Abstract
Background
Spirodela polyrhiza is a species of the order Alismatales, which represent the basal lineage of monocots with more ancestral features than the Poales. Its complete sequence of the mitochondrial (mt) genome could provide clues for the understanding of the evolution of mt genomes in plant.
Methods
Spirodela polyrhiza mt genome was sequenced from total genomic DNA without physical separation of chloroplast and nuclear DNA using the SOLiD platform. Using a genome copy number sensitive assembly algorithm, the mt genome was successfully assembled. Gap closure and accuracy was determined with PCR products sequenced with the dideoxy method.
Conclusions
This is the most compact monocot mitochondrial genome with 228,493 bp. A total of 57 genes encode 35 known proteins, 3 ribosomal RNAs, and 19 tRNAs that recognize 15 amino acids. There are about 600 RNA editing sites predicted and three lineage specific protein-coding-gene losses. The mitochondrial genes, pseudogenes, and other hypothetical genes (ORFs) cover 71,783 bp (31.0%) of the genome. Imported plastid DNA accounts for an additional 9,295 bp (4.1%) of the mitochondrial DNA. Absence of transposable element sequences suggests that very few nuclear sequences have migrated into Spirodela mtDNA. Phylogenetic analysis of conserved protein-coding genes suggests that Spirodela shares the common ancestor with other monocots, but there is no obvious synteny between Spirodela and rice mtDNAs. After eliminating genes, introns, ORFs, and plastid-derived DNA, nearly four-fifths of the Spirodela mitochondrial genome is of unknown origin and function. Although it contains a similar chloroplast DNA content and range of RNA editing as other monocots, it is void of nuclear insertions, active gene loss, and comprises large regions of sequences of unknown origin in non-coding regions. Moreover, the lack of synteny with known mitochondrial genomic sequences shed new light on the early evolution of monocot mitochondrial genomes.
Introduction
Usually, a plant cell contains three genomes: plastid, mitochondrial, and nuclear. In a typical Arabidopsis leaf cell, there are about 100 copies of mitochondrial DNA (mtDNA), about 1,000 copies of chloroplast DNA (cpDNA), and two copies of nuclear DNA (ncDNA) [1].
The mitochondrial genome plays fundamental roles in development and metabolism as the major ATP production center via oxidative phosphorylation [2]. The mitochondrial genetic system in flowering plants exhibit multiple characteristics that distinguish them from other eukaryotes: large genome size with dispersed genes, an incomplete set of tRNAs, trans-splicing, and frequent uptake of plastid DNA or of foreign DNA fragments by horizontal and intracellular gene transfer [2], [3], [4], [5], [6]. Plant mtDNAs are a major resource for evolutionary studies, because coding regions evolve slowly, in contrast to the flexible non-coding DNA. Therefore, the structural evolution and plasticity of plant mtDNAs make them powerful model for exploring the forces that affect their divergence and recombination.
With the emergence of second-generation sequencing technologies, the number of completed plant mitochondrial genomes deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=33090&opt=organelle. Accessed 2012 Sep 11) has increased until August of 2012 to 69. Most are from Chlorophyta (17 of green algae) and seed plants (26 of eudicotyledons). So far, among 11 sequenced monocot mt genomes, 10 are from the Poales, which have been extensively studied and only one, Phoenix, a palm, from the order of Arecale has been sequenced [7]. Obviously, complete mt sequence data will be needed not only from closely but also distant related taxa to give us a broader perspective of mt genome organization and evolution.
Spirodela polyrhiza, with great potential for industrial and environmental applications, is a small, fast growing aquatic plant in the Araceae family of the Alismatales order [8], [9]. There are 14 families, 166 genera, and about 4,500 species in this order. The early diverging phylogenetic position of Alismatales offers a broader view at features of monocot mt genomes. Plant mitochondria could also open a strategy for transgenes with high expression level and biological containment because of their maternal inheritance [10]. Here, we demonstrate the de novo assembly of a complete mt genome sequence from total leaf DNA using the SOLiD sequencing platform and a genome copy number-sensitive algorithm that can filter chloroplast and nuclear sequences. Indeed, comparative analysis of this genome provides us with unique features and new insights of this class of plants that differ from other monocots.
