Abstract
X-linked hypohidrotic ectodermal dysplasia (XLHED) is a heritable disorder of the ED-1 gene disrupting the morphogenesis of ectodermal structures. The ED-1 gene product, ectodysplasin-A (EDA), is a tumor necrosis factor (TNF) family member and is synthesized as a membrane-anchored precursor protein with the TNF core motif located in the C-terminal domain. The stalk region of EDA contains the sequence -Arg-Val-Arg-Arg156-Asn-Lys-Arg159-, representing overlapping consensus cleavage sites (Arg-X-Lys/Arg-Arg↓) for the proprotein convertase furin. Missense mutations in four of the five basic residues within this sequence account for ≈20% of all known XLHED cases, with mutations occurring most frequently at Arg156, which is shared by the two consensus furin sites. These analyses suggest that cleavage at the furin site(s) in the stalk region is required for the EDA-mediated cell-to-cell signaling that regulates the morphogenesis of ectodermal appendages. Here we show that the 50-kDa EDA parent molecule is cleaved at -Arg156Asn-Lys-Arg159↓- to release the soluble C-terminal fragment containing the TNF core domain. This cleavage appears to be catalyzed by furin, as release of the TNF domain was blocked either by expression of the furin inhibitor α1-PDX or by expression of EDA in furin-deficient LoVo cells. These results demonstrate that mutation of a functional furin cleavage site in a developmental signaling molecule is a basis for human disease (XLHED) and raise the possibility that furin cleavage may regulate the ability of EDA to act as a juxtacrine or paracrine factor.
X-linked hypohidrotic ectodermal dysplasia (XLHED) is a human heritable disorder that results in the impaired formation of hair, teeth, and sweat glands during fetal development (1). When this condition goes unrecognized, there is a 30% mortality rate due to hypothermia during the first 2 years of life (2). The XLHED locus, ED-1, encodes a multidomain transmembrane protein, ectodysplasin-A (EDA) (1, 3–6). A similar X-linked disorder also exists in mice and is due to spontaneous mutations of the murine ortholog of EDA (3, 4). The tissues affected by this disorder are all derived from epithelia, which extend into the underlying mesenchyme, proliferate, and then differentiate. Consistent with a role in development of these tissues, EDA mRNA and protein have been detected in epithelial cells of the epidermis, sweat glands, hair follicles, and tooth bud (1, 7, 8).
EDA is a member of the tumor necrosis factor (TNF) family of ligands (7, 9–13), suggesting that mutations in ED-1 could lead to a failure of epithelial signaling required for the normal development of these tissues. In support of this idea, an autosomal gene (EdaR, DL) for XLHED has been cloned in both mouse and human (9, 14). This gene, also expressed in the epithelium, encodes ectodysplasin-A receptor (EdaR), a transmembrane protein with domains homologous to the TNF receptor family. The TNF homologies of the two molecules suggest that EDA and EDA receptor (EDAR) function as a receptor–ligand pair (14), for which there is recent biochemical evidence (15, 16)
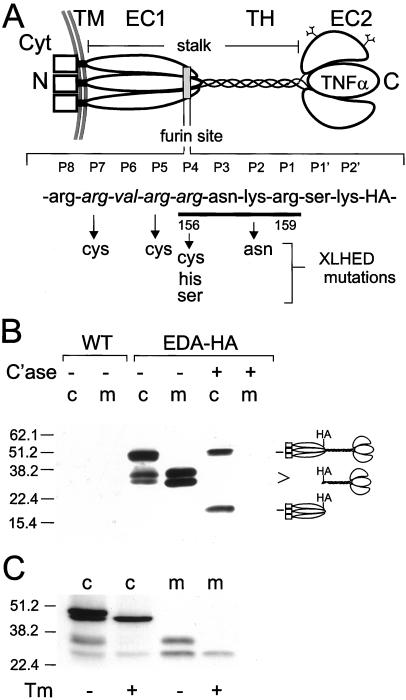
Like other members of the TNF family (17, 18), EDA is a trimeric type II transmembrane protein (see Fig. 1A; refs. 3–5). Two extracellular (EC) domains are separated by a collagenous segment of 19 Gly-X-Y triplets, and the TNF motif resides in the most carboxy-terminal extracellular domain, EC2. The triple helical domain and EC1 compose a stalk region separating the TNF ligand domain from the cell surface. EDA shows extensive alternative splicing, but only the two largest forms, EDA-A1 (391 aa) and EDA-A2 (389 aa), contain the EC2 domain with its TNF motif (6). Although the presence of a collagen domain in the stalk region of EDA is unusual in the context of TNF cytokines, this overall structure is similar to that of the adipocyte protein ACRP30 and characteristic of the structure of C1q family proteins, which together form a TNF-C1q superfamily (11, 19).
Figure 1.
