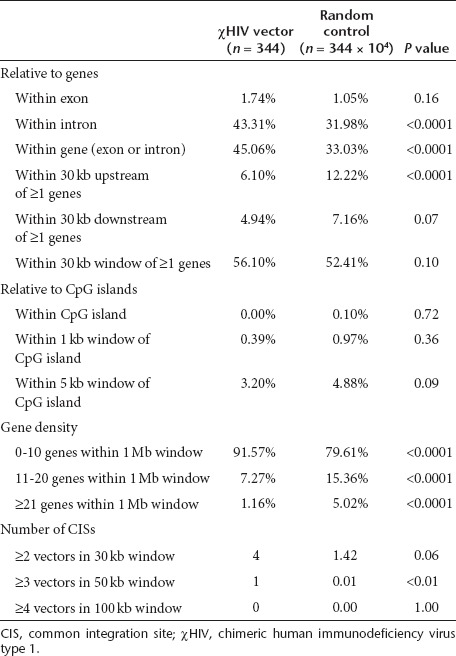
Figure 6.
Distribution of chimeric human immunodeficiency virus type 1 (χHIV) vector integration sites in a 30 kb window around transcription start sites (TSSs). We evaluated the distribution of χHIV vector integration sites from TTSs in peripheral blood cells of the animal RQ7387, and compared these to sites from a random control. The χHIV vector demonstrated a tendency towards more integrations inside of genes and fewer integration upstream of the TSSs.
Discussion
Previously, we demonstrated our χHIV vector allows escape from species-specific restriction in rhesus macaque cells, as judged by the efficient transduction of both rhesus and human CD34+ cells for 3–6 months.22 In the present study, the χHIV vector demonstrated long-term multi-lineage transgene expression comparable with that achieved with the SIV vector, which had previously shown high marking levels in the rhesus HSC transplantation model.21,27Erythroid cells, in particular, showed better transgene expression rates with the χHIV vector in two transplanted rhesus macaques than with the SIV vector. The high level of GFP marking allowed us to recognize the biconcave shape of RBCs using only fluorescence microscopy, and was confirmed by three dimension images using confocal microscopy. Unlike SIV vectors, the χHIV vector system can use HIV1-based therapeutic vector plasmids, which have been extensively tested in mouse disease models. Thus the χHIV vector may be more suitable for preclinical evaluation of transgene expression in rhesus RBCs in vivo.
We previously reported three rhesus macaques that received transplantation of CD34+ cells transduced with the SIV vector alone.21,27 In these animals, transgene expression rates plateaued around 6 months after transplantation, and transgene expression in peripheral blood cells (4–12% in granulocytes and lymphocytes) continued at the same levels for more than 4 years. Our current study also demonstrated stable transgene expression (up to 40% in granulocytes and lymphocytes) from both χHIV and SIV vectors in transplanted rhesus macaques for at least 2 years after arriving at the plateau levels. These data suggest that the transplanted animals acquired immunological tolerance to GFP (and YFP) proteins which were transferred by the χHIV and SIV vectors at the stem cell level. In addition, in the best marking animal in the previously reported recipients of SIV transduced cells, the transgene expression rates were 3% in RBCs and <1% in platelets. In the present study, significantly higher levels (20% in RBCs and 20% in platelets) were observed in the best marking animal (RQ7387).
In the competitive assay animals, stable transgene expression with the GFP-expressing χHIV vector (χHIV-GFP) was observed at 5–10% (RQ7307) and 20–30% (RQ7280) among granulocytes and lymphocytes. These GFP-positive blood cells derived from only half of transplanted HSCs, since one half of transplanted CD34+ cells were transduced with the χHIV vector. Therefore, if we had transduced all rhesus CD34+ cells with the χHIV vector alone for transplantation in these two animals, theoretically the transgene expression rates might have been twice as high at ~40% in granulocytes and lymphocytes and ~20% in RBCs and platelets in animal RQ7280, which were indeed the rates of transgene expression observed in animal RQ7387 (who received cells transduced with the χHIV vector alone).
