ABSTRACT
Fish vitellogenin synthesized and released from the liver of oviparous animals is taken up into oocytes by the vitellogenin receptor. This is an essential process in providing nutrient yolk to developing embryos to ensure successful reproduction. Here we disclose the full length vtgr cDNA sequence for largemouth bass (LMB) that reveals greater than 90% sequence homology with other fish vtgr sequences. We classify LMB Vtgr as a member of the low density lipoprotein receptor superfamily based on conserved domains and categorize as the short variant that is devoid of the O-glycan segment. Phylogenetic analysis places LMB Vtgr sequence into a well-supported monophyletic group of fish Vtgr. Real-time PCR showed that the greatest levels of LMB vtgr mRNA expression occurred in previtellogenic ovarian tissues. In addition, we reveal the effects of insulin, 17beta-estradiol (E2), and 11-ketotestosterone (11-KT) in modulation of vtgr, esr, and ar mRNAs in previtellogenic oocytes. Insulin increased vtgr expression levels in follicles ex vivo while exposure to E2 or 11-KT did not result in modulation of expression. However, both steroids were able to repress insulin-induced vtgr transcript levels. Coexposure with insulin and E2 or of insulin and 11-KT increased ovarian esr2b and ar mRNA levels, respectively, which suggest a role for these nuclear receptors in insulin-mediated signaling pathways. These data provide the first evidence for the ordered stage-specific expression of LMB vtgr during the normal reproductive process and the hormonal influence of insulin and sex steroids on controlling vtgr transcript levels in ovarian tissues.
Keywords: insulin, oocyte development, steroid hormones, vitellogenin, vitellogenin receptor
The sex steroids 17beta-estradiol and 11-ketotestosterone repress insulin-induced vitellogenin receptor mRNA expression in previtellogenic oocytes of largemouth bass.
INTRODUCTION
Control of vertebrate reproduction involves a complex network of hormonal and chemical signals. Two processes that are essential for successful reproduction in oviparous species include the synthesis of vitellogenin (Vtg), a phospholipoglycoprotein that is the precursor molecule for egg yolk protein, and its sequential uptake into developing oocytes. The hormonal driving mechanisms of Vtg synthesis in fish hepatic tissues have been extensively studied and are well demonstrated to be under the control of 17β-estradiol (E2) [1]. 17β-Estradiol classically acts through nuclear estrogen receptor (ER) signaling, which stimulates hepatocytes to synthesize Vtg for release into the bloodstream. Then circulating Vtg is incorporated into oocytes via the vitellogenin receptor (Vtgr) to be proteolytically cleaved into yolk proteins that serve as a nutrient source for developing fish embryos [2].
It has been established that in oviparous vertebrates, Vtgr mediates the entrance of Vtg into oocytes, although in birds and amphibians this receptor may also play a part in the incorporation of very low density lipoproteins (VLDL) [3], and this notion has recently been adopted for fish [4, 5]. With the advent of cloning technology, identification and sequencing of the vgtr has been reported for a handful of fish species, which maintain highly conserved regions; however, two cDNA isoforms have been reported in rainbow trout [3, 6], blue tilapia [7], and sole [8]. The isoforms consist of a larger cDNA sequence containing an O-glycan domain and a shorter one devoid of this region. In ovarian tissue, the predominant isoform is typically the shorter isoform termed a non-O-linked sugar-splicing variant. The larger variant predominantly localizes to somatic tissues [9], and it has been hypothesized that the O-glycan domain controls VLDL incorporation in these tissues [10]. Fish Vtgr is highly homologous with the human very low density lipoprotein receptor (VLDLR), which is part of the LDLR superfamily of cell surface proteins that transports macromolecules into cells through a process known as receptor-mediated endocytosis [11]. In humans, their main function is to maintain cholesterol homeostasis in tissues and organs by mediating access of lipoprotein particles from the bloodstream to the cell cytoplasm [12].
The functionality of Vtgr in fish has been examined in studies with Coho salmon [13], Nile tilapia [14], and seabass [15]; these studies focused on Vtg uptake through implementation of binding assays that demonstrated the presence of surface receptors within oocyte membranes. Furthermore, the existence of a new system of multiple Vtgrs that disparately bind functionally different types of Vtgs in white perch has been suggested [16]. Short-term in vitro studies have reported successful incorporation of radiolabeled Vtg into fish oocytes [17, 18]. Several publications have explored the signals controlling Vtg uptake and suggest control by various hormones including gonadotropin I [19], insulin (INS) and thyroxine [20], a pituitary glycoprotein fraction from salmon [21], 11-ketotestosterone (11-KT), and INS-like growth factor hormone I (IGF-I) [22]. Some of these factors have also been shown to regulate uptake of VLDL and other lipids during early stages of fish ovarian development [22, 23].
In other teleost fish, maximal vtgr expression coincides with previtellogenic stages of oocyte development [5, 6, 8, 24, 25], a stage of oogenesis where RNA is intensively synthesized by the growing oocyte [26, 27]. The mechanisms controlling vtgr synthesis in this window are understudied. Such investigations have been limited to insects [28], which have disparate hormonal pathways compared to vertebrates (i.e., insects use ecdysone, which is not present in fish and other vertebrates) possibly limiting the relevance of fish hormonal contributions to Vtgr regulation. Only one recent study using the medaka model revealed that E2 exposure suppresses vtgr expression in females in concert with potential infertility [29]. Using the largemouth bass (LMB; Micropterus salmoides) model, which have a semisynchronized reproductive cycle that allows monitoring of endocrine biology in several discrete windows of the reproductive process, our goal was to identify and classify the LMB vtgr cDNA using various PCR strategies and phylogenetic analysis. Additionally, we assessed the temporal expression of the LMB vtgr in ovarian tissues during the normal annual reproductive cycle during defined stages of oocyte development. Previous work by our group has focused primarily on estrogen receptors in this model species. Therefore, in an attempt to also investigate androgens, we identified and measured the temporal expression of the LMB androgen receptor (Ar). Finally we investigated the role of sex steroids, E2 and 11-KT, and INS in controlling the expression of vtgr in previtellogenic oocytes using ex vivo ovarian follicle cultures.
Here we reveal the LMB vtgr cDNA sequence and its characteristics that place it in the LDLR gene superfamily. Our phylogenetic analysis indicates that the LMB vtgr falls into a well-supported clade of other sampled fish that lack an O-glycan domain. In addition, we demonstrate that the highest levels of vtgr and ar expression in LMB female gonadal tissues occurred during primary growth (previtellogenic stage) of oocyte development and further determine that transcription of vtgr is induced by INS. Both E2 and 11-KT are able to suppress INS-induced vtgr transcript levels in concert with increasing hormone receptor expression levels.
MATERIALS AND METHODS
Animals
All LMB used for experimentation were maintained in fiberglass tanks at the University of Florida Aquatic Toxicology Facility (Gainesville, FL) in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals. For initial cloning of the vtgr cDNA, immature LMB (>1 yr old) were euthanized by submersion in a water bath containing 100 parts per million MS-222 (Sigma-Aldrich) and killed by a sharp blow to the head followed by abdominal dissection. The ovaries were excised, immediately flash frozen in liquid nitrogen, and stored at −80°C until the RNA was isolated. For ex vivo experiments, juvenile LMB were obtained from Florida Bass Conservation Center (Webster, FL) in June 2011. Fish were acclimated for 1 mo prior to performing ex vivo experiments. All the fish were fed Silver Cup salmon feed daily. Monitoring of oocyte development was carefully recorded biweekly by dissecting female LMB ovaries. All the juvenile LMB were sacrificed as described above. The majority of the ovarian tissues were used for ex vivo experiments, and a small portion was stored in 10% buffered formalin for hematoxylin and eosin (H&E)-stained sections to confirm the stage of oocyte development. For the temporal study, adult female LMB were collected from St. John's River (Welaka, FL) as described previously [30, 31]. Ovarian tissue from each fish was flash frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Ex Vivo Ovarian Follicle Isolation and Exposures
To perform the ex vivo exposures, ovaries from juvenile female fish were excised, carefully cut into 2 × 2 mm pieces and randomly distributed into each well of a 24-well plate. Tissues were cultured by submersion in 1 ml of Leibovitz L-15 phenol-free medium (GIBCO) plus streptomycin (0.1 g/l; Cellgro) and penicillin (100 000 international units [IU]/l; Cellgro) at 20°C in a humidified 5% CO2 cell incubator for 16 h. For the dose-response studies, ovaries from three fish were exposed to 5, 10, 50, or 500 ng/ml of porcine INS (pINS) (Sigma-Aldrich) dissolved in water for 24 h. For subsequent experiments, we used the highest dose of INS to ensure maximal vtgr expression as determined from the dose-response studies (3.2-fold over control). Ovaries from four fish were collected and exposed to 500 ng/ml of pINS, 500 nM of E2 (Sigma-Aldrich), or 500 nM of 11-KT (Sigma-Aldrich) dissolved in ethanol, or the combination of each sex steroid (500 nM) and pINS (500 ng/ml). Control samples were treated with the appropriate vehicles: water, 0.1% ethanol, and a combination of water and 0.1% ethanol. Exposures continued for 24 h followed by RNA isolation and quantitative real-time PCR (qRT-PCR) analysis.