Materials and Methods
DNA Isolation and SOLiD DNA Sequencing
The methods for DNA extraction and DNA sequencing by the SOLiD platform followed a protocol as previously published [11]. Briefly, total genomic DNA was extracted from the clonally grown whole plant tissue of Spirodela polyrhiza. A mate-paired library was made with 1.5 Kb insertions and read length was 50 bp. Since nucleic, mitochondrial and chloroplast sequence all exist in reads from total DNA preparation, copy number between three genomes was significantly different [12], [13], so that it was feasible to de novo assembly both chloroplast and mitochondria genomes using the same dataset but with different coverage cut-off numbers as described previously [11].
Genome Assembly, Finishing and Validation
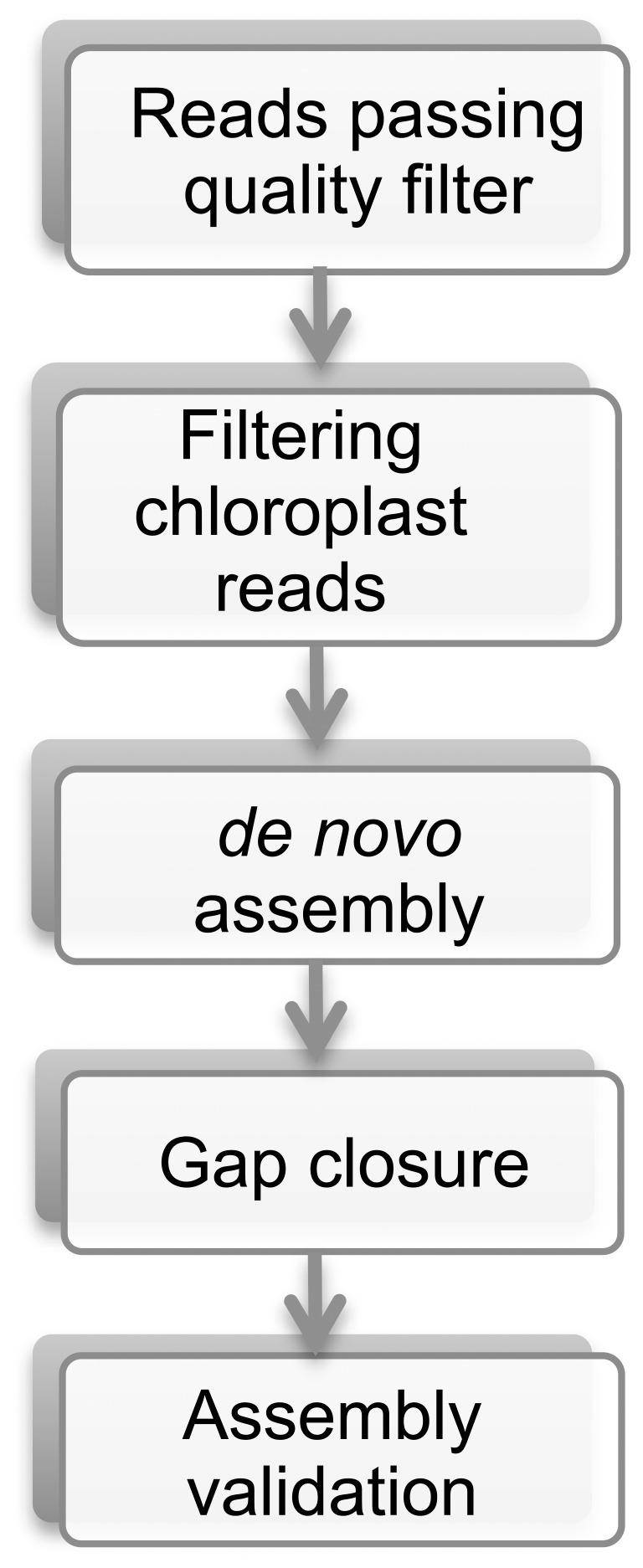
The coverage cut-off was fully utilized to only allow the target organellar genome to be assembled due to obvious differentiation of copy number for three genomes in total reads [12]. Furthermore, low-level contaminating sequences from foreign DNA (mainly nuclear DNA) were discarded by this approach. Quality control and other details were described recently [11]. Before we assembled the mitochondria genome using mate-paired reads, we masked chloroplast reads to reduce effects due to plastid sequence predominance. The detailed pipeline was shown below (Fig. 1).