Structure of EDA and the EDA-HA construct. The EDA protein is a type II transmembrane protein. Cyt, the N-terminal cytoplasmic domain; TM, the transmembrane domain. EC1, the first extracellular domain, contains a compound consensus furin cleavage site, Arg-X-Lys/Arg-Arg (italics, upstream site; underline, downstream site), near its C terminus. TH, the collagenous triple helical domain; EC2, the second extracellular domain, which contains an embedded TNF core motif. This domain also contains two consensus N-linked glycosylation sites, as indicated in the schematic (5). The recombinant EDA-HA molecule (400 aa) used in this study is identical to the EDA-A1 sequence (391 aa), with the addition of the 9-aa hemagglutinin (HA) epitope tag as indicated. The positions of Arg156 and Arg159 are indicated. Mutations within the putative furin site(s) of EDA in patients with XLHED are identified by downward arrows (5, 6, 23, 54) and are abbreviated R153C, R155C, R156S, R156H, R156C, and K156N. The position of each amino acid is given relative to the downstream site (Arg159), P1-P8, N-terminal and P1′-P2′, C-terminal. (B) BSC-40 cells were infected with VV:EDA-HA or wild-type virus (VV:WT) for 16 h, and the cell (c) and medium (m) compartments were analyzed by SDS/PAGE and immunoblotting with HA antibody. Cell and medium samples were also analyzed after digestion with bacterial collagenase (C'ase). A schematic of collagenase digestion products is on the right. After collagenase digestion, the HA epitope tag remains with the N-terminal EC1 domain. The triple helix appears to be more resistant to bacterial collagenase in the context of the parent molecule compared with the 36/27-kDa fragments. (C) BSC-40 cells were infected with VV:EDA-HA for 4 h in the presence (+) or absence (−) of tunicamycin (Tm) to inhibit N-glycosylation. The same proteolytic processing and N-glycosylation patterns were detected when EDA was expressed in other cell lines, HaCaT (epidermal epithelia), 293 (kidney epithelia), COS-7 (kidney fibroblast), and A204 (rhabdomyosarcoma) (data not shown).
The fact that EDA is a membrane protein suggests that it may participate in direct juxtacrine cell–cell signaling through the EDAR receptor. Alternatively, the membrane-attached form may require shedding by specific proteolysis for activation or paracrine signaling. The shedding phenomenon has been observed in a large number of signaling systems involving either the ligand, the receptor, or both (for reviews see refs. 20 and 21). Like some other TNF ligands, such as Baff, Trail, and APRIL (18), EDA contains a consensus site for furin-like proteolytic processing, suggesting that it may also be proteolytically shed or activated.
Furin is a membrane-associated, calcium-dependent, serine endoprotease that cleaves most efficiently at the C-terminal side of an Arg(P4)-X-Lys/Arg-Arg(P1) sequence (22), where the basic residues at the P1 and P4 positions are critical for activity. EDA contains two consecutive consensus furin motifs that overlap by one amino acid, Arg-Val-Arg-Arg-Asn-Lys-Arg159 (upstream, italics; downstream, underlined). The potential compound furin site is located in the first extracellular domain (EC1) near the junction of the collagen sequence (Fig. 1A). Cleavage at the putative furin site in EDA would produce a C-terminal fragment containing the TNF motif (EC2) and its associated collagen segment. This fragment would no longer be membrane bound, and its formation by furin could potentially regulate or be required for EDA signaling between cells.
A role for furin processing in the function of EDA is suggested by the detection of mutations involving four of five basic amino acids in the EDA compound consensus furin site of XLHED patients (5, 6, 23, 54) as shown in Fig. 1A. These mutations, many of which have recurred independently in unrelated families, represent a significant portion (≈20%) of the mutations detected in the ED-1 gene. Mutations affecting Arg156, which is in a key position for both consensus sites (either P1 or P4), are the most frequently observed within the furin site(s) or anywhere in the gene (54).
Here we show that EDA is proteolytically processed during biosynthesis, apparently by furin, which cleaves EDA in the compound consensus site, predominantly at Arg159. We further show that mutations in this furin site found in XLHED patients block the proteolytic processing of EDA, indicating that proteolytic release of the TNF domain is a requirement for proper EDA-EDAR signaling during development.
Materials and Methods
Materials.
Monoclonal hemagglutinin (HA) antibody was obtained from Covance. The vaccinia viral furin construct (VV):fur/f and the adenoviral α1-antitrypsin (Portland) (α1-PDX) construct were prepared as described (24, 25). All restriction endonucleases were obtained from Promega. Unless otherwise indicated, all reagents were purchased from Sigma.
EDA Expression, VV:EDA-HA.
Full-length EDA-A1 encoding 391 aa (5) (accession number: GenBank AF060999) was first cloned into BamHI/SacI sites of pZVneo vector by PCR. Site-direct mutagenesis (CLONTECH) was used to introduce the sequence encoding YPYDVPDYA (the HA epitope tag) after Lys161. The EDA-HA furin site mutants R153C, R156C, R156H, K158N, and R159A (Fig. 1A) were generated by site-direct mutagenesis of the EDA-HA construct (oligonucleotide sequences available on request). Recombinant vaccinia viruses of all EDA-HA constructs were generated by marker transfer as described (26).
Cell Culture and Infection.
BSC-40 and LoVo cells were cultured as described (27, 28), infected for 30 min with recombinant vaccinia virus at a multiplicity of infection of 5, and incubated for 14–16 h as described (27, 29), in medium supplemented with 50 μg/ml ascorbate-2-phosphate (30). Furin processing was inhibited by coexpression of VV:EDA-HA with the α1-PDX construct (25). LoVo cells were infected at a multiplicity of infection of 5 with VV:EDA-HA and coinfected at a multiplicity of infection of 5 with either VV:fur/f (24) or wild-type virus and further cultured for 8 h. Cell layer and media were harvested, and the cells were extracted as described (27, 29) in a volume equal to the medium.
Immunoblotting.
Equal aliquots of cell extract or medium fraction were separated on 10% SDS/PAGE gel and immunoblotted as described (31) with the use of chemiluminescence (ECL, Pierce).
