Abstract
Toll-like receptors (TLRs) function as initiators of inflammation through their ability to sense pathogen-associated molecular patterns and products of tissue damage1,2. Transcriptional activation of many TLR-responsive genes requires an initial de-repression step in which NCoR co-repressor complexes are actively removed from target gene promoters to relieve basal repression3,4. Ligand-dependent SUMOylation of liver X receptors (LXRs) potently suppresses TLR4-induced transcription by preventing the NCoR clearance step5–7, but the underlying mechanisms remain enigmatic. Here, we provide evidence that Coronin 2A (Coro2A), a component of the NCoR complex of previously unknown function8,9, mediates TLR-induced NCoR turnover by a mechanism involving interaction with oligomeric nuclear actin. SUMOylated LXRs block NCoR turnover by binding to a conserved SUMO2/3 interaction motif in Coro2A and preventing actin recruitment. Intriguingly, the LXR transrepression pathway can itself be inactivated by inflammatory signals that induce CaMKIIγ-dependent phosphorylation of LXR, leading to its deSUMOylation by the SUMO protease SENP3 and release from Coro2A. These findings reveal a Coro2A/actin-dependent mechanism for de-repression of inflammatory response genes that can be differentially regulated by phosphorylation and nuclear receptor signaling pathways that control immunity and homeostasis.
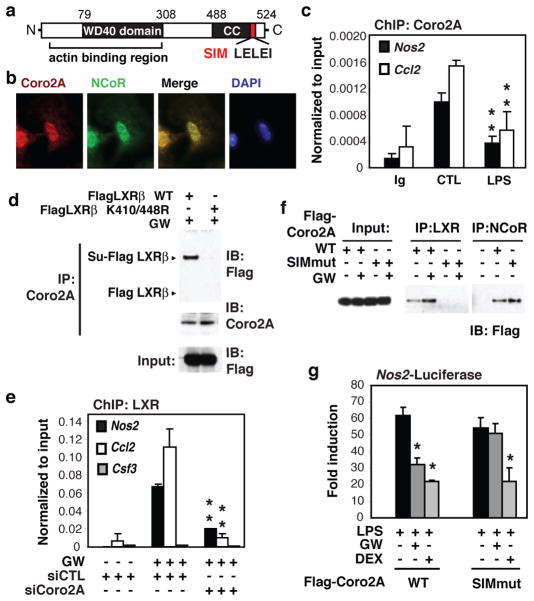
To delineate mechanisms by which SUMOylated LXRs block signal-dependent clearance of NCoR complexes from TLR4-inducible promoters, we searched for potential SUMO interaction motifs (SIMs) within proteins associated with the NCoR complex that might mediate interactions with SUMOylated LXRs. This search identified a conserved motif in the carboxyl-terminus of Coro2A (Fig. 1a), that matches a recently identified SIM that is specific for SUMO2/310. Coro2A is a member of the Coronin family of actin binding proteins that was identified as a component of the NCoR complex in nuclear extracts of HeLa cells8,9 and is highly expressed in cells of the hematopoietic lineage11,12. All other members of the Coronin family have thus far been found to play roles in regulation of the actin cytoskeleton through interactions with F-actin13,14, but functional roles of Coro2A in the NCoR complex have not been established. We confirmed Coro2A/NCoR interactions in primary macrophages by co-immunoprecipitation (Co-IP) assay and found that In contrast to other Coronin family members, Coro2A is primarily localized to the nucleus in primary bone marrow-derived macrophages (BMDMs) (Supplementary Fig. 2a, b). In addition, immunofluoresence microscopy demonstrated that Coro2A co-localizes to NCoR-rich regions of the nucleus (Fig. 1b). Chromatin immunoprecipitation (ChIP) studies further demonstrated that Coro2A is localized to NCoR target promoters such as the Nos2 and Ccl2 promoters in resting macrophages and its occupancy was reduced by treatment with the TLR4 ligand lipopolysaccharide (LPS) (Fig. 1c). Sequential ChIP experiments indicated that Coro2A and NCoR reside together on the Nos2 promoter (Supplementary Fig. 2c). Reduction of NCoR expression in primary macrophages using specific siRNAs resulted in a corresponding reduction in Coro2A occupancy on the Nos2 and Ccl2 promoters without affecting total Coro2A protein expression (Supplementary 2d, e). In contrast, Coro2A was not found on the Csf3 promoter (Supplementary Fig. 2d), which is not a target of NCoR repressison15.