It is possible that the higher in vivo transgene expression rates seen in our current study were improved not only by the χHIV vector system, but also other factors in the present protocol of rhesus HSC transduction and transplantation. In this study, we used a culture condition of 24-hour prestimulation (with cytokines and without vectors) and 24-hour transduction (with cytokines and vectors, single transduction) with the vesicular stomatitis virus G (VSV-G)-pseudotyped χHIV or SIV vectors at a multiplicity of infection 50, whereas the previous study (with SIV vector only) used a culture condition of 36-hour prestimulation and 36-hour transduction (twice transduction of 24 hours and 12 hours) using the SIV vector with an amphotropic envelope at an multiplicity of infection of 1. Perhaps the longer ex vivo culture previously used reduced the repopulating ability of rhesus HSCs, resulting in less efficient engraftment and lower marking levels. This explanation is supported by our recent data which demonstrated that 24-hour ex vivo culture showed higher repopulating ability of human CD34+ cells in humanized xenograft mice, as compared with that of 48-hour culture.30 The other possibility is that the amphotropic envelope might have lower transduction efficiency for long-term hematopoietic progenitor cells as compared with VSV-G envelope, which has been previously suggested in a humanized mouse model using HIV1 vectors.31
CD34 expression is widely used clinically as a HSC marker. However, the CD34+ cell population contains not only HSCs but also early progenitor cells.32,33 Previously, we demonstrated a significant correlation between in vitro %GFP of cultured CD34+ cells before transplantation and in vivo %GFP of rhesus peripheral blood cells 6 months after transplantation.34 In this study, we cultured a small amount of the transduced CD34+ cells to evaluate transduction rates of bulk CD34+ cells (cultured for an additional 2 days) and colony forming units (cultured for an additional 6 days). The cultured CD34+ cells showed higher transduction rates with SIV vectors than with the χHIV vector (P < 0.01), whereas there was no significant difference in transduction rates in colony forming units between the χHIV and the SIV vectors. In rhesus peripheral blood cells, the χHIV vector showed equivalent or higher transgene expression rates as compared with those of the SIV vector. This suggests that the SIV vector transduced early progenitor cells more efficiently than the χHIV vector.
We evaluated integrated vector copy numbers of both χHIV vector and SIV vector in rhesus peripheral blood cells using Southern blot analysis and real time PCR. The χHIV vector showed an estimated three- to fivefold higher vector integration in vivo, as compared with the SIV vector, whereas the χHIV vector had estimated one- to threefold higher transgene expression rates. The χHIV vector resulted in a more efficient transduction of hematopoietic repopulating cells, over the long term. These results suggest that higher transgene expression rates in the χHIV vector might result from increased viral integration. In addition, our real time PCR data revealed a relative overestimation of the average vector copy numbers as compared with Southern blot analysis, especially in the animal RQ7280 who has high transgene expression rates (20–30% in the χHIV vector). We have empirically used Southern blot analysis to calculate average vector copy numbers because PCR techniques are suitable for sensitive detection with a potential risk of overestimation. In our high marking animals, real time PCR might be too sensitive to accurately estimate vector copy numbers.
We then evaluated integration sites of the χHIV vector in rhesus peripheral blood cells using linear amplification-mediated PCR techniques. We found a total of 344 unique integration sites, which suggested polyclonal reconstitution of rhesus peripheral blood cells. Previously, we demonstrated no monoclonal expansion in peripheral blood cells with stable transgene expression rates in χHIV transplanted rhesus macaques.22 In the three rhesus macaques in this study, no clonal expansion of transduced blood cells was also suggested by stable blood counts for 2 years, stable transgene expression rates and marking levels, and no overexpression of LMO2 and HMGA2 genes in peripheral blood mononuclear cells (Supplementary Figure S2 online), which were reported as genes responsible for insertional mutagenesis in previous gene therapy trials.4,35
In the present study, integration analysis showed a propensity of integration of the χHIV vector into gene regions, especially introns, and less integration upstream of genes. No specific relationship was noted for the integration sites in CpG islands (suggesting promoter regions) and the common integration sites with the χHIV vector. When compared with previous data, the integration sites of the χHIV vector were similar to those of both HIV1 and SIV vectors, but they differed from those of γ-retroviral vectors in keeping with previous reports.7,8,9,27,28,29,36 These data are consistent with theoretically predictable results because the χHIV vector system was constructed similarly to the conventional HIV1 vector system in combination with the SIV capsid and the VSV-G envelope.
