Abstract
In eukaryotic cells, lysosomes represent a major site for macromolecule degradation. Hydrolysis products are eventually exported from this acidic organelle into the cytosol through specific transporters. Impairment of this process at either the hydrolysis or the efflux step is responsible of several lysosomal storage diseases. However, most lysosomal transporters, although biochemically characterized, remain unknown at the molecular level. In this study, we report the molecular and functional characterization of a lysosomal amino acid transporter (LYAAT-1), remotely related to a family of H+-coupled plasma membrane and synaptic vesicle amino acid transporters. LYAAT-1 is expressed in most rat tissues, with highest levels in the brain where it is present in neurons. Upon overexpression in COS-7 cells, the recombinant protein mediates the accumulation of neutral amino acids, such as γ-aminobutyric acid, l-alanine, and l-proline, through an H+/amino acid symport. Confocal microscopy on brain sections revealed that this transporter colocalizes with cathepsin D, an established lysosomal marker. LYAAT-1 thus appears as a lysosomal transporter that actively exports neutral amino acids from lysosomes by chemiosmotic coupling to the H+-ATPase of these organelles. Homology searching in eukaryotic genomes suggests that LYAAT-1 defines a subgroup of lysosomal transporters in the amino acid/auxin permease family.
Lysosomes are dense organelles responsible for degrading all four classes of macromolecules. The degradation products are then exported to the cytosol through specific transporters and reused in the cellular metabolism. The physiological importance of lysosomal metabolite efflux is illustrated by the existence of a group of lysosomal storage diseases with transport defects, such as sialic acid storage disorders and nephropathic cystinosis (1). These inherited diseases result from defective efflux of sialic acid and cystine from lysosomes (2–6), respectively, and they have been linked to mutations in the membrane proteins sialin and cystinosin (7, 8), which are believed to represent sialic acid and cystine transporters. However, most lysosomal transporters, although biochemically characterized (1, 9), remain unknown at the molecular level.
Twenty amino acid transporter families have been identified thus far (10). In eukaryotes, most of the characterized transporters have been shown to operate at the level of the plasma membrane (11) or mitochondria. The eukaryotic specific amino acid/auxin permease (AAAP) family (12) differs in this respect because, whereas it was first recognized as a family of H+/amino acid symporters operating at the plasma membrane of plant cells (13, 14), it was later shown to comprise the transporter responsible for the loading of inhibitory amino acids [γ-aminobutyric acid (GABA) and glycine] into synaptic vesicles of animal nerve cells (15, 16) through an H+ antiport mechanism. More recently, additional animal members were identified as the plasma membrane transporters corresponding to system N (17) and system A (18). In this study, we report the characterization of a novel mammalian member of the AAAP family that displays the functional characteristics of an H+/amino acid symporter and that is localized in the lysosomes of brain neurons. This transporter is thus proposed to ensure the export of neutral amino acids from lysosomes.
Methods
cDNA Cloning.
A 495-bp fragment was amplified on the IMAGE (19) cDNA clone number 45237, corresponding to human expressed sequence tag y186d11.r1 (accession number H08076) using primers 5′-AAGCTTGGCACGAGGCGTTTCC (sense) and 5′-TAACCAAGAAGGATCTTATCCC (antisense), and used as a probe to screen a rat hippocampus cDNA library (lambda ZAPII, number 936518, Stratagene) at high stringency. Several partial clones were obtained, and one of them was used to screen again the same library at moderate stringency, thereby allowing the isolation of a 1.824-bp cDNA clone that was subcloned into the pcDNA3 expression vector (Invitrogen) for expression and cRNA synthesis.
Northern Analysis.
A Northern blot with 2 μg of poly(A)+ mRNA from different rat tissues (CLONTECH) was hybridized as recommended by the manufacturer with a 32P-labeled probe obtained by PCR amplification on pcDNA3-LYAAT-1 using primers 5′-ACAGGTGATAGAGGCAGCCAACGG (sense) and 5′-CACTGGTGGAATTGGTAGAGGAGT (antisense).
In Situ Hybridization.
Adult male Sprague–Dawley rats were anesthetized with pentobarbital and perfused transcardially with 4% paraformaldehyde in PBS (pH 7.4). After dissection, brains were postfixed 2 h in the same solution at 4°C, then cryoprotected in 15% sucrose/PBS, frozen, and stored at −80°C until used. Cryosections (14 μm) were hybridized either with 35S-labeled oligonucleotides or digoxigenin-labeled cRNA probes. Three 35S-labeled oligonucleotidic probes, localized either in the coding (positions 184–219 bp; accession number AF361239) or the 3′ noncoding (positions 1574–1634 and 1666–1722 bp) regions of LYAAT-1 cDNA, were hybridized independently according to a standard protocol and visualized by exposure to βmax x-ray film (Amersham Pharmacia). The same mRNA distribution was observed with the three oligonucleotidic probes. Visualization at the cell level was reached by hybridizing brain sections with a cRNA probe labeled with digoxigenin-11-UTP (Promega) using the 1.8-kb LYAAT-1 insert in pcDNA3 as template. Hybridization was performed overnight at 65°C in 1 × SSC/50% formamide/10% dextran sulfate/1 mg/ml rRNA/1 × Denhardt's solution. Sections were then washed twice with 1 × SSC/50% formamide/0.1% Tween 20 (at 65°C) and twice with 1 × TBST (at room temperature). The digoxigenin-labeled hybrids were detected with an alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche Molecular Biochemicals).