Figure 1. SUMO interaction motif of Coronin2A is required for recruiting SUMOylated LXR to NCoR-residing proinflammatory gene promoters.
a, Known and predicted domain structure of mCoro2A. (CC: coiled-coil domain; SIM: SUMO2/3 interaction motif). b, Bone marrow-derived macrophages (BMDMs) were analyzed for Coro2A and NCoR expression and localization using fluorescence microscopy (magnification ×60). c, ChIP for Coro2A-binding to Nos2 and Ccl2 promoters in BMDMs. d, Lysates from HeLa cells transfected with the indicated LXRβ expression vectors and treated with GW3965 were subjected to immunoprecipitation (IP) with Coro2A-specific antibody and analyzed by immunoblotting (IB) against Flag. Su-Flag LXR; expected migration position for SUMO-LXR. e, ChIP for LXR-binding to the Nos2, Ccl2 and Csf3 promoters in BMDMs after siRNA transfection treated with or without GW3965. The Csf3 promoter serves as a negative control. Knockdown efficiencies are provided in Supplemental Fig. 10. f, Lysates from HeLa cells transfected with the indicated vectors and treated with GW3965 were subjected to IP with LXR or NCoR-specific antibodies and analyzed by IB against Flag. g, Luciferase assays were performed in RAW264.7 cells transfected with Nos2-luc reporter plasmids and the indicated Coro2A expression vectors, and stimulated with LPS in the presence of GW3965 or dexamethasome (DEX). (Bar graphs are averages ± s.e.m., **P<0.04 versus control *P<0.02 versus LPS).
To investigate the potential role of Coro2A as a molecular beacon for SUMOylated LXRs, co-immunoprecipitation (Co-IP) studies were performed in which HeLa cells were transfected with Flag-tagged wild type LXRβ or a mutant of LXRβ (K410/448R) that cannot be SUMOylated5 and treated with the synthetic LXR agonist GW3965 (GW). The mutant form of LXRβ did not co-precipitate with Coro2A, whereas wild type LXRβ was co-precipitated with Coro2A and migrated at the expected molecular weight for SUMO-LXRβ (Fig. 1d). This interaction was largely dependent on treatment with GW3965 (Supplementary Fig. 3a). In addition, in vitro transcribed and translated Coro2A preferentially interacted with recombinant GST-LXRβ conjugated to SUMO3 in vitro, as compared to unconjugated GST-LXRβ or de-SUMOylated LXRβ (Supplementary Fig. 3b). Next, ChIP experiments were performed in primary macrophages following transfection with control or Coro2A-specific siRNAs. Knockdown of Coro2A expression resulted in an almost complete loss of recruitment of LXR to the Nos2 and Ccl2 promoters in response to GW3965 (Fig. 1e). Although a recent study reported that GPS2 was required for recruitment of SUMOylated LXRβ to the Crp and Hp promoters in liver7, GPS2 is not present above IgG background on the Nos2 or Il1b promoters as determined by ChIP assay, and GPS2 knockdown had no impact on LXR transrepression of Nos2 or Il1b in primary macrophages (Supplementary Fig. 4a). Point mutations were therefore introduced into the SIM of Coro2A to evaluate its potential importance in recruiting SUMO-LXRs. HeLa cells were transfected with Flag-tagged wild type or SIM mutant versions of Coro2A and treated with vehicle or GW3965. Both WT and SIM mutant Coro2A were immunoprecipitated by anti-NCoR antibody, but only WT Coro2A interacted with LXR, and this interaction was enhanced by treatment with GW3965 (Fig 1f). In addition, a mammalian two-hybrid assay indicated that LXR interacts with Coro2A in a ligand- and SIM motif-dependent manner (Supplementary Fig. 3c). When over-expressed in RAW264.7 macrophages, this Coro2A SIM mutant occupies the NCoR-residing Nos2 promoter (Supplementary Fig. 4b). However, ligand-mediated recruitment of LXR and its repression function was significantly reduced by overexpression of this mutant, consistent with a dominant-negative function (Fig. 1g and Supplementary Fig. 4c). In contrast, dexamethasone repression mediated by the glucocorticoid receptor, via an NCoR and SUMOylation independent mechanism16, remained unaffected (Fig 1g).