Isolation of LMB vtgr and Partial ar cDNA Sequences
Total RNA was extracted from immature LMB ovarian tissue that was primarily in previtellogenic stages using Trizol reagent (Invitrogen Life Technologies) coupled with an RNAeasy Mini Kit (Qiagen) and DNase treatment. Reverse transcription was performed using 1 μg of RNA in a 20 μl reaction mix containing, 50 μM random hexamers, 10 mM deoxyribonucleotide triphosphates (dNTPs) (Applied Biosystems), 5× transcription buffer, and 200 U Superscript II enzyme (Superscript First-Strand kit, Invitrogen Life Technologies). For standard PCR, vtgr primers were designed based on the tilapia vtgr cDNA sequence (Oreochromis aureus, AF514281; Table 1), while primers for ar gene were designed based on Atlantic croaker ar cDNA sequence (Micropogonias undulatus, AY701761.1; Table 1). Briefly, a 25 μl PCR reaction volume was prepared as follows: 12.5 μl master mix (Promega), 8.5 μl nuclease-free water, and 1 μl for each primer (10 μM) were combined. Then, 2 μl of cDNA was added to complete the reaction mix. Thermocycler conditions were: 95°C for 5 min, 35 cycles at 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec. The PCR products were electrophoresed on a 1% agarose/ethidium bromide (EtBr) gel, excised and purified with QIAquick Gel Extraction Kit (Qiagen). The purified cDNA product was ligated into the pGEM-T Easy Vector (Promega), transformed into Escherichia coli DH5α cells (Invitrogen), and a minimum of five positive clones from each target gene were purified and sent to the University of Florida ICBR DNA Sequencing Core Facility for sequence identification. The resulting vtgr and ar cDNA sequences were verified using the National Center for Biotechnology Information (NCBI) BLAST database.
TABLE 1. .
Primer sequences used in this study.
RACE, rapid amplification of cDNA ends; qRT-PCR, quantitative real-time PCR.
The full-length of vtgr cDNA sequence was obtained using the commercially available SMART RACE Kit (Clontech) with specific primers designed with the Integrated DNA Technologies Web site according to manufacturer's protocol (Table 1). PCR conditions for RACE (rapid amplification of cDNA ends) started with five cycles at 94°C for 30 sec and 72°C for 3 min, then five cycles at 94°C for 30 sec, 70°C for 30 sec, and 72°C for 3 min, and 25 cycles at 94°C for 30 sec, 68°C for 30 sec, and 72°C for 3 min. For 3′-RACE, a nested PCR reaction was required. PCR conditions for nested 3′-RACE were as follows: 20 cycles at 94°C for 30 sec, 68°C for 30 sec, and finally 72°C for 3 min. Following PCR, 5′-RACE and 3′-RACE products were visualized in 1.2% agarose/EtBr gel, excised, purified, cloned into the pGEM-T Easy Vector and transformed into JM109 competent cells (Promega). At least four positive clones were purified and sent to the Environmental Genomics Core Facility at the University of South Carolina for sequence identification. Sequence verification was performed as previously described.
Real-Time PCR in Ovarian Tissues
The temporal expression of LMB vtgr and ar mRNA were analyzed in gonadal tissues from female LMB during defined stages of oocyte development using real-time PCR. In addition, the expression of vtgr, esr1, esr2a, esr2b, and ar genes were analyzed for the ex vivo exposure experiments. For all the target genes, RNA was isolated from LMB gonadal tissues using RNAeasy Mini Kit (Qiagen) and individually DNase treated prior to cDNA synthesis. Reverse transcription was performed using 0.2 μg RNA, 0.5 μg/μl random primers, 10 mM dNTPs, 4 μl MgCl2, 2 μl 10× transcription buffer, and 15 U avian myeloblastosis virus reverse transcriptase in a 20 μl volume (Promega). Primers for LMB vtgr mRNA were designed using Primer Express v2.0 software (Applied Biosystems). The set of primers employed for LMB esr1, esr2a, esr2b, ar, and 18S rRNA (rn18) were used as previously reported [32–34].
Quantitative RT-PCR was performed using a 7900HT real-time PCR System (Applied Biosystems) with 2 μl of cDNA for target gene or 1 μl for housekeeping gene in a separate reaction volumes of 25 or 24 μl containing 12.5 μl SYBR Green PCR Master Mix (Applied Biosystems), 7 μl or 6 μl nuclease-free water, and 2.25 μl of each primer (10 μM). Real-time PCR conditions were 50°C for 20 sec, 95°C for 10 min, and a final stage of 40 cycles at 95°C for 15 sec and 60°C for 1 min. For the seasonal ar mRNA amplification, Ct values were calculated following the methodology used in a previous study [31]. All vtgr, esr1, esr2a, esr2b, and ar expression levels were normalized to corresponding 18S rRNA values and presented as fold change in expression using the ΔΔCt method. Validation of the 18S rRNA as an appropriate housekeeping gene in LMB gonadal and other tissues has been conducted previously by our group [30, 33, 35] and in other fish species [5, 36–38].
Staging of Oocyte Development
To confirm oocyte stages for the temporal and ex vivo studies, a portion of gonad tissue from each fish was stored in 10% buffered formalin until processed for histological analysis. H&E stained slides were prepared from each gonad sample by Florida Fish and Wildlife Conservation Commission. Briefly, ovarian tissues were embedded in plastic, cut to 4.0-μm sections, and stained with H&E following standard methods (Grier H., unpublished results). These slides were used for the purposes of sex verification and staging of oocyte development in females. To determine the reproductive stage of our histology slides, we followed the terminology, morphology, and physiology described in Grier et al. [39]. Oocytes were classified into the following six stages: stage I, perinucleolar (PN), and stage II, cortical alveoli (CA), are part of the primary growth phase where oocytes are typically being prepared for Vtg uptake. Stage III, early vitellogenesis, and stage IV, late vitellogenesis, are the secondary growth phase where plasma Vtg is actively incorporated. Finally, the maturation stage was divided into stage V, early oocyte maturation, and stage VI, late oocyte maturation, in which oil droplets fused into oil globules, lipoprotein yolk becomes yolk globules, and migration of germinal vesicle are visible.
Phylogenetic Study
Select vertebrate amino acid sequences belonging to the LDLR gene superfamily were retrieved from the GenBank protein database (Table 2). Sequences were aligned using multiple sequence comparison by log-expectation [40]. The alignment is available from the senior author upon request. Phylogenetic hypotheses were constructed using the maximum parsimony and neighbor-joining algorithms. Parsimony trees were constructed in PAUP (version 4.0b10) using heuristic searches [41]; character states were treated as unordered in all the analyses. Neighbor-joining trees [42] were constructed in MEGA (version 4) [43] using Poisson-corrected distances and the pairwise deletion option for missing data. Bootstrapping (1000 pseudoreplicates) was used to gauge support for nodes of interest in both the parsimony and neighbor-joining analyses [44].
TABLE 2. .
Selected protein sequences retrieved from GenBank to construct phylogenetic tree.
Predicted protein: similar to putative pheromone receptor CPpr8.
Statistical differences among mean Ct values of vtgr and ar transcript levels across ovarian stages for the temporal study and mean Ct values of vtgr, ar, esr1, esr2a, and esr2b transcript levels for hormonal treatments were compared by one-way ANOVA with Tukey multiple comparison test after testing for normality and variance homogeneity. The null hypothesis was rejected at P < 0.05. SigmaPlot version 12.0 (Systat Software Inc.) software for Windows was used for all the statistical analysis.