Transport Assay.
Expression of LYAAT-1 in CV-1 cells using the vaccinia virus/bacteriophage T7 system for transport assays at pH 7.5 was performed according to Povlock and Amara (20). For other experiments, COS-7 cells were transfected with Lipofectin (Life Technologies, Grand Island, NY). Briefly, 1 day before transfection, 50,000 cells/well were plated in 24-well dishes. The day of transfection, cells were washed once with 0.5 ml of serum-free medium and then incubated 16–20 h with 250 μl of serum-free medium containing a complex formed by 3 μl of Lipofectin and 1 μg of pcDNA3 or pcDNA3-LYAAT-1. One milliliter of medium supplemented with FCS was added on the following day, and transport assays were performed 36–48 h after the beginning of transfection. Cells were washed twice with 0.5 ml of Krebs–Ringer (KR) phosphate buffer (146 mM NaCl/3 mM KCl/1 mM CaCl2/1 mM MgCl2/10 mM KH2PO4/K2HPO4) adjusted at pH 7.5 and then incubated for 15 min at 26°C in KR buffer adjusted at pH 5.5, supplemented with 0.5–1 μCi of [3H]GABA and 100 μM GABA unless stated otherwise in the text. Reaction was terminated by two washes with ice-cold KR buffer at pH 7.5. Cells were lysed in 0.1 N NaOH, and their radioactivity was measured after neutralization by scintillation counting in Aquasol (Packard). [3H]GABA, L-[3H]glutamine and L-[3H]glutamate were purchased from Amersham Pharmacia; L-[3H]proline and L-[3H]alanine were purchased from NEN.
Immunocytochemistry.
A polyclonal antibody raised against the peptide SSTDVSPEESPSEGLGC (amino acid residues 15–30) coupled to keyhole limpet hemocyanin was produced in rabbit (Eurogentec, Brussels). For immunological detection of LYAAT-1 on brain sections, the antibody was affinity-purified on the peptide linked to Affigel.
Transfected COS-7 cells were grown on glass coverslips, fixed for 10 min in PBS containing 4% paraformaldehyde, rinsed in PBS, blocked and permeabilized in blocking buffer (PBS/0.2% Triton X-100/5% donkey serum), incubated for at least 1 h with anti-LYAAT-1 serum diluted at 1:500 in the same buffer, washed in PBS, incubated with Cy3-conjugated donkey anti-rabbit IgG (Jackson Immunochemicals) at 1:1,600 in blocking buffer, and rinsed in PBS. Coverslips were mounted on glass slides with Fluoromount-G solution (Southern Biotechnology Associates). All steps were performed at room temperature.
Immunoautoradiographic detection of LYAAT-1 was performed on 14-μm sections of frozen brains from rats anesthetized with pentobarbital and perfused transcardially with 100 ml of 0.9% NaCl/0.1% NaNO2. Sections were fixed for 5 min at 4°C in 4% paraformaldehyde/PBS, preincubated for 1 h in PBS containing 3% BSA and 1% donkey serum (B1 buffer), and incubated overnight at 4°C with affinity-purified anti-LYAAT-1 antibody at 1:250 in B1 buffer. On the following day, sections were washed in PBS, incubated in B1 buffer containing anti-rabbit [125I]IgG (0.2 μCi/ml, 750-3000 Ci/mmol; Amersham Pharmacia), rinsed in PBS, dried, and exposed for 4–5 days.
Immunofluorescence detection of LYAAT-1 was performed on brains from adult rats transcardially perfused with 4% paraformaldehyde in 100 mM potassium phosphate buffer (pH 7.4), postfixed in the same solution for 4 h and cryoprotected in 10% sucrose at 4°C for 2 days. All of the following steps were performed at room temperature. Sections (14 μm) were blocked for 30 min in PBS containing 0.2% gelatin/0.25% Triton X-100 (B2 buffer) and incubated overnight with affinity-purified anti-LYAAT-1 (1:500) and goat anti-cathepsin D antibodies (1:100; Santa Cruz Biotechnology) in B2 buffer. Sections were then washed in B2 buffer, incubated for 90 min with Cy3-conjugated donkey anti-rabbit IgG (1:1,600) and Alexa 488-conjugated donkey anti-goat IgG (1:1,000; Molecular Probes) in B2 buffer, washed in PBS, and mounted with Fluoromount-G solution. Sections were observed under a laser-scanning confocal microscope (Leica TCS 400), and images were generated with Adobe Photoshop 5.0.