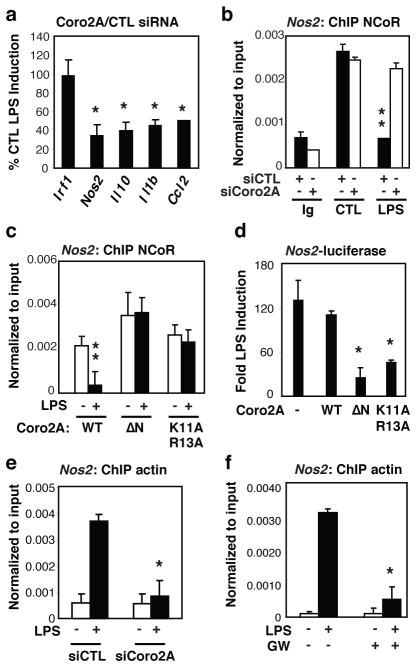
Unexpectedly, knockdown of Coro2A expression resulted in impaired responsiveness of a subset of TLR inducible NCoR target genes, including Nos2, Il10, Il1b and Ccl2, but not of non-NCoR-regulated promoters, such as irf1 (Fig. 2a). ChIP experiments further revealed that knockdown of Coro2A resulted in a failure of LPS to promote clearance of NCoR from the Nos2 and Il1b promoter (Fig. 2b and Supplementary Fig. 5a). As there is increasing evidence for diverse transcriptional roles of oligomeric nuclear actin17, we considered the possible role of nuclear actin in NCoR clearance. We confirmed that Coro2A interacts with actin and using conserved sequence features required for the interaction of murine Coronin1 with actin14,18,19, introduced point mutations in Coro2A (K11A/R13A) that severely compromised actin binding (Supplementary Fig. 5b). Overexpression of either an N-terminal deletion mutant of Coro2A lacking the actin-binding domain (ΔN), or the Coro2A K11A/R13A mutant blocked LPS-induced NCoR clearance and impaired activation of the Nos2 promoter in RAW264.7 macrophages (Fig. 2c, d). ChIP assays using an antibody (2G2) that specifically recognizes nuclear actin20 revealed transient recruitment of actin to the Nos2 (Fig. 2e) and Il1b (Supplementary Fig. 5c) promoters coincident with the timing of LPS-induced NCoR clearance. Actin was not recruited to the TLR4-inducible, NCoR-independent Csf3 promoter or the Abca1 promoter, which is a positive transcriptional target of LXRs that is occupied by NCoR in the absence of LXR agonists4,21 (Supplementary Fig. 5c). Actin recruitment was dependent on both NCoR and phosphorylation of cJun, as it was abolished in primary macrophages by NCoR knockdown and in RAW264.7 macrophages by overexpression of a mutant form of c-Jun that cannot be phosphorylated (Supplementary Fig. 5d-e). LPS-induced actin recruitment to the Nos2 promoter in macrophages was also abolished by siRNA-mediated knockdown of Coro2A (Fig. 2e). Short-term treatment of primary macrophages with latrunculin A, which inhibits actin polymerization22, similarly blocked LPS dependent NCoR clearance and induction of Nos2 mRNA (Supplementary Fig. 6a, b). In contrast, latrunculin A did not affect TLR4 activation of the NCoR-independent Irf1 gene, LXR activation of the Abca1 gene (Supplementary Fig. 6c-e) or TLR4-dependent phosphorylation of cJun (Supplementary Fig. 6f, g). Notably, treatment of primary macrophages with the LXR ligand, GW3965, blocked LPS-induced actin recruitment (Fig. 2f), but not cJun phosphorylation (Supplementary Fig. 6h), suggesting that SUMOylated LXRs may exert anti-inflammatory effects by blocking the actin-dependent step required for NCoR clearance.
Figure 2. The actin-binding domain of Coronin2A is required for LPS de-repression of inflammatory gene promoters.
a, BMDMs were transfected with Coro2A or control siRNAs and treated with or without LPS. Total RNA was used for assay of the indicated transcript levels by Q-PCR. Results are plotted as fold LPS induction in siCoro2A cells/fold LPS induction in siCTL cells × 100. b, ChIP for NCoR-binding to the Nos2 promoter in BMDMs after siRNA transfection followed by 1hr of LPS stimulation. c, ChIP for NCoR-binding to, Nos2-luc promoter in RAW264.7 cells transfected with Nos2-luc reporter plasmids and the indicated expression vectors and stimulated with LPS. d, Luciferase assays were performed in RAW264.7 cells transfected with Nos2-luc reporter plasmids and the indicated expression vectors and stimulated with LPS. Luciferase activities in cells not treated with LPS were normalized to a value of 1.0. e, ChIP for actin-binding to Nos2 promoter in BMDMs after siRNA transfection followed by 5min of LPS stimulation. f, ChIP for actin-binding to the Nos2 promoter in BMDMs treated with LPS for 5min pretreated with or without GW3965. (Averages ± s.e.m., **P<0.05 versus control *P<0.05 versus LPS).
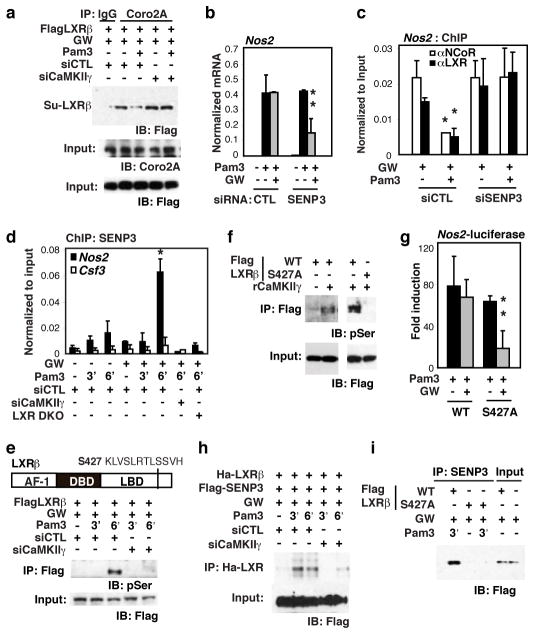
LXRs are unable to suppress transcriptional responses of NCoR target genes to pro-inflammatory signals that induce CaMKII activation, including the TLR1/2 agonist Pam3CSK421, the TLR2/6 agonist FSL-1, and Salmonella typhimurium 23 (Supplementary Fig. 7a, b). Co-IP studies in RAW264.7 macrophages revealed that Pam3CSK (Pam3) signaling significantly reduced interaction of SUMO-LXRβ with Coro2A in a CaMKIIγ-dependent manner (Fig. 3a). Because interaction of LXR with Coro2A requires SUMOylation, we considered the possible involvement of deSUMOylating enzymes in negatively regulating LXR repression functions. Using siRNAs to knock down SUMO proteases expressed in primary macrophages, we found that knockdown of SENP3 restored the ability of LXRs to suppress TLR1/2-dependent Nos2 expression (Fig. 3b), interact with the Nos2 promoter, and prevent NCoR clearance (Fig. 3c). ChIP experiments further demonstrated that SENP3 was rapidly recruited to the Nos2, but not the Csf3, promoter following Pam3 stimulation, and this recruitment was dependent on CaMKIIγ (Fig. 3d). Knockdown of CaMKIIγ also restored LXR transrepression of Il1b and BIC (precursor of miR-155) as well as abolished SENP3 recruitment to these gene promoters (Supplementary Fig. 7c–f).