Figure 1. Pipeline of mitochondrial genome assembly.
Details were described in Methods.
1) Filtering chloroplast reads: we mapped total high quality reads to existing chloroplast genome (GenBank # JN160603) by BWA short-read alignment component with default parameters [14]. Only unmapped reads were used in the next step. 2) de novo assembly: the assembly was executed using the SOLiD™ System de novo Accessory Tools 2.0 (http://solidsoftwaretools.com/gf/project/denovo/) in conjunction with the Velvet assembly engine [15]. 3) Gap closure: since chloroplast reads were pre-removed before mitochondrial assembly, theoretically, any location with chloroplast insertion in mtDNA would create a gap. Using flanking primers bridging 57 gaps, the missing sequences were amplified and sequenced with the ABI 3730×l system, yielding a complete contiguous mtDNA sequence (Table S1). To validate the circularity of the Spirodela mtDNA, PCR products were sequenced with pairs of primers bridging gaps and overlapping with the assembled linear scaffold. 4) Most gaps were small enough for single CE (capillary electrophoresis) sequence reads and overlapping sequences served as a measure for the accuracy of the SOLiD assembly and error rate. Therefore, PCR amplification and CE sequence provided validation of the order of contigs and also revealed sequencing discrepancies between these two platforms.
Genome Annotation and Sequence Analysis
The main pipeline for mitochondrial genome annotation was adapted from other sources [5]. Databases for protein-coding genes, rRNA and tRNA genes were compiled from all previously sequenced seed plant mitochondrial genomes. BLASTX and tRNAscan-SE were the mainly used programs [5]. The boundaries for each gene were manually curated. The sequin file including sequence and annotation was submitted to NCBI GenBank as JQ804980. The graphical gene map was processed by OrganellarGenomeDRAW program [16]. The codon usages for all protein coding genes in Spirodela and Oryza were calculated by using the Sequence Manipulation Suite [17].
Cp-derived tRNAs were identified by aligning all tRNA in annotated cpDNA to mtDNA with 80% of identity, an e-value of 1e-10 and a 50% coverage threshold. All remaining sequences were further scanned by EMBOSS getorf for open reading frames (ORFs) with more than 300 bp [18].
Putative RNA editing sites in protein-coding genes were identified by the PREP-mt Web-based program based on the evolutionary principle that editing increases protein conservation among species (http://prep.unl.edu/. Accessed 2012 Sep 11) [19]. The optimized cut-off value 0.6 was set in order to achieve the maximal accurate prediction. RNA editing sites from four genes were validated by RT-PCR with gene-specific primers (Table S2).
Sequences transferred to mtDNA were found by BLASTN search of mtDNA against the Spirodela chloroplast genome with 80% of identity, e-value of 1e-10 and 50 bp of length threshold. Repeat sequence analysis was predicted by using REPuter web-based interface, including forward, palindromic, reverse and complemented repeats with a cut-off value of 50 bp [20]. The mitochondrial genome was screened by repeatmasker under cross_match search engine (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker. Accessed 2012 Sep 11) for interspersed repeats and low complexity DNA sequences [21].
Phylogenetic Analysis
We aligned 19 homologous protein-coding gene sequences (nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, cob, cox1, cox2, cox3, atp1, atp4, atp6, atp8, atp9 and rps3) from the Spirodela mitochondrial genome and other seven plant organisms (Table S8, Cycad, NC_010303; Phoenix, NC_016740; Spirodela, JQ804980; Oryza, NC_011033; Zea, NC_007982; Boea, NC_016741; Nicotiana, NC_006581; Arabidopsis, NC_001284) and constructed a phylogenetic tree. Annotations were revalidated and sequences were concatenated into a single continuous sequence from 18,537 to 19,041 bp to initiate alignment by MEGA5 [22]. The phylogeny of the mitochondrial genome was estimated by maximum likelihood (ML) with 1,000 Bootstrap of replicates. Cycas was used as the outgroup.