Bacterial Collagenase Digest.
Cell extracts or medium was incubated with bacterial collagenase CLSPA (Worthington; further purified according to the methods in ref. 32) at 37°C for 2 h at 5 mM Ca2+.
Inhibition of N-Linked Glycosylation.
Tunicamycin (10 μg/ml) (Behring) was added to the BSC-40 cell culture media, followed by infection with VV:EDA-HA at a multiplicity of infection of 10, and the cells harvested after 4 h of infection to minimize the toxic effects of tunicamycin.
Preparation of Fluorescence-Labeled Peptide Substrates.
Synthetic peptide, aminobenzoic acid-Glu148-Glu-Glu-Ser-Arg-Arg-Val-Arg-Arg-Asn-Lys-Arg-Ser-Lys-Ser-Asn-Glu-3-nitrotyrosine-Gly165, was prepared by the analytical core facility of the Shriners Hospital, Portland, OR (33). Aminobenzoic acid (fluorenylmethoxycarbonyl- aminobenzoic acid; Bachem) was added to the end of the peptide, and 3-nitrotyrosine (fluorenylmethoxycarbonyl-Nitrotyrosine; Bachem) was inserted between Glu164 and Gly165 to produce an internally quenched peptide for the monitoring of protease activity in kinetic assays (34). Additional peptides harboring XLHED mutations R153C, R155C, R156C, R156H, K158N, and the introduced mutation R159A were prepared in the same way (Fig. 1A).
Assays of Proprotein Convertase (PC) Activity.
In vitro assays of peptide substrate cleavage by furin were conducted as described (35). Fluorescence measurements were performed at 30°C with a substrate concentration of 100 μM in a 1.0-ml final volume, with the use of 10 μl furin (400 nM), prepared as described (35). The final fluorescence value was obtained by the addition of trypsin (1 μg/ml). For N-terminal sequencing, reactions were carried out in a final volume of 100 μl, with 2 μl furin for 2 h at room temperature. Km and kcat were determined as described (25).
Results
Expression of EDA-HA.
To determine whether EDA is proteolytically cleaved during biosynthesis, full-length human EDA corresponding to the EDA-A1 isoform was cloned into vaccinia virus for recombinant expression. The HA epitope tag was incorporated into EDA adjacent to the furin site (Fig. 1A) to facilitate these analyses. After expression of EDA-HA in BSC-40 cells (Fig. 1B), immunoblotting of the cell layer showed a broad band with an apparent molecular mass of about 50 kDa, consistent with a protein of 400 aa including the HA tag. In addition, two smaller bands (Mr 36 kDa and 27 kDa) were detected in the cell extract and were the only immunoreactive products observed in the culture medium. These results suggested the release of a C-terminal fragment(s) of EDA containing the HA tag in a process likely to involve the furin site. Evidence for this conclusion was obtained by digestion of cell and media samples with bacterial collagenase to remove the collagenous sequence (Fig. 1B). Although incomplete, the digestion of the cell material generated a new 18-kDa band consistent with the N-terminal portion of the molecule up to the start of the collagenous sequence and including the HA tag. Collagenase digestion of the medium resulted in the complete loss of the 36-kDa and 27-kDa bands, consistent with processing at the furin site and separation of the HA tag from the C-terminal fragment by removal of the collagen segment (see Fig. 1A).
The broad 50-kDa band (often a doublet) and the appearance of two fragments rather than one suggested the presence of more than one cleavage site or differential posttranslational modification. EDA contains two consensus sites for N-glycosylation in the EC2 domain downstream from the furin consensus site. N-glycosylation of recombinant murine Eda expressed in baby hamster kidney cells has been shown by others (7). The possibility that the 36/27-kDa bands reflect differences in N-glycosylation was examined by treatment of infected BSC-40 cells with tunicamycin, an inhibitor of N-glycosylation (Fig. 1C). The presence of tunicamycin abolished the more slowly migrating band of EDA and the 36-kDa band in the medium and cell layer. This result indicated that one or both of the glycosylation sites are used in the biosynthesis of EDA, and that the multiple EDA fragments are not the result of differential or multiple cleavage site usage.
Evidence for Furin-Like Processing.
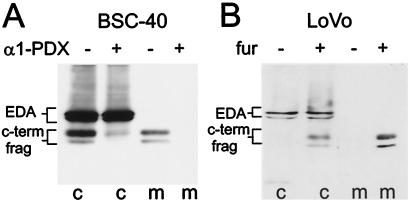
To determine whether the EDA-HA C-terminal fragments arose by furin-like proteolytic processing, two sets of experiments were conducted. First, a protein-based inhibitor, α1-PDX (25), which is highly selective for furin, was used (Fig. 2A). BSC-40 cells were infected with an α1-PDX adenoviral construct and subsequently infected with VV:EDA-HA. In the presence of α1-PDX, full-length 50-kDa EDA-HA protein was detected, whereas little or no 36/27-kDa C-terminal fragments were found in either the cell layer or the medium. Second, EDA-HA was expressed in the colon carcinoma cell line LoVo, in which both alleles of the furin gene are mutated and the furin protein is nonfunctional (36). Only the full-length EDA-HA molecule was detected in the cell layer, and no 36/27-kDa fragments were detected in the medium. Exogenous expression of furin in LoVo cells restored the proteolytic processing of EDA, showing that these cells are otherwise competent for processing of EDA.
Figure 2.