Figure 3. LXR-Coro2A axis is disrupted by CaMKII.
γ phosphorylation and SENP3 deSUMOylation of LXR.
a, RAW264.7 cells were transfected with Flag-LXRβ expression vector and siRNAs as indicated. Cells were pretreated with GW3965 and challenged with Pam3CSK4. Lysates were subjected to IP with Coro2A antibodies and analyzed by IB against Flag. Knockdown efficiencies for CamKIIγ are provided in Supplemental Fig. 10c. b, BMDMs were transfected with control or SENP3 siRNAs and pretreated with or without GW3965 followed by Pam3CSK4. RNA transcript levels were assayed by Q-PCR. Knockdown efficiencies for SENP3 are provided in Supplemental Fig. 10d. c, ChIP for LXR and NCoR-binding to the Nos2 promoter in BMDMs after siRNA transfection. d, ChIP for SENP3 occupancy of the Nos2 promoter in siRNA transfected wildtype or LXRα/β−/− BMDM. Cells were pretreated with GW3965 and then stimulated with Pam3CSK4. e, Domain structure of LXR. (AF-1: Activation Function-1 domain; DBD: DNA binding domain; LBD: ligand binding domain). A predicted CaMKII phosphorylation site at Serine (S) 427 in the LBD is highlighted. RAW264.7 cells were transfected with Flag-LXRβ expression vector and control or CaMKIIγ-specific siRNAs. Cells were treated with GW3965 and challenged by Pam3CSK4 for 3 and 6 min. Lysates were subjected to IP with Flag antibody and IB for phosphoserine (top) or Flag (bottom). f, Flag-LXRβ WT or S427A mutant were expressed in HeLa cells, captured on anti-Flag-agarose, and incubated with 0.5μg of activated rCaMKIIγ. Anti-phospho-serine antibody was used to detect phosphorylated proteins by IB. g, RAW264.7 cells were transfected with Nos2-luc reporter plasmids and the indicated expression vectors and treated with GW3965 followed by Pam3CSK4. Luciferase activity was represented as fold induction compared to untreated cells. h, RAW264.7 cells were transfected with expression vectors for Flag-SENP3 and Ha-LXRβ, and control or CaMKIIγ specific siRNAs as indicated. Lysates were subjected to IP with anti-Ha antibody, and SENP3 was detected by Flag antibody. i, RAW264.7 cells were transfected with the indicated Flag-LXRβ expression vectors, treated with GW3965, and challenged with Pam3CSK4. Lysates were used in IP assays with SENP3 specific antibody, and analyzed by IB against Flag. (Averages ± s.e.m., **P<0.04 versus Pam3 *P<0.04 versus control).
Co-immunoprecipitation experiments suggested a sequence in which Pam3 induced CaMKII-LXR interaction, followed shortly thereafter by SENP3-LXR interaction (Supplementary Fig. 8a). As both LXRα and LXRβ contain a conserved motif that conforms to a CaMKII phosphorylation site (S427 in LXRβ, Fig. 3e), we considered the possibility that LXRs were themselves the targets of CaMKIIγ kinase activity and that phosphorylation of LXR might promote its interaction with SENP3. Consistent with this possibility, TLR1/2 activation was found to result in robust serine phosphorylation of LXRβ in a CaMKIIγ-dependent manner (Fig. 3e). WT-Flag-tagged LXRβ was a substrate for recombinant CaMKIIγ in vitro, but Flag-tagged LXRβ with a serine to alanine point mutation at the 427 residue was not (Fig. 3f and Supplementary Fig. 8b). In addition, the phospho-null mutant of LXR continued to associate with Coro2A-NCoR bound Nos2 promoter and retained transrepression activities upon TLR1/2 signaling (Fig. 3g and Supplementary Fig. 8c), recapitulating the effect of knocking down CaMKIIγ21 and suggesting that LXR S427 is likely the major or only substrate of CaMKII relevant to inactivation of the LXR transrepression pathway. In contrast, a phospho-mimic mutant of LXR (S427D) was not able to interact with Coro2A to promote transrepression of the Nos2 luciferase reporter in RAW64.7 macrophages (Supplementary Fig. 8d, e).