Comparison of Global Genome Structure
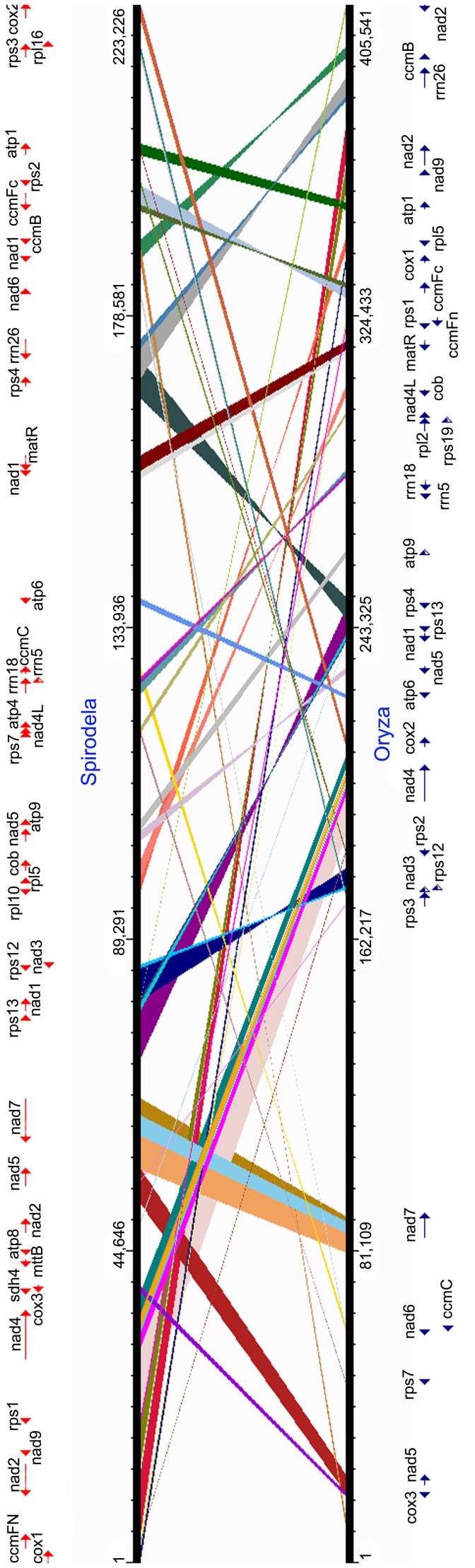
The conserved regions for protein-coding and rRNA genes were identified between Spirodela and Oryza sequences by BLASTN. The synteny together with the annotation file were uploaded to a web-based genome synteny viewer GSV [23]. The relative ordering of a set of homologous genes was illustrated in Fig. 2.
Figure 2. Comparison of synteny in conserved gene loci of Spirodela and Oryza mitochondrial genomes.
The annotated protein-coding genes were indicated for Spirodela and Oryza. Major conserved regions were bridged by lines. The visualized genome synteny was performed by GSV: a web-based genome synteny viewer [23].
Results and Discussion
The de novo Assembly of SOLiD Reads
The optimal parameter of the SOLiD™ System de novo Accessory Tools 2.0 for the assembly of the Spirodela mitochondrial genome has a hash length of 25 and coverage cut-off of 45. Under these conditions, assembly of SOLiD reads from total leaf DNA resulted in 15 scaffolds and 88 contigs, of which three scaffolds were mitochondrial (173,697, 47,896, 1824 bp) (Table 1). As expected, the other scaffolds were mainly copies of ribosomal RNA genes and retroelements of the nuclear genome because their copy number was comparable to the copies of mitochondrial genomes per leaf cell. To validate the assemblies, gaps were amplified with PCR for dideoxy sequencing with the CE ABI 3730×l system. With this information the order of the three scaffolds were resolved. Furthermore, after the SOLiD short read assembly was aligned with the CE long read sequences, only 0.036% discrepancy was found within 19 Kb sequence of overlaps, demonstrating high consistency between the two platforms. When we mapped the total reads back to the complete mtDNA, a total of 467-fold coverage was calculated. Considering the 5,474-fold chloroplast coverage, we found 41-fold coverage of nuclear genome sequences (Table 1). This level of coverage from assembled sequences was consistent with the expected representation of the three genomes in total leaf DNA, yielding chloroplast, mitochondria, and nuclei with the approximate ratio of 100∶10:1.