Interestingly, the present study showed that the vast majority of integration sites occur in regions of average gene density (0–10 genes per Mb), whereas other studies of SIV and γ-retroviral vectors showed integration into regions of higher gene density.27,28 This difference may be explained by our shorter ex vivo transduction protocol.30 These data also suggest that our transduction protocol can potentially induce less gene activation in long-term progenitor cells, resulting in lower gene density around integration sites.
Lentiviral vectors thus appear well suited for the hemoglobin disorders, and clinical trials have already been initiated. An HIV1 vector system was used recently in a β-thalassemia gene therapy trial in which two patients received autologous bone marrow CD34+ cells that were transduced with a SIN-HIV1 vector containing the β-globin gene under the control of the β-globin locus control region, the β-globin promoter, and the 3′-untranslated region.35 The first patient experienced engraftment failure and received HSC rescue by back-up CD34+ cells, whereas the second patient became transfusion independent at 1 year after transplantation. This transfusion independence was, however, due in equal parts to a rise in endogenous hemoglobin F, endogenous hemoglobin E, and vector derived β-globin. In addition, the rise of vector derived β-globin was associated with an increase of a dominant clone that had an activation of the HMGA2 gene. These results, although encouraging for the field, also demonstrate that significant progress will be necessary before this approach can be expected to benefit those with other genotypes in which there is no production of the β-chain or especially, production of a pathologic β-chain such as that characterizing sickle cell disease.
In summary, this study demonstrates that the χHIV vector can efficiently transduce rhesus long-term hematopoietic progenitor cells at levels comparable with SIV-based vectors. This χHIV vector system should allow preclinical testing of a broad range of HIV1-based therapeutic vectors in large animal models, especially for RBC diseases.
Methods
Lentiviral vector preparation. We previously developed the χHIV vector system to allow for efficient transduction of both human and rhesus hematopoietic cells.22,37 The χHIV vector and SIV vector were prepared by four plasmid cotransfections of 293T cells, as previously described.21,30,38,39,40 The GFP-expressing χHIV vector (χHIV-GFP) was prepared by χHIV Gag/Pol plasmid, HIV1 Rev/Tat plasmid, VSV-G envelope plasmid, and GFP-expressing SIN-HIV1 vector plasmid (pCL20cMpGFP).22 The YFP-expressing SIV vector (SIV-YFP) was prepared by SIV Gap/Pol plasmid, SIV Rev/Tat plasmid, and VSV-G envelope plasmid, YFP-expressing SIN-SIV vector plasmid (pCL20c SLFR MSCV-YFP) which was constructed by exchanging GFP to YFP in pCL20c SLFR MSCV-GFP.21 In both vectors, the transgenes were driven by a murine stem cell virus promoter.