Results
cDNA Isolation.
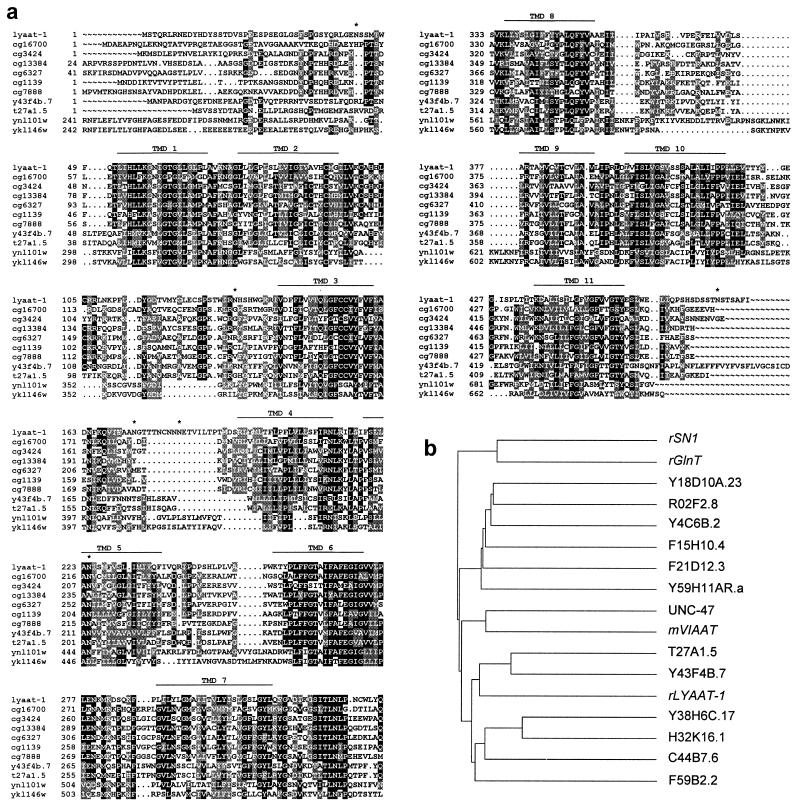
Homology searching (21) in public databases using the amino acid sequence of the synaptic vesicle inhibitory amino acid transporter [VIAAT (15) or VGAT (16)], a member of the AAAP protein family (12), revealed several homologous human brain expressed sequence tags. The IMAGE clone (19) corresponding to one of these expressed sequence tags (accession no. H08076) was used to isolate from a rat hippocampus cDNA library a 1,824-bp cDNA containing a 1,425-bp ORF, with an in-phase STOP codon 120-bp upstream of the initiation codon. The corresponding 475-aa protein, named LYAAT-1, is predicted to comprise 11 transmembrane domains, a cytosolic N-terminal domain, and 3 consensus N-glycosylation sites in predicted extracytosolic loops (Fig. 1a). Pairwise sequence alignment, using the Besfit software of the GCG 10.0-UNIX package, indicated that LYAAT-1 is distantly related to previously characterized members of the AAAP family, such as the mouse VIAAT (15) (24% identity over 408 aa residues), the rat plasma membrane glutamine transporters SN1 (17) (22% over 190) and GlnT (18) (24% over 402), and the plant amino acid permease AAP1 (13) (26% over 457). Interestingly, homology searching in totally sequenced eukaryotic genomes revealed that LYAAT-1 represents a mammalian homologue for a subgroup of functionally uncharacterized members of the AAAP family, as illustrated in Fig. 1b for the nematode worm Caenorhabditis elegans. The six putative proteins of this C. elegans AAAP family subgroup exhibit 29–36% identity to LYAAT-1 over 379–469 aa. In the fruit fly Drosophila melanogaster, seven predicted proteins (CG16700, CG3424, CG13384, CG6327, CG1139, CG7888, and CG8785) are 36–46% identical to LYAAT-1. In the yeast Saccharomyces cerevisiae, two of the seven AAAP family members, YNL101w (40% over 311) and YKL146w (32% over 466), display closer homology to LYAAT-1. Alignment of sequences from this AAAP family subgroup in several species, including rat LYAAT-1, reveals a high conservation in putative transmembrane domains 1, 3, 6, and 10, as well as in the putative cytosolic loop connecting transmembrane domains 7 and 8 (Fig. 1a). Analysis of high throughput genomic sequence databases shows a cluster of human contigs (accession nos. AC025433, AC008520, AC034205, AC011337, AC008552, AC008385, and AC011391) located at chromosome 5q31-33, which allows reconstitution, within ten exons, of a human protein sharing 83% identity and 94% similarity with the rat LYAAT-1.