Table 1. de novo assembly statistics for the Spirodela mitochondrial genome.
| Statistical list | Number |
| Number of scaffolds | 15 |
| N50 scaffolds (bp) | 173,697 |
| Number of contigs | 88 |
| N50 contigs (bp) | 6,528 |
| Sum contig length (bp) | 240,987 |
| Hash length | 25 |
| Expected coverage | 90 |
| Coverage cut-offa | 45 |
| Total reads (X10∧6) | 153 |
| Aligned reads (%) | 1.4 |
| Average chloroplast coverageb | 5,474 |
| Average mitochondrial coverage | 467 |
| Average nuclear coverage | 41 |
Coverage cut-off: minimum coverage required to form a contig.
Average chloroplast coverage was cited from Spirodela chloroplast genome assembly [11].
Here, we applied a layered approach of sequencing organelle genomes without fractionation from total leaf DNA. Thanks to an assembly algorithm of sequence reads that is sensitive to the differential copy number of organelle and nuclear genomes, we did not physically need to fractionate plastid, mitochondrial, and nuclear DNA for deep sequencing. Therefore, we first assembled the complete Spirodela chloroplast genome from ABI SOLiD and gap-closure 3730xl reads, which permitted us to mask all plastid DNA reads before assembling mitochondrial DNA, which is in access of nuclear DNA but not as abundant as plastid DNA [11], [24]. Furthermore, we can take advantage of the ratios of these genomes to limit the value of coverage cut-off with identical dataset of SOLiD reads, which is taken in consideration for the assembly algorithm to distinguish between plastid, mitochondrial, and nuclear genome sequence reads [12], [13]. Assemblies were validated like in the case of chloroplast DNA by PCR and gap sequencing of long reads with the traditional ABI 3730×l sequencing system. Following this protocol, we obtained a complete mitochondrial genome from an aquatic plant in a very cost-efficient way, which can serve as a reference for future mt genomics.
Features of the Spirodela Mitochondrial Genome
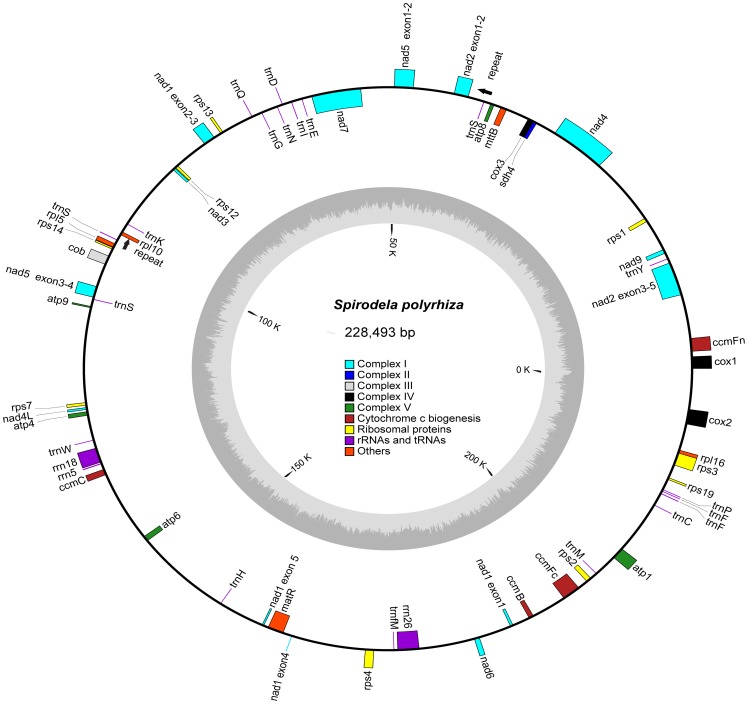
The mitochondrial genome was assembled into a 228,493 bp master circle (Fig. 3), which makes it the smallest genome of all sequenced monocots, much smaller than the 715,001 bp of Phoenix dactylifera [7], 490,520 bp of Oryza sativa [25], or 569,630 bp of Zea mays mitochondria [26]. Because Spirodela diverged at a very early stage in the monocot lineage, it suggests that either the common ancestor of monocots had a relatively compact genome, with a series of independent expansions by accumulation of chloroplast and nuclear sequences or proliferation of pairs of repeats, leading to the large genomes in rice and maize [5], [25], [26], or a number of size contractions happened in Spirodela from the large genome of their ancestor. The GC content in the mtDNA was 45.7%, slightly higher than 43.8% of Oryza and 43.9% of Zea [25], [26]. The coding sequences covered 31% of the mitochondrial genome compared with 57.4% of the chloroplast genome [11] (Table 2). There were 57 functional genes and 4 pseudogenes in total, encoding 35 proteins, 19 tRNAs and 3 rRNAs (Table S3). Therefore, it gave rise to a density of 4.0 Kb per gene. Noticeably, eight genes (ccmFc, cox2, nad1, nad2, nad4, nad5, nad7, rps3) had 15 cis-spliced group II introns, whereas nad1, nad2 and nad5 were disrupted by 6 trans-splicing sites (Table 2 and S1). Previous studies suggested that trans-splicing had evolved before the emergence of hornworts [27]. In general, the numbers and locations of introns in the Spirodela mtDNA were rather well conserved in other sequenced monocot genomes.