Rhesus hematopoietic stem cell transplantation. We previously developed the autologous peripheral blood stem cells transplantation model in rhesus macaques by granulocyte colony-stimulating factor (kindly provided by Amgen, Thousand Oaks, CA) and stem cell factor (kindly provided by Swedish Orphan Biovitrum AB, Stockholm, Sweden) mobilization (Figure 1a).41 After immunoselection of rhesus CD34+ cells, as previously described,22 the CD34+ cells were cultured in fibronectin CH-296-coated (RetroNectin; TaKaRa, Otsu, Shiga, Japan) flasks using serum-free X-VIVO10 media (Lonza, Allendale, NJ) containing stem cell factor, FMS-related tyrosine kinase 3 ligand, and thrombopoietin (all 100 ng/ml; R&D Systems, Minneapolis, MN). After 24-hour prestimulation, half of the cells were transduced at multiplicity of infection 50 with the χHIV-GFP vector in the fresh media with the same concentration of cytokines and the other half of the cells were transduced with SIV-YFP vector, using the same conditions, for both animals (RQ7307 and RQ7280). In an additional animal (RQ7387), the CD34+ cells were transduced with the χHIV-GFP vector alone at multiplicity of infection 50. After 24-hour transduction, the transduced cells were infused into rhesus macaques, which had undergone myeloablative conditioning therapy of split dose total body irradiation (5Gyx2, total 10Gy). The in vitro transduction efficiency was evaluated by flow cytometry (FACSCalibur; BD Biosciences, Franklin Lakes, NJ) using a small number of transduced rhesus CD34+ cells following culture in liquid media for an additional 2 days. In addition, the transduced rhesus CD34+ cells were cultured in methylcellulose media (1,000 cells in 1.5 ml) for an additional 5–6 days. The transduction efficiency in colony forming units was calculated by GFP (or YFP) positive and negative colony counts, which were evaluated by fluorescence microscope.22 After engraftment, we evaluated the complete blood counts, transgene expression by fluorescence microscope and flow cytometry (Figure 2c), and integration site analysis. In addition, we evaluated GFP expression in RBCs by confocal microscope (Carl Zeiss, Thornwood, NY) under ultraviolet light. Three dimension images were produced by LSM Image Browser Rel. 4.2 (Carl Zeiss).
Southern blot analysis. Viral integration into genomic DNA was evaluated by digesting 10 µg total DNA extracted from peripheral blood mononuclear cells with MfeI and ClaI restriction enzymes (New England Biolabs, Ipswich, MA) (Figure 4a). The signals were detected by the murine stem cell virus-promoter probe (the 457b MluI-Tth111I fragment from pCL20c MpGFP) using chemifluorescent reactions. The two vector signals were separated by size (2.0 k bands for the χHIV vector and 2.5 k bands for the SIV vector). The density of vector signals was evaluated by ImageJ (National Institutes of Health, Bethesda, MD) and these were compared with that of a monoclonal cell line in which a single copy of HIV1 vector was integrated. The vector signals were divided by the control signal to calculate average integration vector copy numbers.
Real time PCR. Viral gene marking levels in peripheral blood cells were evaluated by real time PCR. Genomic DNA was extracted form granulocytes and lymphocytes in transplanted animals, which were separated by mononuclear cell separation. The χHIV-GFP and SIV-YFP vector signals were detected by GFP specific probe/primers and YFP specific probe/primers, as previously described.22 Average vector copy numbers were calculated by as compared with those of a monoclonal cell line in which a single copy of HIV1 vector was integrated.
Gene expression levels of LMO2 and HMGA2 in peripheral blood cells were evaluated by real time reverse transcription PCR. Total RNA was extracted from peripheral mononuclear cells in transplanted animals, and cDNA was synthesized by a reverse transcriptase (SuperScript III Reverse Transcriptase; Life Technologies, Carlsbad, CA). The gene signals were detected by specific probe/primers targeting rhesus LMO2 exons 4 and 5 and HMGA2 exons 2 and 3(TaqMan Gene Expression Assay; Life Technologies) and standardized by 18S ribosomal RNA (TaqMan Ribosomal RNA Control Reagents; Life Technologies).