Figure 1.
Alignment of LYAAT-1 to a subgroup of the AAAP family. (a) Multiple alignment of the LYAAT-1 sequence with those of D. melanogaster putative proteins CG16700, CG3424, CG13384, CG6327, CG1139, and CG7888, C. elegans putative proteins Y43F4B.7 and T27A1.5, and S. cerevisiae predicted proteins YNL101w and YKL146w was performed with the PILEUP software of the GCG 10.0-UNIX package. Black boxes indicate identical residues, and gray boxes indicate conservative substitutions. Potential transmembrane domains and sites for N-glycosylation are indicated above alignments by lines and stars, respectively. (b) Dendrogram for the 13 members of the AAAP family in C. elegans, as well as the four functionally characterized mammalian members (in italics). LYAAT-1 shows a closer homology to the worm proteins T27A1.5, Y43F4B.7, Y38H6C.17, H32K16.1, C44B7.6, and F59B2.2.
Distribution of LYAAT-1 mRNA and Protein.
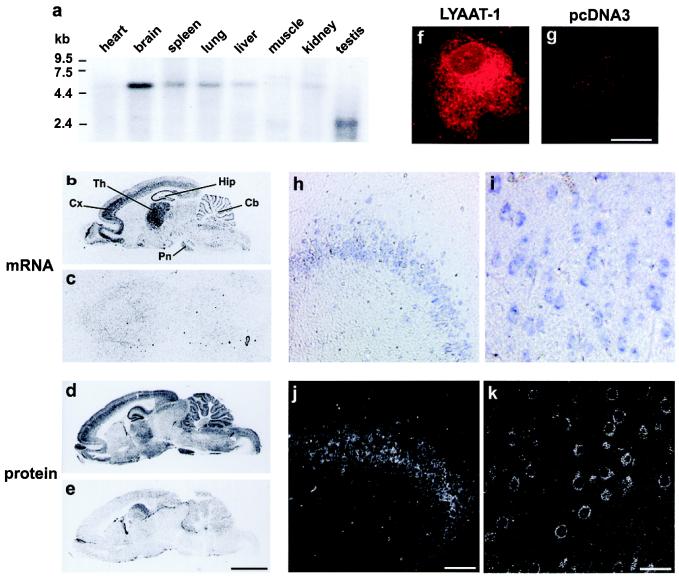
Northern blot analysis revealed that LYAAT-1 is largely expressed in rat tissues and is most abundant in the brain (Fig. 2a). In situ hybridization on brain sections showed that LYAAT-1 mRNA is abundantly found in hippocampus, cerebral cortex, cerebellum, and the thalamic and pontine nuclei, regions known to be rich in neurons using either glutamate or GABA as neurotransmitter (Fig. 2b). Indeed, we confirmed at the cellular level that LYAAT-1 transcript is expressed in glutamatergic neurons such as pyramidal cells in the cerebral cortex and hippocampus, thalamic and pontine nuclei neurons, and GABAergic neurons such as Purkinje cells in the cerebellum (Fig. 2 h and i, and data not shown). Immunodetection of LYAAT-1 on brain sections using an antibody directed against a peptide located in the N-terminal domain of the transporter revealed that the protein, although widely present throughout the encephalon, is more abundant in regions expressing high levels of LYAAT-1 mRNA (Fig. 2d). At the cellular level, immunocytochemistry confirmed that LYAAT-1 is mainly localized in the somatodendritic domain of neurons (Fig. 2 j and k). By contrast, neither LYAAT-1 mRNA nor protein could be detected in glial cells, suggesting that LYAAT-1 expression in brain is restricted to neurons.
Figure 2.
Northern analysis, in situ hybridization, and immunocytochemistry show that LYAAT-1 is predominantly expressed in the adult rat brain, where it is present in glutamatergic and GABAergic neurons. (a) Northern blot analysis of poly(A)+ mRNA with a radiolabeled probe against LYAAT-1 shows a 5.2-kb transcript in all tissues tested, except testis and muscle, where doublets around 2.4 kb and 6 kb are detected, respectively. (b and c) 35S-labeled antisense oligonucleotide probe (b) and the corresponding sense probe (c) were hybridized with brain parasagittal sections. LYAAT-1 mRNA is widely distributed throughout the brain, with higher expression in the cortex, thalamus, pyramidal cells of the hippocampus, and Purkinje cells of the cerebellum, with no significant hybridization by the sense probe. Cb, cerebellum; Cx, cortex; Hip, hippocampus; Pn, pontine nuclei; Th, thalamus. (f and g) A rabbit anti-LYAAT-1 antibody detected by immunofluorescence (red) a protein mainly expressed in intracellular compartments in COS-7 cells transfected with pcDNA3-LYAAT-1 (f) but not in mock-transfected cells (g). (d) Immunoautoradiographic detection of LYAAT-1 on parasagittal brain sections using the anti-LYAAT-1 antibody revealed that the protein is enriched in regions expressing high levels of the corresponding mRNA. (e) Preincubation of the antibody with the peptide used to produce this antibody abolishes the immunoreactivity on an adjacent parasagittal section. (h–k) Comparison of LYAAT-1 mRNA and protein at the cellular level showed that LYAAT-1 is mainly expressed in the somatodendritic domain of neurons. Cellular distribution of LYAAT-1 mRNA was detected with a digoxigenin-labeled antisense probe (h and i), and LYAAT-1 protein was detected by immunofluorescence and laser-scanning confocal microscopy (j and k) in pyramidal cells of Ammon's horn (CA3) of the hippocampus (h and j) and pyramidal cells of the frontal cortex (i and k). Scale bars in e, g, j, and k indicate 5 mm, 35 μm, 105 μm, and 42 μm and apply to b–e, f and g, h and j, and i and k, respectively.