RESULTS
Cloning of LMB vtgr and ar cDNAs
The coding region for LMB vtgr was obtained from ovarian tissue using standard PCR and RACE technologies. Multiple rounds of PCR and RACE were performed resulting in the complete 3768 bp LMB vtgr sequence (GenBank HQ32624), which is represented graphically in Figure 1, A and B. The putative transcriptional start site is located 71 bp upstream of the first encoded methionine (translational start site) representing the 5′ untranslated region (UTR). The 3′ UTR consists of the 1169 bp sequence downstream from the coding region (2600 to 3768). Translation of the 2529 bp open reading frame (ORF) using the Translate Tool Web site service (Swiss Institute of Bioinformatics) revealed a protein sequence consisting of 843 amino acids with a predicted molecular mass of 92.62 kDa. The following conserved domains were identified using the Protein Blast-specialized service at NCBI: eight cysteine-rich ligand binding domains (LBDs), three epidermal growth factor (EGF) repeats, five propeller domains (YWTDs), one transmembrane domain (T), and one cytoplasmic domain (C). Arrangement of these domains is shown in Figure 1C. These characteristics place the LMB vtgr cDNA sequence in the LDLR gene superfamily.
FIG. 1. .
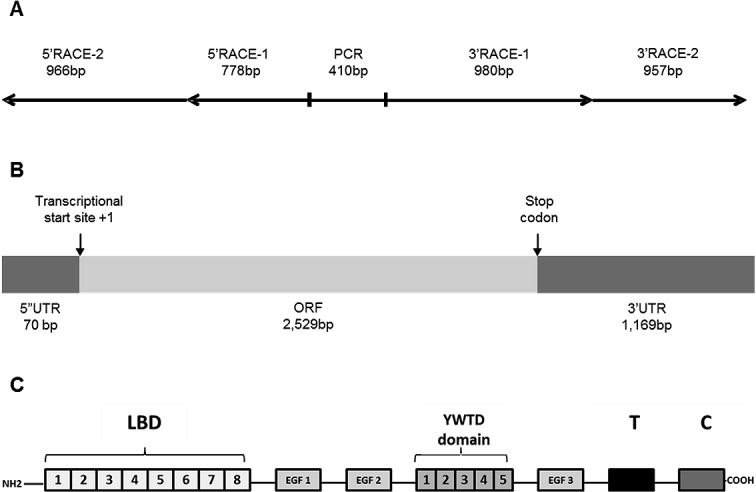
Illustration of LMB vtgr cDNA and its predicted protein domains. A) Fragments obtained by PCR and RACE. B) Open reading frame (ORF) and 5′ and 3′ untranslated regions (UTR). C) Amino acid predicted domains: (LBD) ligand binding domain, (EGF) epidermal growth factor-like, propeller domains (YWTD), transmembrane (T) domain, and cytoplasmic (C) domain.
Analysis using the NCBI Translated BLAST (BLASTX) database revealed the putative LMB Vtgr protein shares amino acid identity with Vtgr sequences from white perch (96%), blue tilapia (94%), medaka (91%), and Senegalese sole (88%). In Figure 2, the amino acid sequence alignment shows a comparison of LMB Vtgr, white perch Vtgr, and two sole sequences, a short Vldlr sequence missing the O-glycan domain and a long Vldlr variant containing this domain (the alignment was constructed using Multalin Web service) [45]. These results demonstrate the specific lack of the O-linked sugar domain in the ovarian LMB Vtgr.
FIG. 2. .
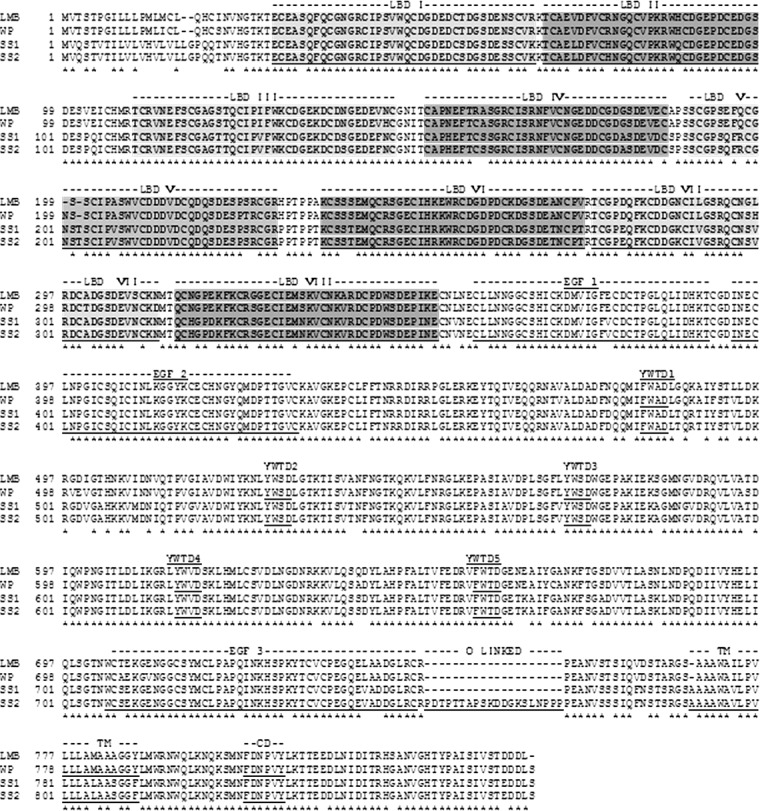
Alignment of predicted amino acids for largemouth bass (LMB) Vtgr, white perch (WP) Vtgr, and two sole Vldlr isoforms, SS1 (short) and SS2 (long). The O-linked domain is doubled underlined in SS2. Ligand-binding domain composed of eight cysteine-rich repeats (LBDs), three epidermal growth factor-like domains (EGF 1, 2, and 3), five YWTD propeller domains, transmembrane domain (TM), and cytoplasmic domain (CD) are underlined. Asterisks indicate identical amino acids among all the sequences.
The partial 699-bp LMB ar cDNA sequence contains an incomplete DNA-binding domain, hinge domain, and a partial LBD. Translation and analysis of this cDNA sequence using BLASTX revealed high amino acid identity with other teleost ar sequences: Large yellow croaker (91%), Atlantic croaker (91%), European seabass (90%), and Orange-spotted grouper (90%). We could not determine whether our LMB ar cDNA sequence was either ar1 or ar2 as the identified domains are very similar for both subtypes. Therefore, ar primers designed for qRT-PCR from this sequence likely measured mRNA expression for both LMB ar subtypes. We are currently attempting to extend this sequence to better distinguish between ar isoforms.
Phylogenetic Analysis
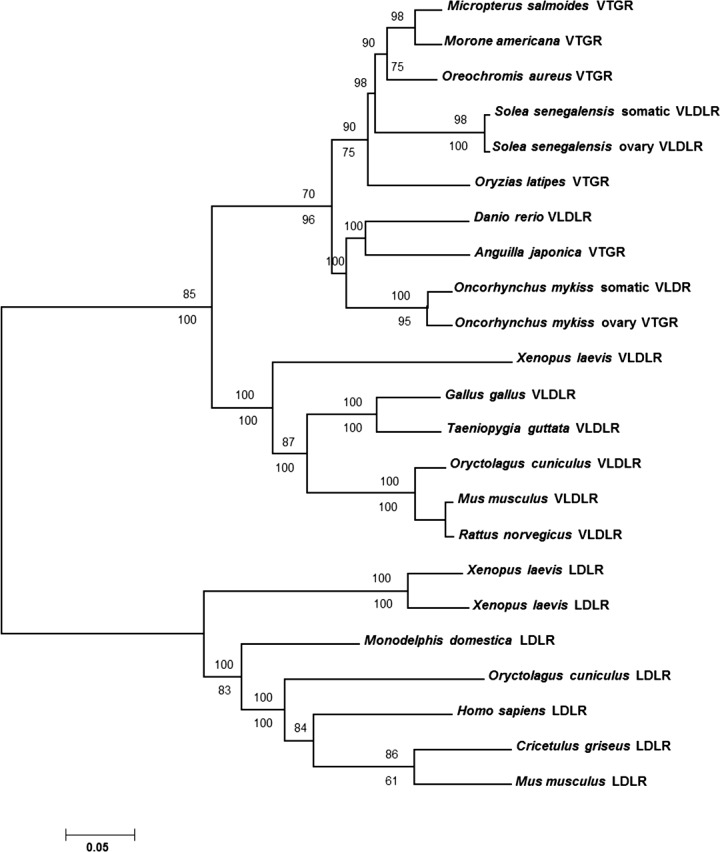
Analyses using the neighbor-joining algorithm yielded a single topology that included several well-supported clades, including a monophyletic group of Vtgr sequences from fishes that were sisters to a grouping that included VLDLR proteins from tetrapod vertebrates (Fig. 3). The LMB sequence was included within the fish Vtgr clade as a sister to a Vtgr sequence sampled from white perch (Morone americana). This clade was supported strongly by bootstrap analysis (98%). A nontraditional pairing of eel Vtgr and zebrafish Vldlr sequences was recovered and strongly supported by bootstrap analysis, although this grouping was not well-supported under the criterion of maximum parsimony (see below).