Inhibition of EDA-HA proteolytic processing. (A) BSC-40 cells were first infected with an adenoviral construct, α1-PDX, a potent inhibitor of furin activity, and subsequently infected with VV:EDA-HA. (B) LoVo cells were coinfected with VV:EDA-HA and either the furin construct, VV:fur/f (fur +), or, in control cultures, wild-type virus (fur −). Cell and media fractions were analyzed by SDS/PAGE and immunoblot with the anti-HA antibody.
Identification of the Furin Cleavage Site.
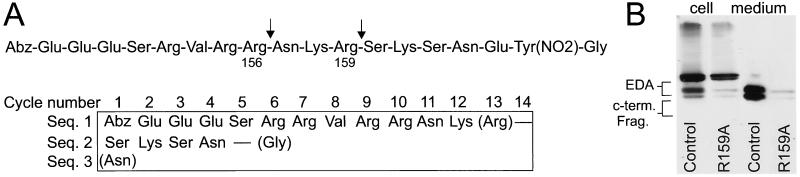
To identify which potential cleavage site(s) is used in EDA, a 19-residue fluorogenic peptide substrate, pEDA (Glu148 to Gly165), which encompasses the consensus, was prepared and cleaved with furin in vitro, and the reaction products were sequenced directly (Fig. 3). Cleavage of pEDA by furin was complete and occurred predominantly at Arg159, producing two major products, aminobenzoic acid-Glu-Glu-Glu-Ser-Arg-Arg-Val-Arg-Arg-Asn-Lys-Arg159 and Ser-Lys-Ser-Asn-Glu-3-nitrotyrosine-Gly. In several repetitions of this assay, detection of Asn in the first cycle, indicative of cleavage at the upstream Arg156 site, was less than 10%. To establish that Arg159 was the major furin cleavage site for EDA, Arg159 (P1) was altered to Ala, and the resultant VV:EDA-HA(R159A) was expressed in BSC-40 cells (Fig. 3B). The amount of the 36/27-kDa fragments produced was markedly reduced relative to the control recombinant EDA. The residual 36/27-kDa fragment observed in the expression of R159A may be due to low-efficiency cleavage at Arg156, as observed in the cleavage of pEDA peptide by furin.
Figure 3.
Determination of the site of EDA processing by furin. (A) Synthetic peptide, pEDA (EDA residues 148–165) (100 μM), was digested with furin, and an aliquot was subjected directly to Edman sequencing. The results for each cycle are shown. The multiple sequence was ordered from the sequence of the starting peptide. Residues in parentheses were present in trace amounts. (B) EDA-HA and EDA-HA with the R159A mutation were expressed in BSC-40 cells and analyzed by SDS/PAGE and immunoblotting.
Effects of Mutations on Furin Processing in Vitro.
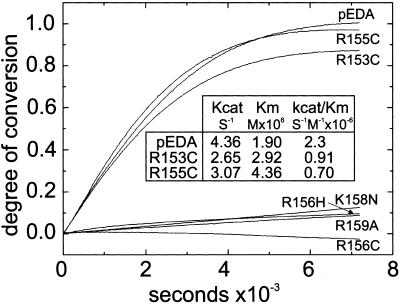
The finding that EDA is cleaved by furin at Arg159 suggested that mutations that disrupted the furin recognition site caused XLHED by abolishing proteolytic processing of EDA. Therefore, the effect of these mutations on furin processing was assessed with the use of the internally quenched fluorescent synthetic peptide pEDA (see Fig. 3A) and pEDA containing corresponding XLHED mutations (Fig. 4). In contrast to pEDA, which was completely cleaved within the incubation period, XLHED mutations involving either the P2 site (pK158N) or the P4 site (pR156C, pR156H) significantly inhibited furin cleavage, as did the substitution of alanine for Arg159. The effect included both a lower initial velocity and a decreased extent of cleavage and reflects the importance of the P1, P2, and P4 basic residues for the proteolytic activity of furin. Cleavage of the mutant peptides by furin was also analyzed by direct N-terminal sequencing of reaction products to verify the processing site (data not shown). The P1 and P2 mutant peptides, pR159A and pK158N, were cleaved at the Arg156 site, but at low efficiency. Cleavage of the P4 mutants, pR156C and pR156H, at Arg159 was detected in trace amounts.
Figure 4.
Kinetics of furin cleavage of normal and mutant peptide substrates. The internally quenched control peptide pEDA, containing the furin site (Fig. 3) as well as mutated versions of this peptide, pR153C, pR155C, pR156H, pR156C, pK158N, and pR159A (Fig. 1A), were digested with furin (Materials and Methods), and the time course of proteolysis was followed by increased fluorescence at 400 nm and presented as the degree of conversion (Materials and Methods). (Inset) kcat, Km, and kcat/Km for pEDA, R153C, and R155C cleavage by furin.
Unlike the P1–P4 mutations, the pR153C and pR155C peptides were cleaved by furin in vitro at the Arg159 site. These results were consistent with the identification of Arg159 as the major cleavage site, inasmuch as Arg153 and Arg155 are outside of the minimal furin consensus motif, Arg156-Asn-Lys-Arg159. However, this finding contrasts with the observation that R153C and R155C cause XLHED that is phenotypically indistinguishable from patients with mutations at the furin P2 and P4 sites (5, 54). More detailed kinetic analysis (Fig. 4 Inset) showed a moderate 2- to 3-fold reduction in catalytic efficiency for the two mutants compared with pEDA. To reconcile these results, the effects of the furin site mutations on the biosynthesis and proteolytic processing of recombinant EDA were examined.