Functional Characterization.
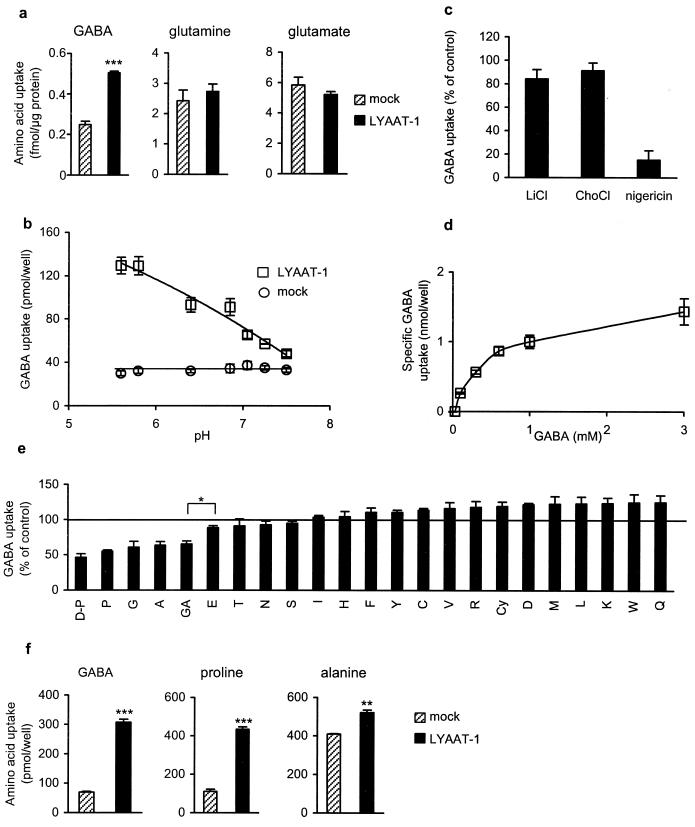
To investigate the functional properties of LYAAT-1, uptake experiments were performed using transiently transfected fibroblasts. Although most recombinant protein was expressed in the intracellular compartment (Fig. 2f), intact cells overexpressing the transporter accumulated twice as much [3H]GABA as mock-transfected cells at neutral pH (Fig. 3a). However, no specific uptake of L-[3H]glutamine or L-[3H]glutamate could be observed under the same conditions (Fig. 3a). LYAAT-1 mediated accumulation of [3H]GABA into transfected fibroblasts was time-dependent, remaining linear for ≈15 min (data not shown) and saturable (Fig. 3d) with a Km of 499 ± 135 μM and a Vmax of 2.5 ± 0.5 pmol/min/μg protein (Eadie–Hofstee analysis, n = 3). LYAAT-1 activity was strongly pH-dependent: a 5-fold increase in [3H]GABA specific uptake was observed at an extracellular pH of 5.5 compared with 7.5 (Fig. 3b). Disruption of the artificially imposed pH gradient with the ionophore nigericin, which exchanges extracellular protons for internal potassium ions (22), abolished the specific uptake of [3H]GABA, suggesting a direct role of protons as cosubstrate (Fig. 3c). By contrast, replacement of sodium ions by either lithium or choline in the uptake buffer had no effect on LYAAT-1-mediated [3H]GABA accumulation, showing that LYAAT-1 does not depend on external Na+ (Fig. 3c). Among natural amino acids, only glycine, L-alanine and L-proline substantially inhibited LYAAT-1-mediated [3H]GABA accumulation, suggesting that they represent additional substrates of the transporter (Fig. 3e). D-Proline competed with [3H]GABA as efficiently as did L-proline (Fig. 3e). Accumulation of L-[3H]proline and L-[3H]alanine into fibroblasts overexpressing LYAAT-1 at acidic pH directly confirmed that the transporter is able to accept several small neutral amino acids as substrate (Fig. 3f).