Integration site analysis. We evaluated the integration sites of the χHIV vector in rhesus long-term repopulating cells by performing linear amplification-mediated PCR using peripheral blood samples from animal RQ7387 at 0.5–1.5 years after transplantation, as previously described.7,21,38,42 Briefly, the HIV3-I primer (5′-biotin-TTT TGC CTG TAC TGG GTC TCT CTG-3′) targets the LTR sequence and it can be used for linear amplification of the 3′-end junction. The amplified DNA was digested by Tsp509I restriction enzyme, and it was ligated to asymmetric DNA-linker (LC-Tsp-a: 5′-ACT GAC AGC GGA GAT AAT CGG TGC GAG TAG CAT ACT AGA G-3′, LC-Tsp-b: 5′-AAT TCT CTA GTA TGC TAC TCG CAC CGA TTA TCT CCG CTG TCA GT-3′). The ligated DNA of 3′LTR-genomic DNA-linker was amplified by nested PCR using HIV3-II (5′-TCT CTG GCT AAC TAG GGA AC-3) and LC-1 (5′-ACT GAC AGC GGA GAT AAT CG-3′) primers for first PCR, and HIV3-III (5′-GCC TTG AGT GCT TCA AGT AGT G-3′) and LC-2 (5′-GTG CGA GTA GCA TAC TAG AG-3′) primers for second PCR. The HIV3-II and HIV3-III primers target the LTR sequence, and the LC-1 and LC-2 primers target the linker sequence. Integration site sequences were validated with the intact LTR-genomic junction. We aligned the 344 unique integration sites to the rhesus macaque genome assembly (Mmul 1.0, January 2006) and determined their genomic context by comparing their locations to those of the 21,905 protein coding genes in Ensembl 61 (February 2011). To evaluate the statistical significance of our findings, we created 10,000 random datasets, each with 344 mock integration sites, as previously described.27,28,29 We evaluated common integration sites with the χHIV vector using three criteria: second order: two or more integration sites within a 30 kb window, third order: three or more integration sites within a 50 kb window, and fourth order: four or more integration sites within a 100 kb window.28
Statistical analysis. Statistical analyses were performed using JMP 9 software (SAS Institute, Cary, NC). Transduction rates of two vectors were evaluated by the student's t-test. A P value of <0.01 was deemed significant. Standard errors of the mean are shown as error bars.
SUPPLEMENTARY MATERIAL Figure S1. Peripheral blood counts after transplantation. Figure S2. LMO2 and HMGA2 gene expression in peripheral blood cells of transplanted rhesus macaques.
Acknowledgments
This work is supported by the intramural research programs of the National Institute of Diabetes, Digestive, and Kidney Diseases, the National Heart, Lung, and Blood Institute, and the National Human Genome Research Institute at the National Institutes of Health. We thank the animal care staff and technicians at 5 Research Court for their excellent care and handling of the animals. We would also like to thank William DeGraff and Dr Jim Mitchell of the Radiation Biology Branch, National Cancer Institute, for the use of the Eldorado Cobalt-60 irradiator. We would like to thank Heidi Dorward, Medical Genetics Branch, National Human Genome Research Institute,for the use of the confocal microscope. The authors have declared that no conflict of interest exists.
Supplementary Material
Peripheral blood counts after transplantation.
LMO2 and HMGA2 gene expression in peripheral blood cells of transplanted rhesus macaques.
REFERENCES
- Schwarzwaelder K, Howe SJ, Schmidt M, Brugman MH, Deichmann A, Glimm H.et al. (2007Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo J Clin Invest 1172241–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiuti A, Slavin S, Aker M, Ficara F, Deola S, Mortellaro A.et al. (2002Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning Science 2962410–2413. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L.et al. (2009Gene therapy for immunodeficiency due to adenosine deaminase deficiency N Engl J Med 360447–458. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E.et al. (2008Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1 J Clin Invest 1183132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E.et al. (2003A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency N Engl J Med 348255–256. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U.et al. (2006Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1 Nat Med 12401–409. [DOI] [PubMed] [Google Scholar]