To investigate whether CaMKII phosphorylation of LXRβ created a docking site for SENP3, RAW264.7 macrophages were transfected with expression vectors for HA-LXRβ, Flag-SENP, and control or CaMKIIγ-specific siRNAs and treated with GW3965 in the absence or presence of Pam3. Immunoprecipitation assays demonstrated that TLR1/TLR2 activation induced rapid interaction of SENP3 with LXRβ that was dependent on CaMKIIγ (Fig. 3h). This interaction was not observed with the LXRβ S427A mutant (Fig. 3i).
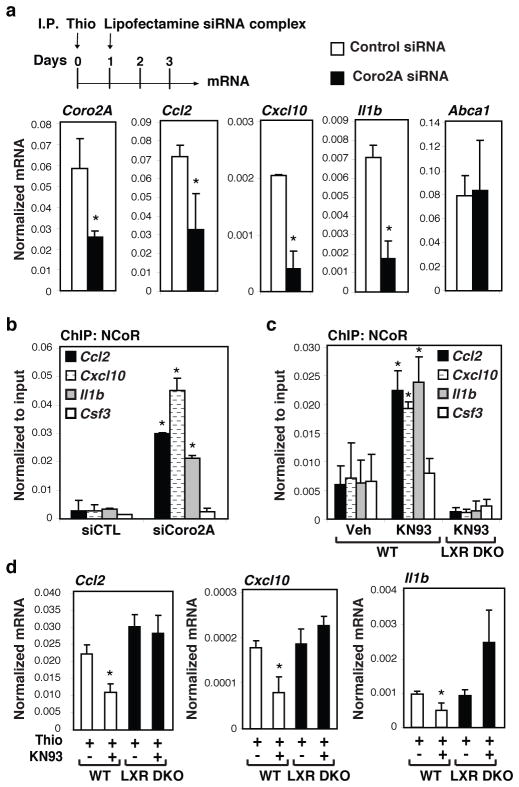
We next used a thioglycollate-induced sterile peritonitis model 24 to investigate potential roles of the Coro2A-NCoR clearance pathway in vivo. Injection of thioglycollate into the peritoneal cavity leads to massive infiltration of activated macrophages, which by three days following injection account for more than 90% of the total peritoneal cavity cell population. In vivo knockdown of Coro2A expression in this macrophage population using two independent methods significantly reduced expression of Ccl2, Il1b, and Cxcl10, but not Abca1 (Fig. 4a and Supplementary Fig. 9a) and resulted in significantly increased NCoR occupancy (Fig. 4b). The presence of phosphorylated (active) CaMKII in the elicited macrophage population (Supplementary Fig. 9b), suggested that an endogenous LXR repression pathway in these cells might be inactivated. Consistent with this possibility, CoIP studies demonstrated that LXRα/β were not associated with Coro2A in elicited macrophages in control animals, but became associated with Coro2A in animals that were treated with the CaMKII inhibitor, KN93, (Supplementary Fig. 9b). In addition, KN93 treatment increased NCoR occupancy on the Ccl2, Cxcl10 and Il1b promoters in macrophages in WT mice, but not in LXRα/β double knockout mice (Fig. 4c). Finally, KN93 treatment reduced the expression of Ccl2, Cxcl10 and Il1β in wild type mice, but not in LXR double knockout mice (Fig. 4d), suggesting that pharmacological inhibition of CaMKII ‘rescues’ endogenous LXR transrepression activity in this inflammatory context.
Figure 4. Function of the Coro2A-LXR-CaMKII pathway in thioglycollate-elicited peritonitis.
a, Sterile peritonitis was initiated by intraperitoneal (I.P.) injection of thioglycollate (Thio) on day 0. Mice were injected with lipofectamine-siRNA complexs on day one, and peritoneal exudate cells were recovered for analysis of Coro2A, Ccl2, Cxcl10, Il1β and Abca1 mRNA on day 3 (n=4 per condition). (Values are normalized to Gapdh. Averages ± s.e.m., *P<0.05 versus siCTL). b, ChIP for NCoR-binding to the Ccl2, Cxcl10, Il1b, and Csf3 promoters in peritoneal exudate cells described in a (n=3 per condition). (Averages ± s.e.m., *P<0.05 versus siCTL). c, ChIP for NCoR-binding to the Ccl2, Cxcl10, Il1b, and Csf3 promoters in peritoneal exudate cells derived from wild type (WT) or LXRα/β double knockout (DKO) mice treated with KN93 as indicated (n=3 per condition). (Averages ± s.e.m., *P<0.05 versus non-KN93 treated wildtype animals). d, Expression of Ccl2, Cxcl10, and Il1b expression in peritoneal exudate cells derived from WT (n=5 per condition) or LXRα/β−/− (n=3 per condition) mice treated with KN93 as indicated. (Averages ± s.e.m., *P<0.01 versus non-KN93 treated wildtype animals). Statistical significance was determined by a two-tailed Student’s t-test.