Figure 3.
LYAAT-1 is a small amino acid/proton symporter. (a) CV-1 cells transiently expressing LYAAT-1 (black bars) using the vaccinia virus expression system accumulate significantly more [3H]GABA (57 nM) but not l-[3H]glutamine (91 nM) or l-[3H]glutamate (208 nM) than mock-transfected cells (dashed bars). (b) Accumulation of [3H]GABA into COS-7 expressing LYAAT-1 (□) strongly increases at acidic pH, whereas transport into mock-transfected cells is not affected (○). pH was controlled by 10 mM K2HPO4/KH2PO4 in the uptake buffer. (c) At pH 5.5, the replacement of NaCl by an equivalent concentration of either lithium chloride (LiCl) or choline chloride (ChoCl) in the transport buffer had no effect on the 3H-amino acid specific accumulation. By contrast, preincubation of the cells for 10 min with 5 μM nigericin in a potassium-free KR buffer strongly inhibited the LYAAT-1-mediated accumulation of GABA into COS-7 cells. Results are expressed as percent of controls performed in standard (LiCl, ChoCl) or potassium-free (nigericin) KR buffer. (d) Specific LYAAT-1-mediated GABA accumulation (mock-subtracted) into COS-7 cells at pH 5.5 is saturable. (e) Transport of GABA is inhibited by l- and d-proline, glycine, and l-alanine. Transport of GABA (0.5 μCi [3H]GABA + 30 μM unlabeled GABA) was measured in the absence (100%) or the presence of 0.5 mM amino acid at pH 5.5. Amino acids, named with the one letter code, were of the l-form except d-proline (D-P). GA, GABA; Cy, cystine. (f) l-Proline and l-alanine are substrates of LYAAT-1. At pH 5.5, COS-7 cells expressing LYAAT-1 accumulated specifically 238 ± 14, 322 ± 23, and 112 ± 17 pmol/well of 100 μM [3H]GABA, l-[3H]proline, and l-[3H]alanine, respectively. Each panel shows one representative (a, b, d, and f) or the mean (c and e) of at least three experiments performed on cells from independent transfections. Transport measurements were performed in triplicate on LYAAT-1-transfected wells and on paired mock-transfected wells. Error bars represent the SEM. ***, P < 0.0001; **, P < 0.002; *, P < 0.02.
Colocalization with a Lysosomal Marker.
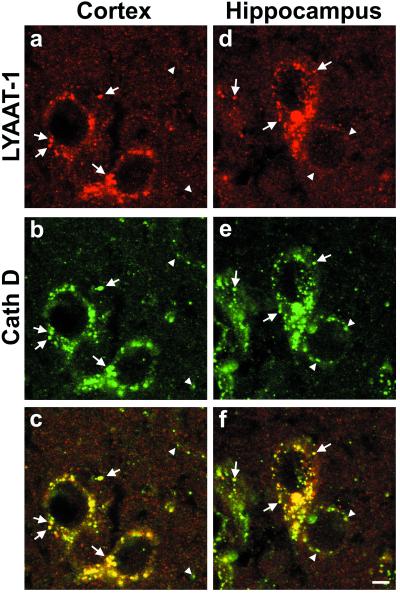
Because LYAAT-1 appeared as a proton/amino acid symporter mostly active at acidic pH, we reasoned that it might be involved in the efflux of small neutral amino acids from acidic organelles. Lysosomes, which produce amino acids by proteolysis, thus appeared as choice candidate organelles for LYAAT-1 function. To test this hypothesis, we compared the intracellular distribution of native LYAAT-1 to that of cathepsin D (EC 3.4.23.5), a lysosomal aspartyl protease (23). Laser-scanning confocal microscopy on brain sections revealed that LYAAT-1 immunoreactivity massively colocalized with that of cathepsin D (Fig. 4). Quantification of LYAAT-1-immunoreactive puncta indicated that 95% and 92% of them coexpressed cathepsin D in pyramidal neurons of hippocampus and cortex, respectively (analysis of 529 and 451 puncta on 20 randomly selected neuronal somata in each region). Conversely, 97% and 94% of cathepsin D-positive puncta also expressed LYAAT-1 (analysis of 516 and 438 puncta on 20 hippocampal and cortical pyramidal neurons, respectively).
Figure 4.
Double immunofluorescence labeling and laser-scanning confocal microscopy on brain sections show that the native LYAAT-1 colocalizes with the lysosomal protease cathepsin D. In pyramidal neurons of frontal cortex (a–c) and of Ammon's horn (CA3) (d–f), LYAAT-1 immunoreactivity (red) is shown in a and d, and cathepsin D immunoreactivity (green) is shown in b and e. In the superimposed images (c and f), the yellow color indicates the colocalization of the two markers (arrows); arrowheads indicate cathepsin D-positive, LYAAT-1-negative puncta. Scale bar indicates 5 μm and applies to a–f.