Effect of Mutations on Recombinant EDA Processing.
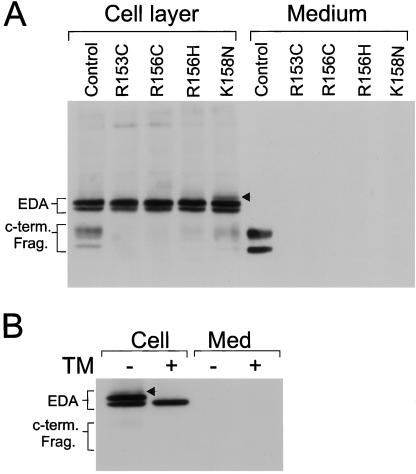
Mutations corresponding to R153C, R156C, R156H, and K158N (Fig. 1A) were incorporated into the VV:EDA-HA construct (Materials and Methods) and expressed in the BSC-40 cell line (Fig. 5A). All of the mutations greatly reduced the level of the 36/27-kDa C-terminal fragments observed in the culture medium and the cell layer. The mutations of Arg156 in the P4 site effectively blocked proteolytic cleavage of EDA and any potential cleavage at Arg156 itself (P1 for the upstream site). Mutation of the P2 Lys158 also effectively blocked the appearance of the 36/27-kDa furin cleavage products, although residual processing could be detected in the cell layer likely involving the Arg156 site, as observed with the pK158N peptide. The K158N mutation generated a new consensus N-linked glycosylation site, -Asn-Arg-Ser-, adjacent to the furin cleavage site at Arg159 and resulted in the appearance of an additional band above intact EDA (Fig. 5, arrowhead). This band was abolished by tunicamycin (Fig. 5B), suggesting the glycosylation of Asn158.
Figure 5.
Expression of recombinant EDA harboring mutations in the furin cleavage site. (A) The biosynthesis of recombinant EDA harboring XLHED mutations in the putative furin cleavage site (see Fig. 1A). (B) VV:EDA-HA(K158N) was expressed in the presence or absence of tunicamycin (10 μM). All constructs were expressed in BSC-40 cells, and the media and cell extracts were analyzed by SDS/PAGE and immunoblotting with HA antibody.
Even though cleavability of pR153C and pR155C was only moderately reduced in vitro, the furin processing of recombinant R153C was effectively blocked by this mutation. Thus, the pathology of the R153C mutation (and likely also that of R155C) appears to result from the inhibition of proteolytic processing at Arg159.
Discussion
Typically generation of a “soluble” ligand from a membrane-associated protein is achieved through a proteolytic event to release a portion of the extracellular domain (20, 21). TNFα and its receptors are cleaved by TNFα converting enzyme, one of the ADAMS group of cell-surface proteases (ADAMS 17) (37–39), or potentially by PCs, such as furin (18). We have shown here that recombinant EDA-A1 is cleaved by furin-like activity to release a C-terminal fragment containing the TNF domain. Failure to detect cleavage products in a previous study (7) may have been because of the fact that the C-terminal furin fragment could not be detected with the antibody used. The N-terminal 18-kDa fragment was detected in another study with N-terminal domain antibodies (10).
Furin is implicated in the biosynthesis of EDA by several aspects of the data. In vitro, furin cleaves EDA synthetic peptide substrates at Arg-Asn-Lys-Arg159, a consensus proprotein convertase site. Cleavage of EDA in culture was inhibited by the presence of α1-PDX, a highly selective inhibitor of furin. EDA expressed in LoVo cells that lack functional furin enzyme was not processed, but cleavage was restored by coexpression with active furin. Finally mutations of the basic amino acids at positions P1, P2, and P4, key to the activity of furin (40), blocked cleavage of corresponding peptides by furin and proteolytic processing of recombinant EDA in cell culture. The fact that these P2 and P4 mutations are ones identified in patients with XLHED (5, 6, 23, 54) further implicates furin in the biological processing of EDA.
Our data point strongly toward furin as the enzyme responsible for the proteolytic processing of EDA. However, additional members of the PC family could play a role. The PC family contains six enzymes in addition to furin: PC1, PC2, PC4, PACE4, isoforms PC6A and PC6B, and PC7 (for reviews see refs. 41 and 42). The expression patterns of the mammalian PCs during development are distinct from one another and yet overlap in many tissues. Furin and PC7 are expressed ubiquitously at low levels and are thought to process constitutively secreted proteins (42). PC6 and PACE4 show dramatic changes in expression levels in certain tissues during development and may be associated with processing of bone morphogenic protein growth factors (35, 43). PACE4 is expressed in both the dental epithelium and mesenchyme at early developmental stages (44) and overlaps with the Eda expression in the outer dental epithelium of the bud stage tooth (16). Despite this overlap, EDA was not processed in LoVo cells that are reported to express PACE4 (28), and EDA processing was not rescued by overexpression of PACE4 in LoVo cells (data not shown). Nevertheless, definitive identification of the EDA processing PC enzyme(s) must await in situ characterization of EDA cleavage in tooth and skin during development.