Figure 3. The gene map of Spirodela polyrhiza mitochondrial genome.
Genes indicated as closed boxes on the outside of the circle are transcribed clockwise, whereas those on the inside were transcribed counter-clockwise. Pseudogenes were indicated with the prefix “Ψ”. The biggest repeat pair was also marked by arrows. The genome coordinate and GC content are shown in the inner circle.
Table 2. Summary of general features for Spirodela mitochondrial genome.
| Feature | Value |
| Genome size (bp) | 228,493 |
| GC content (%) | 45.7 |
| Coding sequences (%)a | 31.4 |
| Protein coding gene # | 35 |
| ORFs # | 39 |
| cis−/trans-intron # | 15/6 |
| tRNA gene # | 19 |
| rRNA gene # | 3 |
| Chloroplast-derived (%) | 4.1 |
| Gene density (bp) | 4009 |
coding sequences include identified mitochondrial genes, pseudogenes, ORFs and cis-spliced introns.
Protein Genes and Transcript Editing
The content of key protein coding genes in Spirodela mtDNA is highly conserved with other angiosperms [26], [28], [29], [30]. There were nine subunits of the oxidative phosphorylation complex I (nad1, 2, 3, 4, 4L, 5, 6, 7 and 9); one subunit of complex II (sdh4); one subunit of complex III (cob); three subunits of complex IV (cox1, cox2 and cox3); five subunits of complex V (atp1, 4, 6, 8 and 9); and four subunits of a complex involved in cytochrome c biogenesis (ccmB, ccmC, ccmFn and ccmFc). Other genes encoding maturase (matR) and transport membrane protein (mttB) were also present in Spirodela mtDNA. As in maize [26], the matR gene in Spirodela also resided in the intron 4 of nad1, which is trans-spliced after transcription. In Spirodela, there were ten functional ribosomal genes and two pseudogenes of rps14 and rps19 with early stop codons, whereas rice had a functional rps19 and a non-functional rps14 [25] and both were missing in maize (Table S3) [26]. All annotated genes and coordinates were listed in Table S3 and shown in a graphical map (Fig. 3).
Post-transcriptional editing occurs in nearly all plant mitochondria, which results in altered amino acid sequences of the translated protein by converting specific Cs into Us in their transcripts. We used the program of the predictive RNA editor of plant mitochondrial genomes (PREP-mt) to predict the location of RNA editing sites, which are based on well-known principles that plant organelles maintain the conservation of protein sequences across many species by editing mRNA [19]. By setting the cut-off value to 0.6 within the 35 protein-coding genes of Spirodela mtDNA 600 sites were predicted as C-to-U RNA editing sites (Table S4). To validate the accuracy of this prediction, we compared RNA transcripts from atp9, nad9, cox3 and rps12 by RT-PCR with the corresponding genomic sequences yielding a confirmation for 90.8% of the predicted sites. Considering a level of about 10% artificial predictions, we estimate about 540 RNA editing sites, a number that lies between the 441 of protein-coding genes of Oryza [25] and 1,084 of Cycas [29].
It is generally accepted that RNA editing is essential for functional protein expression as it is required to modify amino acids to maintain appropriate structure and function [31], or to generate new start or stop codons [32]. Indeed, the abundance of RNA editing sites in Spirodela mtDNA might have increased genome complexity and pace of divergence. We summarized the number of potentially modified codons of Spirodela mtDNA in Table S5. Three edited codons (TCA (S) = >TTA (L); TCT (S) = >TTT (F); CCA (P) = >CTA (L)) were found most frequently, whereas three editing events from two codons (CAA (Q) = >TAA (X); CAG (Q) = >TAG (X)) resulted in stop codons (Table S5). Even though three new stop codons are located close at the carboxyterminal end of proteins (ccmC, rps1 and rpl16), it is not clear whether these small truncations affect their functions or not, which would require experimental evidence.