FIG. 3. .
Phylogenetic tree of vertebrate LDLRs showing LMB Vtgr position. Bootstrap values based on 1000 pseudoreplicates are shown on the nodes. The values above the nodes are for neighbor joining, and the values below the nodes are for maximum parsimony analysis.
Searches under the criterion of maximum parsimony recovered eight equally parsimonious trees of length 2086 steps (consistency index = 0.822). Differences among these eight trees were found primarily within the fish Vtgr clade and involved the position of the zebrafish and eel sequence, placement of the tilapia and medaka sequences, and the position of the rabbit LDLR sequence (not shown). The nontraditional grouping of eel and zebrafish was recovered in four of eight equally parsimonious trees as in the neighbor-joining analyses, but this grouping was found in only 33% of 1000 bootstrap pseudoreplicates. As in the neighbor-joining analysis, the LMB sequence was recovered as a sister to the white perch Vtgr, and this grouping received moderate support (75%). The two fish sequences containing sugar domains were recovered as sister to their respective sugar domain-deficient proteins, and both groupings received high bootstrap support.
LMB vtgr and ar Temporal Expression Throughout Reproduction
To determine the temporal patterns of vtgr and ar expression throughout a discrete reproductive season of LMB, mRNA transcript levels of these genes were measured in female ovarian tissues by real-time PCR. Ovarian tissues utilized in the study were classified into specific stages of oocyte development (I–VI) as determined by histological analysis. Each tissue sample was microscopically examined and classified into stages based on distinct morphological criteria observed on H&E-stained sections as shown in Figure 4. During primary growth, stage I PN follicles start to form, oocytes begin to expand, and the appearance of CA defines progression to stage II. In stages III (early vitellogenesis) and IV (late vitellogenesis), uptake of plasma Vtg and other lipids is evident by the presence of the red-stained ooplasmic inclusions. The follicle size also increases in conjunction with Vtg uptake in stage IV, and the germinal vesicle is centrally located. Finally, the oocyte maturation stages V (early) and VI (late) are distinguished by the presence of oil globules and migration of the central germinal vesicle.
FIG. 4. .
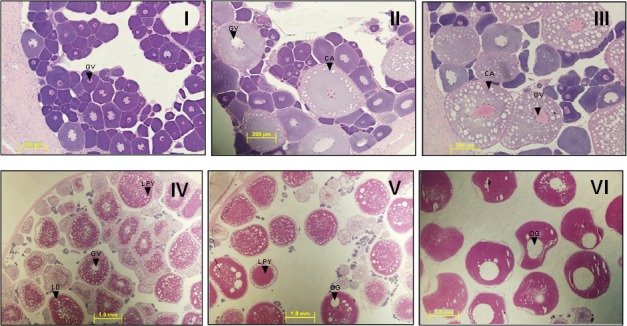
Ovarian follicle development in LMB. Stages I to VI are as follows: (I) primary growth, (II) cortical alveoli, (III) early vitellogenesis, (IV) late vitellogenesis, (V) early oocyte maturation, and (VI) late oocyte maturation. Abbreviations are cortical alveoli (CA), germinal vesicle (GV), lipid droplet (LD), lipoprotein yolk (LY), and oil globule (OG). Bars = 200 μm (I, II, and III), and 1.0 mm (IV, V, and VI).
Analysis of the temporal expression of LMB vtgr revealed that the highest transcript levels were present during previtellogenic growth, primarily during stages I and II, carrying a 28- and 20-fold increase in expression, respectively, as compared to the lowest measured value for all the samples that occurred during stage IV (Fig. 5A). As oocyte development ensues, vtgr expression declines in subsequent stages (III, IV, V, and VI), presenting a low abundance of transcripts during Vtg uptake and maturation processes. These levels are relatively constant as values are not statistically significant among the later stages. Similarly, the greatest expression of ar occurred during primary growth stages I and II, reaching levels approximately 10- and 12-fold, respectively, compared to the lowest values measured (stage V) (Fig. 5B).
FIG. 5. .
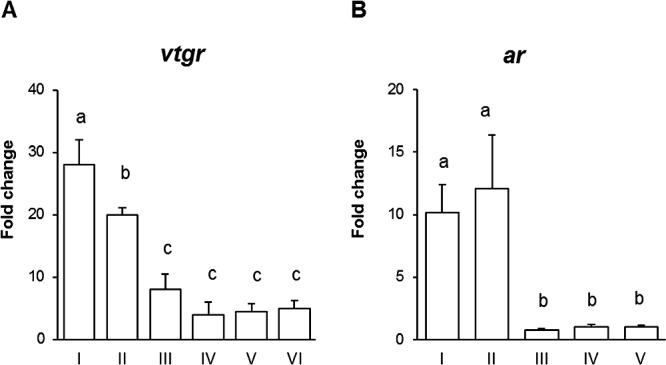
Temporal expression of LMB vtgr (A) and ar (B) during oocyte development. Open bars represent expression levels detected by real-time PCR compared to the lowest measured value for all the samples (stage IV). Data is plotted as mean ± SEM (n = 6). Ovarian stages are perinucleolar (I), cortical alveoli (II), early vitellogenesis (III), late vitellogenesis (IV), early oocyte maturation (V), and late oocyte maturation (VI). Significant differences (P < 0.05) among stages are represented by letters.
Effects of Hormones on LMB vtgr mRNA Expression in Ovarian Tissue
To gain a better understanding regarding hormonal signals that control vtgr transcript levels, LMB previtellogenic ovarian tissues were exposed to pINS and the sex steroids 11-KT and E2. The results of these experiments show that both 50 and 500 ng/ml of pINS treatment for 24 h elicited a positive effect on vtgr expression, resulting in 3.0- and 3.2-fold increases compared to control unexposed tissues (Fig. 6, A and B). In contrast, exposure of the ovarian tissues to either E2 or 11-KT (500 nM) alone did not alter vtgr mRNA expression (Fig. 6B). However, combination treatments of each sex steroid with the highest dose of pINS (500 ng/ml) resulted in a significant decrease in vtgr mRNA levels when compared to the response elicited by pINS treatment alone. Specifically, the mixture of pINS with either 11-KT or E2 resulted in approximately a 40% decrease in mRNA levels compared to pINS. These results suggest a role for both INS and sex steroids in modulation of LMB vtgr transcript levels in ovarian tissues. Gonadal stage of LMB ovaries is illustrated by representative micrographs of histological sections (Fig. 6C). The presence of nucleoli around the inner membrane of the germinal vesicle confirmed that ovarian tissues used in our ex vivo experiment were primarily in stage I.
FIG. 6. .