Discussion
Homology searching in expressed sequence tag databases allowed us to identify a novel mammalian member of the AAAP family, named LYAAT-1. This 475-aa protein belongs to a subgroup of the AAAP family, consisting of noncharacterized putative transporters revealed by the sequencing of eukaryotic genomes. LYAAT-1 is predominantly expressed in the brain, where both mRNA and protein could be detected in neurons but not in glial cells. When overexpressed in fibroblasts, the protein is able to accumulate some neutral amino acids into the cells, through an H+/amino acid symport driven by an artificially imposed pH gradient. Finally, the colocalization of LYAAT-1 with cathepsin D, a lysosomal protease, in brain revealed that the native protein is localized on lysosomes. Taken together, these data support a physiological role of LYAAT-1 as an amino acid transporter involved in the efflux of lysosomal proteolysis products, such as L-proline, L-alanine, or glycine, from the organelle lumen to the cytosol. Each of these amino acids is present at a ≈50 μM concentration in the lumen of liver lysosomes (24), thus amounting to an overall substrate concentration of 150 μM, which is in good agreement with a Km value of ≈500 μM (Fig. 3 d and e).
Its H+ symport mechanism (Fig. 3c) implies that LYAAT-1-mediated amino acid efflux is actively driven by the lysosomal H+-ATPase, possibly to allow net flux against high concentrations of these amino acids in the cytosol. It should be noted that our functional assay, which takes advantage of a partial expression of LYAAT-1 at the plasma membrane of transfected fibroblasts, allows measurement of LYAAT-1-mediated transport in the same direction as the proposed LYAAT-1-mediated lysosomal efflux because the extracellular medium is topologically equivalent to the lysosomal lumen. Two transport systems for L-proline, systems f and p, have been biochemically described in lysosomes purified from fibroblasts (25), but none seems to correspond to LYAAT-1, as they appeared to be pH-independent and stereospecific for L-proline. LYAAT-1 may thus be absent from fibroblasts, as it is from glial cells in the brain. GABA, which should be considered as a model substrate rather than an actual physiological substrate, may be recognized by LYAAT-1 because it adopts a cyclic conformation structurally similar to proline, as proposed in the case of another member of the AAAP family, ProT2, which transports L-proline, D-proline, and GABA with similar efficiencies (26).
LYAAT-1 shares no sequence homology with the putative lysosomal transporters sialin or cystinosin, and it appears as the first characterized member of a novel subgroup of proteins in the AAAP family, present in diverse eukaryotic species (Fig. 1). The identification of LYAAT-1 could thus open an avenue for the isolation of other lysosomal amino acid transporters and, possibly, for the characterization of novel lysosomal storage diseases.
The restricted expression of LYAAT-1 in neurons indicates that lysosomal transporters, beyond their general role in cellular metabolism, may be involved in specialized cellular functions. A previously documented example is lysosomal transport system h, which recognizes and recycles monoiodotyrosine in thyroid epithelial cells, thereby contributing to a more efficient synthesis of thyroid hormones (27). It will thus be interesting to examine in the future whether LYAAT-1 also participates to specific neuronal functions, including neurotransmission.
Acknowledgments
We thank P. Gaspar for help with immunofluorescence on brain sections, B. Goud and L. Johannes for allowing access to the vaccinia virus facility, J. L. Popot for prediction of the secondary structure, H. Boenisch for fruitful discussions, and C. Betancur for careful reading of the manuscript. This work is supported in part by The Association France Parkinson and the European Economic Community (to C.S.), Hoechst Marion Roussel fellowship (to C.A.) and funding (to B. Giros), the Fondation Singer Polignac (to C.A.), Institut National de la Santé et de la Recherche Médicale (to B. Giros and M.H.), and Centre National de la Recherche Scientifique (to B. Gasnier and P.R.).
Abbreviations
- GABA
γ-aminobutyric acid
- AAAP
amino acid/auxin permease
Note Added in Proof.
After submission of the manuscript, a study reported the functional characterization of S. cerevisiae AAAPs YNL 101w and YKL 146w (28).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF361239).