The compound consensus furin site of EDA, Arg-Arg-Val-Arg-Arg-Asn-Lys-Arg-Ser Lys161, could conceivably be cleaved at either Arg156 or Arg159. However, cleavage of the pEDA peptide occurred predominantly at Arg159. In addition to the requirement for arginine at the P1 site of the furin consensus sequence, Arg-X-Lys/Arg-Arg, the basic amino acids at P2 and P4 are energetically important for furin activity (40) and participate in substrate binding (45). Therefore, the furin site mutations detected in patients with XLHED at the P2 and P4 positions relative to Arg159 should directly block proteolysis at the downstream site, as was observed in vitro and in cell culture. Mutations of Arg156, the most common furin consensus site mutation, would affect both potential sites (P1 upstream, P4 downstream), and pR156C is virtually inert as a furin substrate. Considering that mutations of the P2 and P4 sites result in XLHED, it is somewhat surprising that no spontaneous mutations have been identified in the P1 Arg codon in XLHED patients. The Arg159 codon is AGA, and the other mutated arginine codons are CGT or CGC. These contain CpGs, which, when methylated, can produce a C-to-T transversion and are therefore highly mutable (46, 47). Even though mutation of Arg159 effectively blocks furin cleavage, spontaneous mutation of the AGA codon likely occurs at a lower rate during gametogenesis.
Because they do not involve the consensus basic residues relative to Arg159, mutation of Arg153 and Arg155 have a relatively small effect on cleavage of the corresponding peptide by furin. However, mutation of Arg153 blocked processing of the recombinant protein in cell culture and produced a phenotype of severity equal to those of the other mutations. Possibly the R153C mutation (and likely R155C) blocks furin activity at Arg159 by perturbing the local structure of EDA around the furin site, in combination with a small direct effect on catalytic activity. It is unlikely that these mutations caused a gross corruption of EDA structure, inasmuch as R153C did not cause a dramatic increase in degradation of the recombinant protein. Moreover, cysteine residues are rarely found in the vicinity of known furin sites (48).
Proteolytic processing at the furin consensus site could be a target for the temporal and spatial modulation of EDA activity. Furin enzyme is dynamically sorted between various cellular compartments in a regulated manner (49). Processing and activity of EDA could depend on the colocalization of EDA and furin in the appropriate compartment or on the interplay between the utilization of the major Arg159 site and the low-level cleavage at Arg156. Modulation of EDA processing could also involve additional members of the PC family, perhaps by cleavage at the alternative Arg156 site.
It is now apparent that the primary defect in the XLHED results from a failure of an epithelial signaling system required for the normal development of teeth and skin. The emerging model has the EDA ligand, a transmembrane protein, binding its receptor, EDAR (15, 16, 50), and signaling to the target cell through activation of the NF-κB system of transcriptional regulation (50, 51). During tooth development in mouse, signaling between EDA and its receptor, EDAR, is required to establish and maintain enamel knot morphology and function (16, 52). The earliest expressions of Eda and EdaR are in adjacent and only partially overlapping domains of the thickened dental epithelium (16). As the dental epithelium proliferates and extends into the underlying mesenchyme to form the tooth bud, EdaR expression accompanies the leading edge of the epithelial ingrowth and is ultimately confined to the enamel knot. On the other hand, expression of Eda remains with the outer enamel epithelium (3, 16, 52, 53), resulting in spatial segregation of ligand and receptor. Together with our results, these observations suggest that the EDA transmembrane protein is a precursor that is proteolytically processed during biosynthesis and intracellular transport and that signaling is paracrine rather than autocrine or juxtacrine in nature. These conclusions provide a rationale for signaling between EDA and EDAR requiring a soluble (or at least non-membrane-attached) form of EDA. Indeed, the fact that mutations in the EDA furin site that might discriminate between paracrine and juxtacrine signaling produce pathology equivalent to that of functional knockouts (large deletions and mutations of the TNF region) (1) strongly points to a predominantly paracrine mode of signaling. Similar considerations implicate paracrine signaling between EDA and EDAR in the development of hair follicles and sweat glands (8, 14, 15).
In conclusion, furin cleaves EDA at a consensus site in the stalk region, liberating the receptor-binding TNF fragment and releasing it to the extracellular space. This proteolytic step is essential for the paracrine signaling of EDA in tooth and skin development. These mutations of the ED-1 gene are one of the few examples of a heritable disorder being associated with a failure of furin-processing sites (reviewed in refs. 41 and 49). Our findings extend this list to include intercellular signaling molecules primarily involved in differentiation and development.
Acknowledgments
We thank Bruce Donaldson for peptide purification and Larry David, School of Dentistry, Oregon Health Sciences University, for mass spectroscopy. The authors also acknowledge Shriners Hospital (N.P.M.), the National Institutes of Health (Grant DK37274 to G.T. and Grant DE11311 to B.F. and J.Z.), and the National Foundation for Ectodermal Dysplasias (J.Z.) for their support of this work.