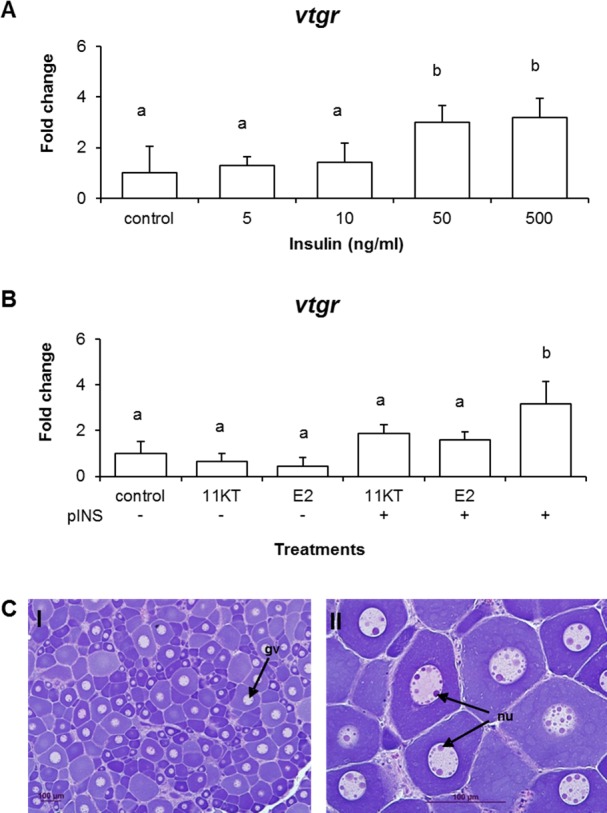
LMB vtgr expression in previtellogenic oocytes after hormonal treatments. A) Dose response effects of pINS on vtgr expression. Data is plotted as mean ± SEM (n = 3). B) Effects of a single hormone or hormone combination treatment on vtgr expression. Open bars represent expression levels detected by real-time PCR. Data is plotted as mean ± SEM (n = 4). C) Histology sections stained with H&E showing LMB previtellogenic oocytes from ex vivo exposure. I) Oocytes are mostly in perinucleolar stage (PN). II) Section showing nucleoli (nu) around the inner germinal vesicle membrane of the germinal vesicle (gv). Significant differences (P < 0.05) among treatments are represented by letters. Treatments: 11-KT (500 μM), E2 (500 μM), pINS (500 ng/ml), pINS (500 ng/ml) + 11KT (500 μM), pINS (500 ng/ml) + E2 (500 μM). Bars = 100 μm.
Effects of Hormones on LMB esr1, esr2a, esr2b, and ar mRNA Expression in Ovarian Tissues
As nuclear receptors are activated by estrogens and androgens to drive downstream gene transcriptional events, we measured the effect of E2, 11-KT, and pINS on the three esr and ar in ovarian follicles. Individual treatments of E2, 11-KT, and pINS did not modulate mRNA levels of esr1, esr2a, esr2b, or ar (Fig. 7A–D) in tissues exposed for 24 h, but the combined treatment of pINS and E2 increased expression of esr2b by 2.6-fold and pINS and 11-KT increased ar mRNA levels 2.3-fold compared to control cells, respectively.
FIG. 7. .
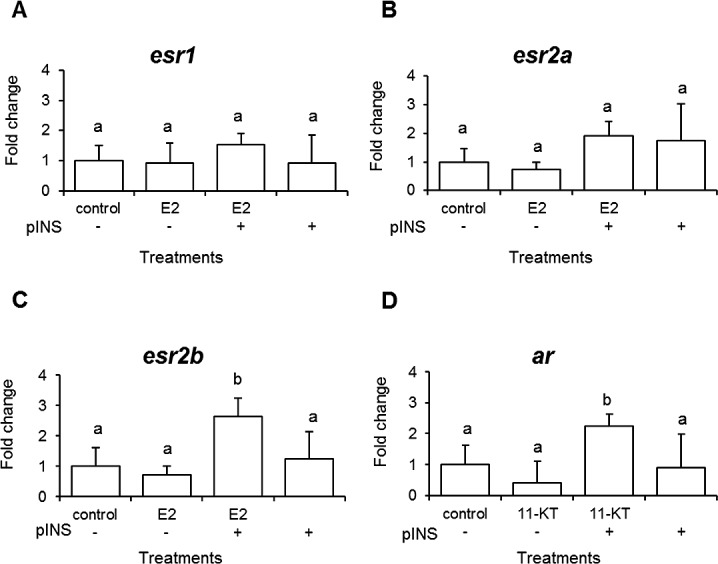
Expression of LMB steroid receptors in previtellogenic oocytes after hormonal treatments (A–D). Open bars represent expression levels detected by real-time PCR. Data is plotted as mean ± SEM (n = 4). Significant differences (P < 0.05) among treatments are represented by letters. Treatments 11-KT (500 μM), E2 (500 μM), pINS (500 ng/ml), pINS (500 ng/ml) + 11KT (500 μM), pINS (500 ng/ml) + E2 (500 μM).
DISCUSSION
The uptake of Vtg into developing oocytes is an essential event necessary for successful reproduction, as it is processed into yolk, which provides a valuable source of nutrients for growing embryos postfertilization. The receptor-mediated incorporation of Vtg and other lipoproteins into developing oocytes occurs via Vtgr [4]. The process of hepatic vitellogenesis is fairly well understood where it is primarily under the control of E2 through activation of cognate nuclear receptors; however, much less is known about the molecular control of vtgr expression, an equally vital event that contributes to the reproductive process. In LMB, we have previously characterized the temporal expression of vtg, esr1, esr2a, and esr2b mRNA levels in various tissues (liver and ovary) and the circulating plasma levels of vitellogenin and steroids (E2, T, and 11-KT) [34, 46]. We determined that all three esr isoforms are highly expressed in ovarian tissues during previtellogenic stages of oocyte development (I and II), particularly esr2a and esr2b, which decline as plasma levels of the steroids begin to rise. The greatest concentrations of E2 occur in concert with hepatic vitellogenin production and uptake of vitellogenin into oocytes stages (III and VI). To begin to characterize vtgr transcriptional expression and regulation in LMB, we successfully cloned the full length of LMB vtgr cDNA sequence, revealing an open-reading frame consistent with characteristics of lipoprotein receptors that belong to the LDLR gene superfamily, including eight cysteine-rich LBDs, three EGF domains, five YWTDs, a TM, and a CD. Being a member of the LDLR family of cell-surface receptors, many of these domains are essential for extra- and intracellular regulation of circulating lipoproteins. For example, the LBD repeats mediate the interaction between receptor and lipoproteins in mammals [47, 48]. In tilapia, it has been reported that the first three ligand-binding repeats of Vtgr interact with the amino-terminal region of Vtg [7]. In general, Vtgr has eight LBD repeats (LR8), one additional repeat than in human LDLRs; however, vtgr cDNAs from fish are more highly similar in structure to mammalian VLDLRs [49], which contain eight LBD repeats. It has been suggested that VLDLR may function as an intermediate density lipoprotein receptor with unique ligand specificity in peripheral tissues in concert with lipoprotein lipase in mammals [48].
Investigation of other piscine ovarian Vtgr, including rainbow trout [6], blue tilapia [7], and sole [8], reveal two Vtgr splice variants, one that contains an O-linked sugar domain and one that lacks this region. Classification of these variants was created based on structural features rather than ligand specificity [5] taking as a model the chicken Vtgr that has two similar Vldlr variants [50]. Vtgr that contain the O-linked sugar domain has been termed LR8+, and sequences lacking this domain are named LR8−. In rainbow trout, blue tilapia, and sole, both of these isoforms have been reported, and here we report the LMB sequence in its LR8− form based on similar criteria.
The functional difference between these variants may be driven by tissue-specific localization. The dominant type in ovarian tissue is the shorter isoform lacking the O-linked sugar region (LR8−). In cell-based studies, it is reported that the larger form (LR8+) containing the O-linked sugar domain controls the incorporation of VLDL in somatic tissues whereas the loss of the O-linked domain confers stable expression of the VLDLR on the cell surface [10]. These observations imply that the two forms have tissue-selective expression that may relate to the preference for certain ligands. After scrutinizing many bacterial colonies containing vtgr cDNA sequences, we could not identify a nucleotide region that represents the O-linked sugar domain. It is possible that the LR8+ form is minimally expressed in LMB ovarian tissue or that expression occurs in this tissue during discrete timeframes that were not analyzed in this study.