Collectively, these findings provide evidence that Coro2A functions as an NCoR exchange factor that is required for de-repression of TLR target genes in the macrophage (Supplementary Fig. 1). Recent findings providing evidence for roles of nuclear actin in movements of specific chromosomal loci to transcriptional ‘hubs’ 25,26 raise a number of interesting possibilities with respect to dynamic regulation of the localization of repression machinery. The ability of other Coronins to mediate disassembly of actin polymers in the cytoplasm14,27 suggests that Coro2A may function to release inflammatory response genes from localized actin networks dedicated to repression functions. The integrated functions of Coro2A as both a docking site for SUMOylated LXRs and as an actin-dependent NCoR exchange factor suggest a parsimonious transrepression model in which SUMOylated LXRs prevent NCoR turnover and repress responses to LPS by blocking actin recruitment. The ability of CaMKIIγ to inactivate the LXR transrepression pathway by creating a docking site for SENP3 further identifies a new transcriptional role for SUMO proteases and reveals an entry point for negative regulation of the LXR repression pathway that could be engaged by numerous other signaling pathways that mobilize intracellular calcium, such as purinergic receptors21. It is notable that the SUMO2/3-specific SIM in Coro2A and the CaMKIIγ phosphorylation sites in LXRs are highly conserved across vertebrate species, suggesting evolutionary pressure to maintain these regulatory pathways. While such pressures presumably relate to key functions of the innate immune system in responding to infection and injury, signal-induced inactivation of the LXR transrepression pathway may also contribute to pathological forms of inflammation.
Methods Summary
BMDM were generated from 6-wk old wildtype C57BL/6 (Harlan) and LXRα/β−/− mice as described in 15. RAW264.7 cells were cultured as described in 5. LPS, Pam3CSK4, and GW3965 were used at a concentration of 100ng/ml, 300ng/ml, and 1μM respectively. GW3965 was used to pre-treat macrophages for 1hr. LPS and Pam3CSK4 treatment was 1hr for ChIP assays and 6hrs for luciferase and mRNA expression analysis unless otherwise noted in the figure. ChIP assays were performed as described in detail in 15. Proximal promoter regions of Nos2, Ccl2, Il1b, Csf3, Abca1, and BIC were amplified by real-time PCR. mRNA values determined by real time PCR are normalized to GAPDH mRNA content. Data are represented as mean +/− SD of three independent experiments in duplicates. RAW264.7 or HeLa cells were transfected with wildtype or mutant expression vectors for LXR and Coro2A as described in 21. For RNAi experiments in cultured primary macrophages, scrambled control or smart-pool siRNAs (Dharmacon) against Coronin2A, SENP3, CaMKIIγ, and NCoR were transfected into primary macrophages using lipofectamine 2000 (Invitrogen) as described 15. For in vitro CaMK kinase assays, 0.5μg recombinant CaMKIIγ (Upstate) was incubated with bacterially purified GST-LXRβ (full-length) fusion proteins bound on glutathione beads. Thiogycollate elicited sterile peritonitis was induced as previously described 28. Briefly, 2ml of thiogycollate medium was delivered to in each animal by i.p. injection and elicited macrophages in the peritoneal cavities were harvested with 10ml of PBS after three days. For in vivo RNAi experiments, 100μg scrambled or Coronin2A specific siRNA were complexed in lipofectamine 2000 and serum depleted medium in a final volume of 1ml and delivered to animals by i.p. injection as previously described in 29. Alternatively, GeRP capsulated scrambled and Coronin2A specific siRNA were prepared and delivered to animals by i.p. injection for 5 consecutive days as described in 30.
Methods
Reagents and plasmids
LPS and Flag M2 antibodies were obtained from Sigma. Pam3CSK4 was from InvivoGen. GW3965 was kindly provided by GlaxoSmithKline. 22R and T0901317 were purchased from Sigma and 25EC from Biomol. KN93 was purchased from CalBiochem. Goat anti-Coro2A, rabbit anti-Coro2A, CaMKII, SENP3, LXRa/b, Myc, HDAC4, goat anti-NCoR and normal rabbit/goat antibodies were from Santa Cruz Biotech. Rabbit anti-NCoR antibodies were from ABR. α-tubulin and phospho-CaMKII antibodies were from Cell Signalling and phosphor-serine antibodies were from Chemicon. The reporter plasmid nos2-luc has been previously described4 . Expression vector for VP16LXRb was obtained from Xceptor. Expression constructs, Myc-SUMO2, FlagLXRb, HaLXRb had been previously described5. Mutations in expression vectors were made using the QuickChange site-directed mutagenesis kit (Stratagene).