Abbreviations
- α1-PDX
α1-antitrypsin (Portland)
- EDA (human) and Eda (mouse)
ectodysplasin-A
- EDAR (human) and EdaR (mouse)
EDA receptor
- XLHED
X-linked hypohidrotic ectodermal dysplasia
- TNF
tumor necrosis factor
- EC
extracellular
- VV
vaccinia virus
- HA
hemagglutinin
- PC
proprotein convertase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kere J, Srivastava A K, Montonen O, Zonana J, Thomas N, Ferguson B, Munoz F, Morgan D, Clarke A, Baybayan P, et al. Nat Genet. 1996;13:409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- 2.Clarke A, Phillips D I, Brown R, Harper P S. Arch Dis Child. 1987;62:989–996. doi: 10.1136/adc.62.10.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srivastava A K, Pispa J, Hartung A J, Du Y, Ezer S, Jenks T, Shimada T, Pekkanen M, Mikkola M L, Ko M S, et al. Proc Natl Acad Sci USA. 1997;94:13069–13074. doi: 10.1073/pnas.94.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson B M, Brockdorff N, Formstone E, Ngyuen T, Kronmiller J E, Zonana J. Hum Mol Genet. 1997;6:1589–1594. doi: 10.1093/hmg/6.9.1589. [DOI] [PubMed] [Google Scholar]
- 5.Monreal A W, Zonana J, Ferguson B. Am J Hum Genet. 1998;63:380–389. doi: 10.1086/301984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayes M, Hartung A J, Ezer S, Pispa J, Thesleff I, Srivastava A K, Kere J. Hum Mol Genet. 1998;7:1661–1669. doi: 10.1093/hmg/7.11.1661. [DOI] [PubMed] [Google Scholar]
- 7.Mikkola M L, Pispa J, Pekkanen M, Paulin L, Nieminen P, Kere J, Thesleff I. Mech Dev. 1999;88:133–146. doi: 10.1016/s0925-4773(99)00180-x. [DOI] [PubMed] [Google Scholar]
- 8.Montonen O, Ezer S, Saarialho-Kere U K, Herva R, Karjalainen-Lindsberg M L, Kaitila I, Schlessinger D, Srivastava A K, Thesleff I, Kere J. J Histochem Cytochem. 1998;46:281–289. doi: 10.1177/002215549804600301. [DOI] [PubMed] [Google Scholar]
- 9.Monreal A W, Ferguson B M, Headon D J, Street S L, Overbeek P A, Zonana J. Nat Genet. 1999;22:366–369. doi: 10.1038/11937. [DOI] [PubMed] [Google Scholar]
- 10.Ezer S, Bayes M, Elomaa O, Schlessinger D, Kere J. Hum Mol Genet. 1999;8:2079–2086. doi: 10.1093/hmg/8.11.2079. [DOI] [PubMed] [Google Scholar]
- 11.Copley R R. J Mol Med. 1999;77:361–363. doi: 10.1007/s001090050362. [DOI] [PubMed] [Google Scholar]
- 12.Eck M J, Sprang S R. J Biol Chem. 1989;264:17595–17605. doi: 10.2210/pdb1tnf/pdb. [DOI] [PubMed] [Google Scholar]
- 13.Jones E Y, Stuart D I, Walker N P. Nature (London) 1989;338:225–228. doi: 10.1038/338225a0. [DOI] [PubMed] [Google Scholar]
- 14.Headon D J, Overbeek P A. Nat Genet. 1999;22:370–374. doi: 10.1038/11943. [DOI] [PubMed] [Google Scholar]
- 15.Yan M, Wang L C, Hymowitz S G, Schilbach S, Lee J, Goddard A, de Vos A M, Gao W Q, Dixit V M. Science. 2000;290:523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- 16.Tucker A S, Headon D J, Schneider P, Ferguson B M, Overbeek P, Tschopp J, Sharpe P T. Development (Cambridge, UK) 2000;127:4691–4700. doi: 10.1242/dev.127.21.4691. [DOI] [PubMed] [Google Scholar]
- 17.Banner D W, D'Arcy A, Janes W, Gentz R, Schoenfeld H J, Broger C, Loetscher H, Lesslauer W. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 18.Schneider P, MacKay F, Steiner V, Hofmann K, Bodmer J L, Holler N, Ambrose C, Lawton P, Bixler S, Acha-Orbea H, et al. J Exp Med. 1999;189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shapiro L, Scherer P E. Curr Biol. 1998;8:335–338. doi: 10.1016/s0960-9822(98)70133-2. [DOI] [PubMed] [Google Scholar]
- 20.Hooper N M, Karran E H, Turner A J. Biochem J. 1997;321:265–279. doi: 10.1042/bj3210265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werb Z, Yan Y. Science. 1998;282:1279–1280. doi: 10.1126/science.282.5392.1279. [DOI] [PubMed] [Google Scholar]
- 22.Molloy S S, Bresnahan P A, Leppla S H, Klimpel K R, Thomas G. J Biol Chem. 1992;267:16396–16402. [PubMed] [Google Scholar]
- 23.Aoki N, Ito K, Tachibana T, Ito M. J Invest Dermatol. 2000;115:329–330. doi: 10.1046/j.1523-1747.2000.00065-1.x. [DOI] [PubMed] [Google Scholar]
- 24.Molloy S S, Thomas L, VanSlyke J K, Stenberg P E, Thomas G. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jean F, Stella K, Thomas L, Liu G, Xiang Y, Reason A J, Thomas G. Proc Natl Acad Sci USA. 1998;95:7293–7298. doi: 10.1073/pnas.95.13.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.VanSlyke J K, Thomas L, Thomas G. In: Peptidases and Neuropeptide Processing. Smith A I, editor. Vol. 23. San Diego: Academic; 1995. pp. 45–64. [Google Scholar]