References
- 1.Mancini G M, Havelaar A C, Verheijen F W. J Inherited Metab Dis. 2000;23:278–292. doi: 10.1023/a:1005640214408. [DOI] [PubMed] [Google Scholar]
- 2.Gahl W A, Tietze F, Bashan N, Steinherz R, Schulman J D. J Biol Chem. 1982;257:9570–9575. [PubMed] [Google Scholar]
- 3.Havelaar A C, Mancini G M, Beerens C E, Souren R M, Verheijen F W. J Biol Chem. 1998;273:34568–34574. doi: 10.1074/jbc.273.51.34568. [DOI] [PubMed] [Google Scholar]
- 4.Mancini G M, Verheijen F W, Galjaard H. Hum Genet. 1986;73:214–217. doi: 10.1007/BF00401229. [DOI] [PubMed] [Google Scholar]
- 5.Renlund M, Tietze F, Gahl W A. Science. 1986;232:759–762. doi: 10.1126/science.3961501. [DOI] [PubMed] [Google Scholar]
- 6.Tietze F, Seppala R, Renlund M, Hopwood J J, Harper G S, Thomas G H, Gahl W A. J Biol Chem. 1989;264:15316–15322. [PubMed] [Google Scholar]
- 7.Town M, Jean G, Cherqui S, Attard M, Forestier L, Whitmore S A, Callen D F, Gribouval O, Broyer M, Bates G P, et al. Nat Genet. 1998;18:319–324. doi: 10.1038/ng0498-319. [DOI] [PubMed] [Google Scholar]
- 8.Verheijen F W, Verbeek E, Aula N, Beerens C E, Havelaar A C, Joosse M, Peltonen L, Aula P, Galjaard H, van der Spek P J, Mancini G M. Nat Genet. 1999;23:462–465. doi: 10.1038/70585. [DOI] [PubMed] [Google Scholar]
- 9.Pisoni R L, Schneider J A. In: Mammalian Amino Acid Transport. Kilberg M S, Häussinger D, editors. New York: Plenum; 1992. pp. 89–99. [Google Scholar]
- 10.Saier M H., Jr Microbiol Mol Biol Rev. 1999;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palacin M, Estevez R, Bertran J, Zorzano A. Physiol Rev. 1998;78:969–1054. doi: 10.1152/physrev.1998.78.4.969. [DOI] [PubMed] [Google Scholar]
- 12.Young G B, Jack D L, Smith D W, Saier M H., Jr Biochim Biophys Acta. 1999;1415:306–322. doi: 10.1016/s0005-2736(98)00196-5. [DOI] [PubMed] [Google Scholar]
- 13.Frommer W B, Hummel S, Riesmeier J W. Proc Natl Acad Sci USA. 1993;90:5944–5948. doi: 10.1073/pnas.90.13.5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortiz-Lopez A, Chang H, Bush D R. Biochim Biophys Acta. 2000;1465:275–280. doi: 10.1016/s0005-2736(00)00144-9. [DOI] [PubMed] [Google Scholar]
- 15.Sagné C, El Mestikawy S, Isambert M F, Hamon M, Henry J P, Giros B, Gasnier B. FEBS Lett. 1997;417:177–183. doi: 10.1016/s0014-5793(97)01279-9. [DOI] [PubMed] [Google Scholar]
- 16.McIntire S L, Reimer R J, Schuske K, Edwards R H, Jorgensen E M. Nature (London) 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhry F A, Reimer R J, Krizaj D, Barber D, Storm-Mathisen J, Copenhagen D R, Edwards R H. Cell. 1999;99:769–780. doi: 10.1016/s0092-8674(00)81674-8. [DOI] [PubMed] [Google Scholar]
- 18.Varoqui H, Zhu H, Yao D, Ming H, Erickson J D. J Biol Chem. 2000;275:4049–4054. doi: 10.1074/jbc.275.6.4049. [DOI] [PubMed] [Google Scholar]
- 19.Lennon G, Auffray C, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 20.Povlock S L, Amara S G. Methods Enzymol. 1998;296:436–443. doi: 10.1016/s0076-6879(98)96031-1. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Pressman B C. Fed Proc. 1968;27:1283–1288. [PubMed] [Google Scholar]
- 23.Whitaker J N, Terry L C, Whetsell W O., Jr Brain Res. 1981;216:109–124. doi: 10.1016/0006-8993(81)91281-6. [DOI] [PubMed] [Google Scholar]
- 24.Vadgama J V, Chang K, Kopple J D, Idriss J-M, Jonas A J. J Cell Physiol. 1991;147:447–454. doi: 10.1002/jcp.1041470310. [DOI] [PubMed] [Google Scholar]
- 25.Pisoni R L, Flickinger K S, Thoene J G, Christensen H N. J Biol Chem. 1987;262:6010–6017. [PubMed] [Google Scholar]
- 26.Breitkreuz K E, Shelp B J, Fischer W N, Schwacke R, Rentsch D. FEBS Lett. 1999;450:280–284. doi: 10.1016/s0014-5793(99)00516-5. [DOI] [PubMed] [Google Scholar]
- 27.Andersson H C, Kohn L D, Bernardini I, Blom H J, Tietze F, Gahl W A. J Biol Chem. 1990;265:10950–10954. [PubMed] [Google Scholar]
- 28.Russnak, R., Konzcal, D. & McIntire, S. L. (March 26, 2001) J. Biol. Chem., 10.1074/jbc.M008028200. [DOI] [PubMed]