Plasmid and siRNA Transfection
Transient transfections in RAW264.7 cells were performed as described in5 using Superfect reagent (Qiagen). For siRNA experiments in RAW264.7 cells, transfections were carried out with (100nM) using Superfect reagent for 48 h before activation with TLR ligands. Transfection data are represented as mean +/− SD of three independent experiments in triplicates. For RNAi experiments in cultured primary macrophages, scrambled control or smart-pool siRNAs (Dharmacon) against Coronin2A, SENP3, CaMKIIγ, and NCoR were transfected into primary macrophages using lipofectamine 2000 (Invitrogen) as described15.
siControl Non-targeting siRNA Pool(Dharmacon, D-001206-13): AUGAACGUGAAUUGCUCAA, UAAGGCUAUGAAGAGAUAC, AUGUAUUGGCCUGUAUUAG, UAGCGACUAAACACAUCAA; siGenome SMART pool for mouse Coro2A (Dharmacon, D-058157): GAAAGUUGGACCCUCACUA, GCAAGAUUCGGAUUGUUGA, GGAUCGGUAUCAUGCCAAA, GAGGGAACGUCUUGGACAU; siGenome SMART pool for mouse SENP3 (Dharmacon, D-057149): GGACCACAGUGCCAACUAG, CGACGUACCAUCACCUAUU, GCUCUCCGACCCUCUCAUA, CAAACUCCGUCGUCAGAUC; siGenome SMART pool for mouse CaMKIIg (Dharmacon, D-040009): GGAAAGAUCCCUAUGGAAA, GUAGAGUGCUUACGCAAAU, GGAGUUGUUUGAAGACAUU, AAACAUCUACGCAGGAAUA; siGenome SMART pool for mouse NCoR1 (Dharmacon, D-058556): GAAAUCCCACGGCAAGAUA, CCAGGUCGAUGACAAGUGA, GCAGUGGAAGGAAGUAUAA, GAUCAUCACCCGGCAAAUU. Note that siRNA in pools of four were used in in vitro studies and the siRNAs underlined were used for the in vivo experiments.
Preparation of GeRPs containing Coro2A siRNA
β1,3-d-glucan particles were purified from baker’s yeast by a series of alkaline and solvent extractions, hydrolysing outer cell wall and intracellular components and yielding purified, porous 2 4 μm β1,3-d-glucan particles, as previously described30. To load siRNA into glucan shells for 1 mouse injection, 5 nmoles Coro2A siRNA was incubated with 50nmoles Endo-Porter peptide (Gene Tools) in acetate buffer pH 4.8 for 15 min at 21 degrees C. The siRNA/Endo-Porter solution was added to 1 mg glucan shells and then vortexed and incubated for 1 hour. TRIS-EDTA buffer pH 8.4 was added to the particles and incubated for 15 min at RT to adjust the pH. The resulting glucan encapsulated siRNA Particles (GeRPs) were resuspended in PBS for intraperitoneal (i.p.) injections.
RNA isolation and real time PCR
Total RNA (isolated by RNeasy kit, Qiagen) was prepared from MEFs or macrophages treated as indicated in each legend. One μg of total RNA was used for cDNA synthesis, and 1 μl of cDNA was used for real time PCR analysis was performed on an Applied Biosystems 7300 Real-time PCR system. Values are normalized with GAPDH content. Data are represented as mean +/− SD of at least three independent experiments in duplicates.
Primers for mRNA-QPCR analysis
Gapdh (AATGTGTCCGTCGTGGATCT, CATCGAAGGTGGAAGAGTGG); Nos2 (AGCCTTGCATCCTCATTGG, CACTCTCTTGCGGACCATCT); Irf1 (AGGCCGATACAAAGCAGGAGA, GCTGCCCTTGTTCCTACTCTG); Il1b (GCAACTGTTCCTGAACTCAACT, ATCTTTTGGGGTCCGTCAACT); Ccl2 (CCCAATGAGTAGGCTGGAGA, TCTGGACCCATTCCTTCTTG); Senp3 (TTTGACTCCCAGCGAACTCT, AAAGAGACTATACAGGGGACCG); Coro2A (AGGGGATCGGTATCATGCCAA, CATGGACACTGGCTCGATGAG); NCoR (CTGTGCATGAGAAGCAGGAC, TGGTGAAGAATGAAGGCAAG); Abca1 (AAAACCGCAGACATCCTTCAG, CATACCGAAACTCGTTCACCC); BIC (AAACCAGGAAGGGGAAGTGT, GTCAGTCAGAGGCCAAAACC); CaMKIIγ (AACAAAAACAGTCTCGTAAGCCC, GCCCTTGATCCCATCTGTAGC) Gps2 (AGGCGAAAGGAACAGAGTGA, GAGTACCTGGGCGATTGTGT).
Primers for promoter ChIP-QPCR analysis
Nos2 (GGAGTGTCCATCATGAATGAG, CAACTCCCTGTAAAGTTGTGACC); Il1b (GGACAATTGTGCAGATGGTG, CCTACCTTTGTTCCGCACAT); BIC (GCTGGAGACAAGTCCCTTGA, TCCTCCAGTGATTCCTTTGG); Ccl2 (TCCAGGGTGATGCTACTCCT, AGTGAGAGTTGGCTGGTGCT); Hp (CACTAAGGCAGCATGGTTGA, TGGGAGAGGGTGTTGGTTAG); Csf3 (CCTACCTAGGGTGCTGTGGA, GGACAAACATCCCGAGAGAA), Abca1 (GGGGCTGAGCAAACTAACAA, TTGTGGGTGTTGCTTTTTGA).