Phylogenetic analyses of select vertebrate LDLR cell surface proteins placed LMB Vtgr into a monophyletic group of fish Vtgr, which also include Vldlr proteins. The phylogenetic tree rendered two different groups, one that contains vertebrate LDLRs and a second that contains vertebrate VLDLRs. The VLDLR cluster splits into two significant internal subgroups: Vldlr and Vtgr. This suggests that these proteins share a common gene ancestor. Based on strong bootstrapping analysis (98%), LMB Vtgr was included in a clade as sister to white perch Vtgr; these proteins share 96% amino acid sequence identity. As expected, these two proteins formed a cluster with other fish Vtgr lacking the O-linked domain. Fish species that reported two Vtgr isoforms, rainbow trout and sole (100% bootstrap value), formed respective sister clades containing both sequences, suggesting that these protein isoforms arose independently. In addition, neighbor-joining and maximum parsimony analysis recovered a nontraditional pairing of zebrafish Vldlr (LR8+) with Japanese eel Vtgr (LR8−), although this clade was not strongly supported by bootstrap analyses. Both proteins were reported in different fish tissues: kidney in zebrafish and ovary in Japanese eel. However, it seems that they appeared to share a close relationship that may be explained by the fact that both species are included in older teleost orders.
The study of the temporal expression of vtgr throughout the reproductive cycle showed maximal expression in the ovary prior to hepatic vtg mRNA synthesis and uptake of Vtg into oocytes. Since we did not identify an LR8+ isoform from LMB tissues, we hypothesize that vtgr expression in the LMB ovary is predominantly the LR8− form. It is not surprising that Vtgr is produced prior to Vtg synthesis because the oocytes need to be fully prepared to accept and traffic Vtg internally. Similar observations have recently been reported in a handful of other teleost fish species [5, 6, 8, 24, 25]. A current theory has been proposed to explain the early synthesis of Vtgr, which proposes that recycling of the receptor is essential to controlling the expression and function of this receptor [6]. The mechanism of Vtg internalization by Vtgr likely occurs by clathrin-mediated endocytosis through an organized set of intermediate subreactions in endosomes [51], and it is in these compartments that cleavage of Vtg from the Vtgr occurs followed by shuttling of the receptor back to the plasma membrane. The conserved timing of expression in early developing follicles in multiple fish species implies that molecular control of vtgr expression and cycling is likely conserved.
The molecular signals that control ovarian expression of vtgr mRNA during reproduction have not been elucidated. Data available to date have been derived from mosquitoes (Aedes aegypti) revealing that vtgr expression is highly correlated with ecdysteroid concentrations [52, 53]. Ecdysone receptors (EcRs) are steroid receptors that regulate the growth, molting, and reproduction processes in arthropods. Although controversial, it is postulated that ERs and EcRs evolved from a common steroid ancestral receptor and perform redundant functions in arthropods and vertebrates, respectively [54, 55]. We previously observed that patterns of expression for LMB esr2a and esr2b [34] share a similar trend to vtgr expression during the reproductive cycle in the ovary with maximal expression in previtellogenic tissues. Because both estrogens and androgens have been suggested to contribute to primary growth of follicles in other teleosts [22, 23, 56], we also cloned the ar cDNA and determined its temporal expression profile. We reveal here for the first time that the mRNA levels of LMB ar follow the same trend as esr and vtgr. In female LMB, esr and ar transcript levels decrease after oocyte primary growth (stages I and II), while plasma concentrations of E2, testosterone (T), and 11-KT begin to rise and peak during vitellogenic stages (III and IV), which coincides with hepatic vitellogenin production and uptake by oocytes [34, 46].
As E2 and 11-KT bind and activate their corresponding receptors, it is intuitive that localized production of steroids in ovarian tissues is modulating receptor activity during early oocyte growth when receptor levels are maximal. Although the functional role of nuclear hormone receptors in the teleost ovaries is not well characterized, a few studies have investigated the contribution of estrogens and androgens during primary oocyte growth and showed that sex steroids affect sex differentiation, cell growth, and gene expression changes related to steroidogenesis [22, 23, 56–59]. Here we set out to determine whether E2 or 11-KT could modulate the transcriptional levels of vtgr in early growth stage follicles. In these studies, neither E2 nor 11-KT significantly altered vtgr expression levels compared to control unexposed follicles (Fig. 6B). Although we are unaware of any studies that have assessed expression of vtgr in isolated follicles after hormone treatment, a recent study found that aqueous exposure of medaka to E2 caused a decrease in female vtgr mRNA levels in concert with increased esr2b mRNA expression in gonad tissues [29]. We failed to see modulation of expression for any esr subtypes by E2 nor did we see changes in ar expression following the 11-KT treatment. However, we did observed INS-induced levels of vtgr mRNA were decreased by E2 following exposure of follicles to both agents. The levels of esr2b also increased what was observed in E2-exposed medaka. Because INS concentrations was not measured in this study, it is difficult to determine whether endogenous levels contributed to repression of vtgr by E2.
In addition to sex steroids, INS has been suggested to stimulate the incorporation of Vtg in oocytes of rainbow trout at doses ≥50 ng/ml [20] and contribute to oocyte growth, which led us to investigate the role of INS in vtgr expression. A novel observation of our study was the increase of vtgr expression in previtellogenic oocytes in response to pINS (Fig. 6A) where the highest dose tested (500 ng/ml) resulted in a 3.2-fold increase over controls. Minimal information exists describing normal seasonal plasma levels of INS in teleosts, and few studies have revealed that circulating levels fluctuate during the reproductive cycle, presenting the highest concentrations prior to vitellogenesis [60]. The precise functions of INS during these time frames, particularly during primary ovarian growth, have been described by measuring DNA synthesis and binding receptor studies [61, 62]. INS and INS-like growth factor-I (IGF-I) have been shown to bind to fish ovaries primarily in previtellogenic oocytes of carp [62], and recombinant human IGF-I increased the diameter of shortfinned eel previtellogenic oocytes [22]. These studies support a role for INS in directing primary growth events in developing oocytes, and our data further suggest a specific role for controlling vtgr mRNA production during this window.
Surprisingly, both E2 and 11-KT had a suppressive effect on INS-induced vtgr transcript levels, decreasing the levels by approximately 40%. This observation supports the notion that sex steroids play a role in regulating the vtgr gene either by directly repressing transcription at the level of promoter or by altering mRNA degradation pathways. Crosstalk between ERs and INS-signaling systems has been well accepted by the mammalian communities and, although not as well studied, a few studies support this notion in fish. For example, treatment of tilapia with a strong estrogen 17α-ethinylestradiol (EE2) significantly elevated igf-1 expression after 100 days posthatching, implying estrogens interact with the autocrine/paracrine part of INS growth factor-driven networks [63]. The upregulation of esr2b and ar transcript levels by the combination of pINS and E2 or pINS and 11-KT, but not by either agent alone, implies a complex signaling network that involves hormone- and INS-mediated receptors. Our results are consistent with the decrease in vtgr mRNA in medaka exposed to E2 and further support a requirement for INS to participate in the modulation of vtgr during previtellogenic oocyte stages. It is possible that as transcription factors, esr2b and/or ar interact directly with the vtgr promoter in repressing expression or by modulating vtgr mRNA levels through more indirect means such as enhancing transcript turnover. Our results set the stage for more in-depth molecular control studies to elucidate transcriptional control of vtgr by INS and sex steroid-signaling networks.
It is surprising that both E2 and 11-KT elicited similar effects because they are both final products of steroid metabolism that interact specifically with their cognate receptors. Although a number of biotansformation enzymes may be modulated at the level of transcription or activity, conversion of 11-KT to E2 cannot occur, which is the case for T conversion by aromatase. It is inherently possible that T could be converted to 11-KT, which would enhance the AR-driven response. Recently, clusters of steroid-producing cells in the tunica of fish ovary have been detected as the site for biosynthesis of 11-KT [64] in support of this possibility. In addition, crosstalk between the AR and ER pathways has been suggested, through soluble and membrane receptors as a mechanism for dual involvement of both steroids. VLDL and lipid incorporation, as measured by oocyte diameter increase, is the only related process that has been shown to be under the control of 11-KT, and it is possible that it dually regulates repression of vtgr mRNA while enhancing receptor cycling and function. The addition of IGF-I combined with 11-KT enhanced oocyte growth, which additionally suggests that multiple factors likely control these processes [22, 23].
In conclusion, our study identifies and characterizes the temporal expression of vtgr and ar genes, and presents evidence for reproductive stage-specific expression in female LMB. Furthermore, this is the first report revealing INS as a key hormone involved in the transcriptional upregulation of the vtgr gene in previtellogenic oocytes and decrease in INS-induced mRNA levels by E2 and 11-KT. These data expand our current knowledge of teleost reproductive biology and highlights the potential interplay between metabolic and sex steroid-regulated pathways. To achieve a more complete grasp of oocyte yolk sequestration in teleosts and the molecular mechanisms controlling this process, it is necessary to gain a systematic understanding of the factors that control Vtgr production and activity, including signaling crosstalk, which is the focus of our current and future research efforts.