- Hematti P, Hong BK, Ferguson C, Adler R, Hanawa H, Sellers S.et al. (2004Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells PLoS Biol 2e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC.et al. (2004Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences PLoS Biol 2E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B., and, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Ryu BY, Evans-Galea MV, Gray JT, Bodine DM, Persons DA., and, Nienhuis AW. An experimental system for the evaluation of retroviral vector design to diminish the risk for proto-oncogene activation. Blood. 2008;111:1866–1875. doi: 10.1182/blood-2007-04-085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May C, Rivella S, Callegari J, Heller G, Gaensler KM, Luzzatto L.et al. (2000Therapeutic haemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin Nature 40682–86. [DOI] [PubMed] [Google Scholar]
- Pestina TI, Hargrove PW, Jay D, Gray JT, Boyd KM., and, Persons DA. Correction of murine sickle cell disease using gamma-globin lentiviral vectors to mediate high-level expression of fetal hemoglobin. Mol Ther. 2009;17:245–252. doi: 10.1038/mt.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivella S, May C, Chadburn A, Rivière I., and, Sadelain M. A novel murine model of Cooley anemia and its rescue by lentiviral-mediated human beta-globin gene transfer. Blood. 2003;101:2932–2939. doi: 10.1182/blood-2002-10-3305. [DOI] [PubMed] [Google Scholar]
- Persons DA, Allay ER, Sabatino DE, Kelly P, Bodine DM., and, Nienhuis AW. Functional requirements for phenotypic correction of murine beta-thalassemia: implications for human gene therapy. Blood. 2001;97:3275–3282. doi: 10.1182/blood.v97.10.3275. [DOI] [PubMed] [Google Scholar]
- Imren S, Payen E, Westerman KA, Pawliuk R, Fabry ME, Eaves CJ.et al. (2002Permanent and panerythroid correction of murine beta thalassemia by multiple lentiviral integration in hematopoietic stem cells Proc Natl Acad Sci USA 9914380–14385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveetil G, Scholes J, Carbonell D, Qureshi N, Xia P, Zeng L.et al. (2004Successful correction of the human beta-thalassemia major phenotype using a lentiviral vector Blood 1043445–3453. [DOI] [PubMed] [Google Scholar]
- Malik P, Arumugam PI, Yee JK., and, Puthenveetil G. Successful correction of the human Cooley's anemia beta-thalassemia major phenotype using a lentiviral vector flanked by the chicken hypersensitive site 4 chromatin insulator. Ann N Y Acad Sci. 2005;1054:238–249. doi: 10.1196/annals.1345.030. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P., and, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H.et al. (2006Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor Proc Natl Acad Sci USA 1035514–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kootstra NA, Munk C, Tonnu N, Landau NR., and, Verma IM. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc Natl Acad Sci USA. 2003;100:1298–1303. doi: 10.1073/pnas.0337541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa H, Hematti P, Keyvanfar K, Metzger ME, Krouse A, Donahue RE.et al. (2004Efficient gene transfer into rhesus repopulating hematopoietic stem cells using a simian immunodeficiency virus-based lentiviral vector system Blood 1034062–4069. [DOI] [PubMed] [Google Scholar]
- Uchida N, Washington KN, Hayakawa J, Hsieh MM, Bonifacino AC, Krouse AE.et al. (2009Development of a human immunodeficiency virus type 1-based lentiviral vector that allows efficient transduction of both human and rhesus blood cells J Virol 839854–9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa H, Hargrove PW, Kepes S, Srivastava DK, Nienhuis AW., and, Persons DA. Extended beta-globin locus control region elements promote consistent therapeutic expression of a gamma-globin lentiviral vector in murine beta-thalassemia. Blood. 2004;104:2281–2290. doi: 10.1182/blood-2004-03-0863. [DOI] [PubMed] [Google Scholar]
- Persons DA, Hargrove PW, Allay ER, Hanawa H., and, Nienhuis AW. The degree of phenotypic correction of murine beta -thalassemia intermedia following lentiviral-mediated transfer of a human gamma-globin gene is influenced by chromosomal position effects and vector copy number. Blood. 2003;101:2175–2183. doi: 10.1182/blood-2002-07-2211. [DOI] [PubMed] [Google Scholar]
- Samakoglu S, Lisowski L, Budak-Alpdogan T, Usachenko Y, Acuto S, Di Marzo R.et al. (2006A genetic strategy to treat sickle cell anemia by coregulating globin transgene expression and RNA interference Nat Biotechnol 2489–94. [DOI] [PubMed] [Google Scholar]