Co-IP assay
RAW264.7 or HeLa cells were transfected with wildtype or mutant expression vectors for LXR and Coro2A. Cells were lysed in 10 mM Tris-HCl ph 7.5, 450 mM NaCl, 0.5% NP40, 1mM EDTA with protease inhibitors and 10uM NEM. Co-IPs were carried out with anti-SENP3, LXR, CaMKII, NCoR, or Flag antibodies and detected by western blotting.
Kinase Assays
For in vitro CaMK kinase assays, 0.5ug recombinant CaMKIIγ (Upstate) was incubated with bacterially purified GST-LXRb (full-length) fusion proteins attached to glutathione beads. The beads were washed, boiled, and then subjected to SDS-PAGE. Serine phosphorylations were detected by anti-p-Serine antibody (Chemicon).
Supplementary Material
Acknowledgments
We thank Shinya Amano for preparation of GeRP delivery spheres. We thank Lynn Bautista for assistance with preparation of the manuscript. W.H. was supported by NRSA 1F31DK083913. These studies were supported by NIH grants CA52599, DK074868 and HC088093 and a Leducq Foundation Transatlantic Network Grant. M.G.R. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions
W.H., S.G., K.S., M.C., B.L.G., M.G.R. and C.K.G. conceived the project and planned experiments and analysis, which were performed by W.H., S.G., K.S., M.G., M.A., G.T., D.Z and J.Y. The entire project was supervised by C.K.G., who wrote the manuscript with W.H.
Competing financial interests: Dr. Michael Czech is a consultant to and an equity holder in RXi Pharmaceuticals Inc, which has licensed the GeRP siRNA delivery method.
References
- 1.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 2.Beutler BA. TLRs and innate immunity. Blood. 2009;113:1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogawa S, et al. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci U S A. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pascual G, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghisletti S, et al. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaschke F, et al. A nuclear receptor corepressor-dependent pathway mediates suppression of cytokine-induced C-reactive protein gene expression by liver X receptor. Circ Res. 2006;99:e88–99. doi: 10.1161/01.RES.0000252878.34269.06. [DOI] [PubMed] [Google Scholar]
- 7.Venteclef N, et al. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXR{beta} in the hepatic acute phase response. Genes Dev. 2010;24:381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon HG, et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon HG, et al. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Ouyang J, et al. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lattin JE, et al. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uetrecht AC, Bear JE. Coronins: the return of the crown. Trends Cell Biol. 2006;16:421–426. doi: 10.1016/j.tcb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Gandhi M, Achard V, Blanchoin L, Goode BL. Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol Cell. 2009;34:364–374. doi: 10.1016/j.molcel.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghisletti S, et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogawa S, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat Rev Mol Cell Biol. 2004;5:410–415. doi: 10.1038/nrm1370. [DOI] [PubMed] [Google Scholar]
- 18.Appleton BA, Wu P, Wiesmann C. The crystal structure of murine coronin-1: a regulator of actin cytoskeletal dynamics in lymphocytes. Structure. 2006;14 :87–96. doi: 10.1016/j.str.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Gandhi M, Jangi M, Goode BL. Functional surfaces on the actin-binding protein coronin revealed by systematic mutagenesis. J Biol Chem. 2010 doi: 10.1074/jbc.M110.171496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonsior SM, et al. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J Cell Sci. 1999;112 ( Pt 6):797–809. doi: 10.1242/jcs.112.6.797. [DOI] [PubMed] [Google Scholar]
- 21.Huang W, et al. Transcriptional integration of TLR2 and TLR4 signaling at the NCoR derepression checkpoint. Mol Cell. 2009;35:48–57. doi: 10.1016/j.molcel.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yarmola EG, et al. Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J Biol Chem. 2000;275:28120–28127. doi: 10.1074/jbc.M004253200. [DOI] [PubMed] [Google Scholar]
- 23.Gewirtz AT, et al. Salmonella typhimurium induces epithelial IL-8 expression via Ca(2+)-mediated activation of the NF-kappaB pathway. J Clin Invest. 2000;105:79–92. doi: 10.1172/JCI8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson RB, Hobbs JA, Mathies M, Hogg N. Rapid recruitment of inflammatory monocytes is independent of neutrophil migration. Blood. 2003;102:328–335. doi: 10.1182/blood-2002-10-3228. [DOI] [PubMed] [Google Scholar]
- 25.Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segal BH, et al. Thioglycollate peritonitis in mice lacking C5, 5-lipoxygenase, or p47(phox): complement, leukotrienes, and reactive oxidants in acute inflammation. J Leukoc Biol. 2002;71:410–416. [PubMed] [Google Scholar]
- 29.Amarzguioui M, et al. Ex vivo and in vivo delivery of anti-tissue factor short interfering RNA inhibits mouse pulmonary metastasis of B16 melanoma cells. Clin Cancer Res. 2006;12:4055–4061. doi: 10.1158/1078-0432.CCR-05-2482. [DOI] [PubMed] [Google Scholar]
- 30.Aouadi M, et al. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature. 2009;458:1180–1184. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.