ACKNOWLEDGMENT
The authors thank employees of the Florida Wildlife Research Institute (FWRI) for sampling fish from the St. John's River, particularly G. Delpizzo, B. Thompson, and H. Smith. They also want to thank N. Perry, T. Piacenza, and Y. Waters of the FWRI for completing the histological procedures. We thank Florida Bass Conservation Center for providing the LMB used for the ex vivo experiment.
Footnotes
1Supported in part by the University of Florida and South Carolina Research Foundations and the National Institute of Environmental Health Sciences RO1 ES015449 to N.D.D.
REFERENCES
- Walllace RA, Bergink EW. Amphibian vitellogenin: properties, hormonal regulation of hepatic synthesis and ovarian uptake, and conversion to yolk proteins. Am Zool 1974; 14: 1159 1175 [Google Scholar]
- Busson-Mallibot S. Endosomes transfer yolk proteins to lysosomes in the vitellogenic oocyte of the trout. Biol Cell 1984; 51: 53 66 [DOI] [PubMed] [Google Scholar]
- Prat F, Coward K, Sumpter J, Tyler C. Molecular characterization and expression of two ovarian lipoprotein receptor in the rainbow trout, Oncorhynchus mykiss. J Biol Reprod 1998; 58: 1146 1153 [DOI] [PubMed] [Google Scholar]
- Hiramatsu N, Hara A, Matsubara T, Hiramatsu K, Sullivan CV. Oocyte growth in temperate basses: multiple forms of vitellogenin and their receptor. Fish Physiol Biochem 2003; 28: 301 303 [Google Scholar]
- Hiramatsu N, Chapman RW, Lindzey JK, Haynes MR, Sullivan CV. Molecular characterization and expression of vitellogenin receptor from white perch (Morone americana). Biol Reprod 2004; 70: 1720 1730 [DOI] [PubMed] [Google Scholar]
- Davail B, Pakdel F, Bujo H, Perazzolo LM, Waclawek M, Schneider WJ, Le Menn F. Evolution of oogenesis: the receptor for vitellogenin from the rainbow trout. J Lipid Res 1998; 39: 1929 1937 [PubMed] [Google Scholar]
- Li A, Sadasivam M, Ding JL. Receptor-ligand interaction between vitellogenin receptor (VtgR) and vitellogenin (Vtg), implications on low density lipoprotein receptor and apolipoprotein B/E. J Biol Chem 2003; 278: 2799 2806 [DOI] [PubMed] [Google Scholar]
- Agulleiro MJ, André M, Morais S, Cerdà J, Babin PJ. High transcript level of fatty acid-binding protein 11 but not of very low-density lipoprotein receptor is correlated to ovarian follicle atresia in a teleost fish (Solea senegalensis). Biol Reprod 2007; 77: 504 516 [DOI] [PubMed] [Google Scholar]
- Bujo H, Lindstedt KA, Hermann M, Dalmau LM, Nimpf J, Schneider WJ. Chicken oocytes and somatic cells express different splice variants of a multifunctional receptor. J Biol Chem 1995; 270: 23546 23551 [DOI] [PubMed] [Google Scholar]
- Magrané J, Casaroli-Marano RP, Reina M, Gåfvels M, Vilaró S. The role of O-linked sugars in determining the very low density lipoprotein receptor stability or release from the cell. FEBS Lett 1999; 451: 56 62 [DOI] [PubMed] [Google Scholar]
- Nykjaer A, Willnow TE. The low-density lipoprotein receptor gene family: a cellular Swiss army knife? Trends Cell Biol 2002; 12: 273 280 [DOI] [PubMed] [Google Scholar]
- Weber L, Boll M, Stampfl A. Maintaining cholesterol homeostasis: sterol regulatory element-binding proteins. World J Gastroenterol 2004; 10: 3081 3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifani S, Menn FL, Rodriguez JN, Schneider WJ. Regulation of oogenesis: the piscine receptor for vitellogenin. Biochim Biophys Acta 1990; 1045: 271 279 [DOI] [PubMed] [Google Scholar]
- Chan SL, Tan CH, Pang MK, Lam TJ. Vitellogenin purification and development of assay for vitellogenin receptor in oocyte membranes of the tilapia (Oreochromis niloticus, linnaeus 1766). J Exp Zool 1991; 257: 96 109 [Google Scholar]
- Mañanos EL. Núñez Rodríguez J, Le Menn F, Zanuy S, Carrillo M. Identification of vitellogenin receptors in the ovary of a teleost fish, the Mediterranean sea bass (Dicentrarchus labrax). Reprod Nutr Dev 1997; 37: 51 61 [DOI] [PubMed] [Google Scholar]
- Reading BJ, Hiramatsu N, Sullivan CV. Disparate binding of three types of vitellogenin to multiple forms of vitellogenin receptor in white perch. Biol Reprod 2011; 84: 392 399 [DOI] [PubMed] [Google Scholar]
- Kanungo J, Petrino TR, Wallace RA. Oogenesis in Fundulus heteroclitus. VI. Establishment and verification of conditions for vitellogenin incorporation by oocytes in vitro. J Exp Zool 1990; 254: 313 321 [DOI] [PubMed] [Google Scholar]
- Tyler CR, Sumpter JP, Bromage NR. An in vitro culture system for studying vitellogenin uptake into ovarian follicles of the rainbow trout, Salmo gairdneri. J Exp Zool 1990; 255: 216 231 [Google Scholar]
- Tyler CR, Sumpter JP, Kawauchi H, Swanson P. Involvement of gonadotropin in the uptake of vitellogenin into vitellogenic oocytes of the rainbow trout, Oncorhynchus mykiss. Gen Comp Endocrinol 1991; 84: 291 299 [DOI] [PubMed] [Google Scholar]
- Shibata N, Yoshikuni M, Nagahama Y. Vitellogenin incorporation into oocytes of rainbow trout, Oncorhynchus mykiss, in vitro: effect of hormones on denuded oocytes. Dev Growth Differ 1993; 35: 115 121 [DOI] [PubMed] [Google Scholar]
- Kayaba T, Sasaki N, Adachi S, Yamauchi K. Effects of pituitary glycoprotein hormones and thyroid hormones on in-vitro vitellogenin incorporation into organ-cultured oocytes in the Japanese eel, Anguilla japonica. Zool Sci 2008; 25: 334 343 [DOI] [PubMed] [Google Scholar]
- Lokman PM, George KAN, Divers SL, Algie M, Young G. 11-Ketotestosterone and IGF-I increase the size of previtellogenic oocytes from shortfinned eel, Anguilla australis, in vitro. Reproduction 2007; 133: 955 967 [DOI] [PubMed] [Google Scholar]
- Endo T, Todo T, Lokman PM, Kudo H, Ijiri S, Adachi S, Yamauchi K. Androgens and very low density lipoprotein are essential for the growth of previtellogenic oocytes from Japanese eel, Anguilla japonica, in vitro. Biol Reprod 2011; 84: 816 825 [DOI] [PubMed] [Google Scholar]
- Perazzolo LM, Coward K, Davail B, Normand E, Tyler CR, Pakdel F, Schneider WJ, Le Menn F. Expression and localization of messenger ribonucleic acid for the vitellogenin receptor in ovarian follicles throughout oogenesis in the rainbow trout, Oncorhynchus mykiss. Biol Reprod 1999; 60: 1057 1068 [DOI] [PubMed] [Google Scholar]
- Luckenbach J, Iliev D, Goetz F, Swanson P. Identification of differentially expressed ovarian genes during primary and early secondary oocyte growth in coho salmon, Oncorhynchus kisutch. Reprod Biol Endocrinol 2008; 6: 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RA, Selman K. Ultrastructural aspects of oogenesis and oocyte growth in fish and amphibians. J Electron Microsc Tech 1990; 16: 175 201 [DOI] [PubMed] [Google Scholar]
- Song JL, Wessel GM. How to make an egg: transcriptional regulation in oocytes. Differentiation 2005; 73: 1 17 [DOI] [PubMed] [Google Scholar]