- Arumugam PI, Scholes J, Perelman N, Xia P, Yee JK., and, Malik P. Improved human beta-globin expression from self-inactivating lentiviral vectors carrying the chicken hypersensitive site-4 (cHS4) insulator element. Mol Ther. 2007;15:1863–1871. doi: 10.1038/sj.mt.6300259. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Kim YS, Larochelle A, Renaud G, Wolfsberg TG, Adler R.et al. (2009Sustained high-level polyclonal hematopoietic marking and transgene expression 4 years after autologous transplantation of rhesus macaques with SIV lentiviral vector-transduced CD34+ cells Blood 1135434–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métais JY, Topp S, Doty RT, Borate B, Nguyen AD, Wolfsberg TG.et al. (2010Feline leukemia virus integrase and capsid packaging functions do not change the insertion profile of standard Moloney retroviral vectors Gene Ther 17799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler T, Cantilena A, Métais JY, Xu X, Nguyen AD, Borate B.et al. (2010No evidence for clonal selection due to lentiviral integration sites in human induced pluripotent stem cells Stem Cells 28687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Hsieh MM, Hayakawa J, Madison C, Washington KN., and, Tisdale JF. Optimal conditions for lentiviral transduction of engrafting human CD34+ cells. Gene Ther. 2011;18:1078–1086. doi: 10.1038/gt.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Wielgosz MM, Hargrove P, Kepes S, Gray J, Persons DA.et al. (2010Transduction of human primitive repopulating hematopoietic cells with lentiviral vectors pseudotyped with various envelope proteins Mol Ther 181310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D. Haematopoietic stem cells. J Pathol. 2002;197:430–440. doi: 10.1002/path.1153. [DOI] [PubMed] [Google Scholar]
- Guo Y, Lübbert M., and, Engelhardt M. CD34- hematopoietic stem cells: current concepts and controversies. Stem Cells. 2003;21:15–20. doi: 10.1634/stemcells.21-1-15. [DOI] [PubMed] [Google Scholar]
- Uchida N, Bonifacino A, Krouse AE, Metzger ME, Csako G, Lee-Stroka A.et al. (2011Accelerated lymphocyte reconstitution and long-term recovery after transplantation of lentiviral-transduced rhesus CD34+ cells mobilized by G-CSF and plerixafor Exp Hematol 39795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F.et al. (2010Transfusion independence and HMGA2 activation after gene therapy of human ß-thalassaemia Nature 467318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa J, Washington K, Uchida N, Phang O, Kang EM, Hsieh MM.et al. (2009Long-term vector integration site analysis following retroviral mediated gene transfer to hematopoietic stem cells for the treatment of HIV infection PLoS ONE 4e4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Princiotta M, Piatak M, Jr, Yuan F, Zhang F, Lifson JD.et al. (2006Generation of simian-tropic HIV-1 by restriction factor evasion Science 31495. [DOI] [PubMed] [Google Scholar]
- Uchida N, Hanawa H, Dan K, Inokuchi K., and, Shimada T. Leukemogenesis of b2a2-type p210 BCR/ABL in a bone marrow transplantation mouse model using a lentiviral vector. J Nihon Med Sch. 2009;76:134–147. doi: 10.1272/jnms.76.134. [DOI] [PubMed] [Google Scholar]
- Uchida N, Washington KN, Lap CJ, Hsieh MM., and, Tisdale JF. Chicken HS4 insulators have minimal barrier function among progeny of human hematopoietic cells transduced with an HIV1-based lentiviral vector. Mol Ther. 2011;19:133–139. doi: 10.1038/mt.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa H, Kelly PF, Nathwani AC, Persons DA, Vandergriff JA, Hargrove P.et al. (2002Comparison of various envelope proteins for their ability to pseudotype lentiviral vectors and transduce primitive hematopoietic cells from human blood Mol Ther 5242–251. [DOI] [PubMed] [Google Scholar]
- Donahue RE, Kirby MR, Metzger ME, Agricola BA, Sellers SE., and, Cullis HM. Peripheral blood CD34+ cells differ from bone marrow CD34+ cells in Thy-1 expression and cell cycle status in nonhuman primates mobilized or not mobilized with granulocyte colony-stimulating factor and/or stem cell factor. Blood. 1996;87:1644–1653. [PubMed] [Google Scholar]
- Schmidt M, Zickler P, Hoffmann G, Haas S, Wissler M, Muessig A.et al. (2002Polyclonal long-term repopulating stem cell clones in a primate model Blood 1002737–2743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Peripheral blood counts after transplantation.
LMO2 and HMGA2 gene expression in peripheral blood cells of transplanted rhesus macaques.