- 27.Bresnahan P A, Leduc R, Thomas L, Thorner J, Gibson H L, Brake A J, Barr P J, Thomas G. J Cell Biol. 1990;111:2851–2859. doi: 10.1083/jcb.111.6.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miranda L, Wolf J, Pichuantes S, Duke R, Franzusoff A. Proc Natl Acad Sci USA. 1996;93:7695–7700. doi: 10.1073/pnas.93.15.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas L, Leduc R, Thorne B A, Smeekens S P, Steiner D F, Thomas G. Proc Natl Acad Sci USA. 1991;88:5297–5301. doi: 10.1073/pnas.88.12.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hata R, Senoo H. J Cell Physiol. 1989;138:8–16. doi: 10.1002/jcp.1041380103. [DOI] [PubMed] [Google Scholar]
- 31.Davies G B, Oxford J T, Hausafus L C, Smoody B F, Morris N P. Dev Dyn. 1998;213:12–26. doi: 10.1002/(SICI)1097-0177(199809)213:1<12::AID-AJA2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 32.Peterkofsky B, Diegelmann R. Biochemistry. 1971;10:988–994. doi: 10.1021/bi00782a009. [DOI] [PubMed] [Google Scholar]
- 33.Mechling D E, Bachinger H P. J Biol Chem. 2000;275:14532–14536. doi: 10.1074/jbc.275.19.14532. [DOI] [PubMed] [Google Scholar]
- 34.Jean F, Basak A, DiMaio J, Seidah N G, Lazure C. Biochem J. 1995;307:689–695. doi: 10.1042/bj3070689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui Y, Jean F, Thomas G, Christian J L. EMBO J. 1998;17:4735–4743. doi: 10.1093/emboj/17.16.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi S, Kasai K, Hatsuzawa K, Kitamura N, Misumi Y, Ikehara Y, Murakami K, Nakayama K. Biochem Biophys Res Commun. 1993;195:1019–1026. doi: 10.1006/bbrc.1993.2146. [DOI] [PubMed] [Google Scholar]
- 37.Moss M L, Jin S L, Milla M E, Bickett D M, Burkhart W, Carter H L, Chen W J, Clay W C, Didsbury J R, Hassler D, et al. Nature (London) 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 38.Peschon J J, Slack J L, Reddy P, Stocking K L, Sunnarborg S W, Lee D C, Russell W E, Castner B J, Johnson R S, Fitzner J N, et al. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 39.Black R A, Rauch C T, Kozlosky C J, Peschon J J, Slack J L, Wolfson M F, Castner B J, Stocking K L, Reddy P, Srinivasan S, et al. Nature (London) 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 40.Krysan D J, Rockwell N C, Fuller R S. J Biol Chem. 1999;274:23229–23234. doi: 10.1074/jbc.274.33.23229. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama K. Biochem J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seidah N G, Chretien M. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 43.Constam D B, Calfon M, Robertson E J. J Cell Biol. 1996;134:181–191. doi: 10.1083/jcb.134.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akamatsu T, Matsuda Y, Tsumura K, Tada J, Parvin M N, Kanamori N, Hosoi K. Dev Dyn. 1999;216:481–488. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<481::AID-DVDY16>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 45.Creemers J W, Siezen R J, Roebroek A J, Ayoubi T A, Huylebroeck D, Van de Ven W J. J Biol Chem. 1993;268:21826–21834. [PubMed] [Google Scholar]
- 46.Huttley G A, Jakobsen I B, Wilson S R, Easteal S. Mol Biol Evol. 2000;17:929–937. doi: 10.1093/oxfordjournals.molbev.a026373. [DOI] [PubMed] [Google Scholar]
- 47.Jones P A, Rideout W M, Shen J C, Spruck C H, Tsai Y C. BioEssays. 1992;14:33–36. doi: 10.1002/bies.950140107. [DOI] [PubMed] [Google Scholar]
- 48.Rholam M, Brakch N, Germain D, Thomas D Y, Fahy C, Boussetta H, Boileau G, Cohen P. Eur J Biochem. 1995;227:707–714. doi: 10.1111/j.1432-1033.1995.tb20192.x. [DOI] [PubMed] [Google Scholar]
- 49.Molloy S S, Anderson E D, Jean F, Thomas G. Trends Cell Biol. 1999;9:28–35. doi: 10.1016/s0962-8924(98)01382-8. [DOI] [PubMed] [Google Scholar]
- 50.Kumar A, Eby M T, Sinha S, Jasmin A, Chaudhary P M. J Biol Chem. 2001;276:2668–2677. doi: 10.1074/jbc.M008356200. [DOI] [PubMed] [Google Scholar]
- 51.Zonana J, Elder M E, Schneider L C, Orlow S J, Moss C, Golabi M, Shapira S K, Farndon P A, Wara D W, Emmal S A, et al. Am J Hum Genet. 2000;67:1555–1562. doi: 10.1086/316914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pispa J, Jung H S, Jernvall J, Kettunen P, Mustonen T, Tabata M J, Kere J, Thesleff I. Dev Biol. 1999;216:521–534. doi: 10.1006/dbio.1999.9514. [DOI] [PubMed] [Google Scholar]
- 53.Laurikkala J, Mikkola M, Mustonen T, Aberg T, Koppinen P, Pispa J, Nieminen P, Galceran J, Grosschedl R, Thesleff I. Dev Biol. 2001;229:443–455. doi: 10.1006/dbio.2000.9955. [DOI] [PubMed] [Google Scholar]
- 54.Schneider, P., Street, S. L., Gaide, O., Hertig, S., Tardivel, A., Tschopp, J., Runkel, L., Alevizopoulos, K., Ferguson, B. M. & Zonana, J. (March 14, 2001) J. Biol. Chem., 10.1074/jbc.M101280200. [DOI] [PubMed]