- Cho K-H, Cheon H-M, Kokoza V, Raikhel AS. Regulatory region of the vitellogenin receptor gene sufficient for high-level, germ line cell-specific ovarian expression in transgenic Aedes aegypti mosquitoes. Insect Biochem Mol Biol 2006; 36: 273 281 [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Katsu Y, Zhou LY, Miyagawa S, Nagahama Y, Iguchi T. Estrogen receptors in medaka (Oryzias latipes) and estrogenic environmental contaminants: an in vitro-in vivo correlation. J Steroid Biochem Mol Biol 2011; 123: 115 121 [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Kroll KJ, Porak WF, Steward C, Grier HJ, Denslow ND. Seasonal relationship between gonadotropin, growth hormone, and estrogen receptor mRNA expression in the pituitary gland of largemouth bass. Gen Comp Endocrinol 2009; 163: 306 317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doperalski NJ, Martyniuk CJ, Prucha MS, Kroll KJ, Denslow ND, Barber DS. Cloning and expression of the translocator protein (18 kDa), voltage-dependent anion channel, and diazepam binding inhibitor in the gonad of largemouth bass (Micropterus salmoides) across the reproductive cycle. Gen Comp Endocrinol 2011; 173: 86 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N, Barber DS, Gross TS, Johnson KG, Sepúlveda MS, Szabo NJ, Denslow ND. Dietary exposure of largemouth bass to OCPs changes expression of genes important for reproduction. Aquat Toxicol 2006; 78: 358 369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum JL, Nyagode BA, James MO, Denslow ND. Effects of the pesticide methoxychlor on gene expression in the liver and testes of the male largemouth bass (Micropterus salmoides). Aquat Toxicol 2008; 86: 459 469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo-Attwood T, Kroll KJ, Denslow ND. Differential expression of largemouth bass (Micropterus salmoides) estrogen receptor isotypes alpha, beta, and gamma by estradiol. Mol Cell Endocrinol 2004; 218: 107 118 [DOI] [PubMed] [Google Scholar]
- Kocerha J, Prucha MS, Kroll KJ, Steinhilber D, Denslow N. Regulation of steroidogenic acute regulatory protein transcription in largemouth bass by orphan nuclear receptor signaling pathways. Endocrinology 2010; 151: 341 349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hu J. Development and validation of endogenous reference genes for expression profiling of medaka (Oryzias latipes) exposed to endocrine disrupting chemicals by quantitative real-time RT-PCR. Toxicol Sci 2007; 95: 356 368 [DOI] [PubMed] [Google Scholar]
- McCurley A, Callard G. Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Biol 2008; 9: 102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filby A, Tyler C. Appropriate ‘housekeeping' genes for use in expression profiling the effects of environmental estrogens in fish. BMC Mol Biol 2007; 8: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier H, Uribe M, Patino R. The ovary, folliculogenesis, and oogenesis in teleosts : Jamieson B. (ed.), Reproductive Biology and Phylogeny of Fishes (Agnathans and Bony Fishes): Phylogeny, Reproductive System, Viviparity, Spermatozoa, Vol. 8a. Enfield, NH: Science Publishers; 2009; 23 84 [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32: 1792 1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* phylogenetic analysis using parsimony (*and other methods) version 4.04beta. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4: 406 425 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24: 1596 1599 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39: 783 791 [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 1988; 16: 10881 10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross TS, Sepulveda M, Wiebe J, Schoeb T, Denslow N. Characterization of annual reproductive cycles for pond-reared Florida largemouth bass Micropterus salmoides floridanus. : Phillip D, Ridgway M. (eds.), Black Bass: Ecology, Conservation, and Management, Vol. Symposium 31. Bethesda, MD: American Fisheries Society; 2002; 205 212 [Google Scholar]
- Russell DW, Brown MS, Goldstein JL. Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J Biol Chem 1989; 264: 21682 21688 [PubMed] [Google Scholar]
- Takahashi S, Sakai J, Fujino T, Miyamori I, Yamamoto TT. The very low density lipoprotein (VLDL) receptor—a peripheral lipoprotein receptor for remnant lipoproteins into fatty acid active tissues. Mol Cell Biochem 2003; 248: 121 127 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Kawarabayasi Y, Nakai T, Sakai J, Yamamoto T. Rabbit very low density lipoprotein receptor: a low density lipoprotein density receptor-like protein with distinct ligand specificity. Proc Natl Acad Sci U S A 1992; 89: 9252 9256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujo H, Hermann M, Kaderli MO, Jacobsen L, Sugawara S, Nimpf J, Yamamoto T, Schneider WJ. Chicken oocyte growth is mediated by and eight ligand binding repeat member of the LDL receptor family. EMBO J 1994; 13: 5165 5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid EM, McMahon HT. Integrating molecular and network biology to decode endocytosis. Nature 2007; 448: 883 888 [DOI] [PubMed] [Google Scholar]
- Sun G, Zhu J, Li C, Tu Z, Raikhel AS. Two isoforms of the early E74 gene, an Ets transcription factor homologue, are implicated in the ecdysteroid hierarchy governing vitellogenesis of the mosquito, Aedes aegypti. Mol Cell Endocrinol 2002; 190: 147 157 [DOI] [PubMed] [Google Scholar]
- Chen L, Zhu J, Sun G, Raikhel AS. The early gene Broad is involved in the ecdysteroid hierarchy governing vitellogenesis of the mosquito Aedes aegypti. J Mol Endocrinol 2004; 33: 743 761 [DOI] [PubMed] [Google Scholar]
- Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A 2001; 98: 5671 5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science 2003; 301: 1714 1717 [DOI] [PubMed] [Google Scholar]
- Kortner TM, Rocha E, Arukwe A. Previtellogenic oocyte growth and transcriptional changes of steroidogenic enzyme genes in immature female Atlantic cod (Gadus morhua L.) after exposure to the androgens 11-ketotestosterone and testosterone. Comp Biochem Physiol A Mol Integr Physiol 2009; 152: 304 313 [DOI] [PubMed] [Google Scholar]
- Nakamura I, Kusakabe M, Young G. Differential suppressive effects of low physiological doses of estradiol-17β in vivo on levels of mRNAs encoding steroidogenic acute regulatory protein and three steroidogenic enzymes in previtellogenic ovarian follicles of rainbow trout. Gen Comp Endocrinol 2009; 163: 318 323 [DOI] [PubMed] [Google Scholar]
- Bhandari RK, Alam MA, Higa M, Soyano K, Nakamura M. Evidence that estrogen regulates the sex change of honeycomb grouper (Epinephelus merra), a protogynous hermaphrodite fish. J Exp Zool 2005; 303A: 497 503 [DOI] [PubMed] [Google Scholar]
- Kortner TM, Rocha E, Silva P, Castro LFC, Arukwe A. Genomic approach in evaluating the role of androgens on the growth of Atlantic cod (Gadus morhua) previtellogenic oocytes. Comp Biochem Physiol D Genomics Proteomics 2008; 3: 205 218 [DOI] [PubMed] [Google Scholar]
- Dickhoff W, Yan L, Plisetskaya E, Sullivan C, Swanson P, Hara A, Bernard M. Relationship between metabolic and reproductive hormones in salmonid fish. Fish Physiol Biochem 1989; 7: 147 155 [DOI] [PubMed] [Google Scholar]
- Srivastava RK, Van Der Kraak G. Regulation of DNA synthesis in goldfish vitellogenic ovarian follicles by hormones and growth factors. J Exp Zool 1994; 270: 263 272 [Google Scholar]
- Maestro MA, Méndez E, Párrizas M, Gutiérrez J. Characterization of insulin and insulin-like growth factor-I ovarian receptors during the reproductive cycle of carp (Cyprinus carpio). Biol Reprod 1997; 56: 1126 1132 [DOI] [PubMed] [Google Scholar]
- Shved N, Berishvili G, Baroiller J-F, Segner H, Reinecke M. Environmentally relevant concentrations of 17α-ethinylestradiol (EE2) interfere with the growth hormone (GH)/insulin-like growth factor (IGF)-I system in developing bony fish. Toxicol Sci 2008; 106: 93 102 [DOI] [PubMed] [Google Scholar]
- Alam MA, Komuro H, Bhandari RK, Nakamura S, Soyano K, Nakamura M. Immunohistochemical evidence identifying the site of androgen production in the ovary of the protogynous grouper Epinephelus merra. Cell Tissue Res 2005; 320: 323 329 [DOI] [PubMed] [Google Scholar]