The rRNA, tRNA Genes and Codon Usage
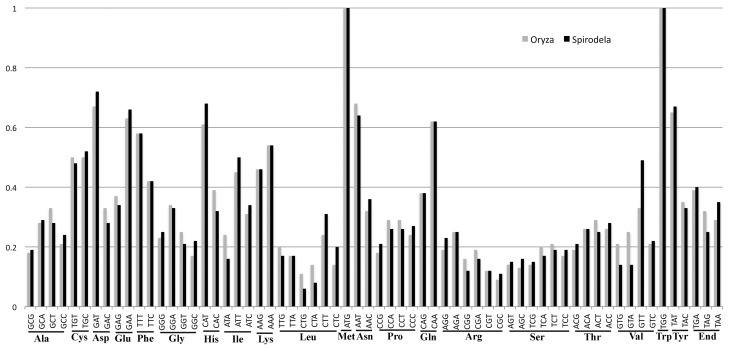
Spirodela mtDNA contains 3 ribosomal RNA genes (rrn5, rrn18, rrn26) and one pseudogene of rrn26. The 19 putatively expressed tRNA genes are specific for 15 amino acids (Table S3). Four of them (trnN-GTT, trnH-GTG, trnM-CAT and trnS-GGA) are probably chloroplast-derived because of high sequence similarity. They are also predicted as chloroplast origin in maize, rice, sugar beet and Arabidopsis except trnS-GGA in maize [26]. Therefore, they were not recently acquired from chloroplast, but more likely an event of horizontal transfer in a common ancestor. One trnH-GTG is considered to be a non-functional pseudogene. Functional tRNA genes for the amino acids Ala, Arg, Leu, Thr and Val are absent. Because all 20 amino acids are required for protein synthesis, and all 64 codons are used in the Spirodela mt genome based on a codon-usage scan (Fig. 4 and Table S6) [17], the missing tRNAs are presumably encoded by the nuclear genome and imported from the cytosol into the mitochondria [33]–[34]. We also found that the two codons for TAT-Tyr and TTT-Phe are highly preferred in Spirodela and Oryza and overall other codon usage is rather similar between the two species (Table S6).
Figure 4. The fraction of each codon usage among the same amino acid in Spirodela compared to that in Oryza.
Black bar was Spirodela and grey was Oryza. The fraction of each codon usage was shown on Y-axis.
ORFs and Intergenic Sequences
Only ORFs encoded by a hypothetical gene with more than 300 bp in length between start and stop codons and no match with a known mt coding sequence were counted. Based on this cut-off, we found 39 mitochondrial ORFs, most of which were not cp migrations and specific to Spirodela (Table S3). We named ORFs using their amino acid numbers. When the same length of ORFs happened, a lower case letter (a, b, c, etc) was added. Given the large amount of intergenic DNA in Spirodela mtDNA, it is not surprising to find an abundance of additional ORFs in its genome. Rarely, ORFs showed conservation to any other plants so that putative ORFs were considered to be spurious prediction [35]. However, orf100a had an ortholog of a NADH-ubiquinone oxidoreductase chain in Nicotiana tabacum (GenBank: YP_717128) and orf257 had sequence similarity to DNA polymerase (GenBank: YP_003875487) found in plant mt plasmids [4]. Some studies found that unidentified ORFs had transcripts in rapeseed [36] or to be actively transcribed in sugar beet [37], but further studies are needed to determine whether they encode functional proteins.
A striking feature of Spirodela mtDNA was that 81% of the intergenic regions were species-specific and showed no sequence similarity to any other known sequence. It seemed that anonymous sequences in intergenic DNA were quite common. For instance, unidentifiable sequences comprised 70% of Beta vulgaris mtDNA [38]. Although they split about 50 million years ago, 76% of rice mtDNA sequences appeared to be highly divergent from maize in intergenic regions [26]. The repetitive DNAs [39], mt plastidal migrations [40] and viral DNA insertions [41] could contribute to the expansion of intergenic regions, but still comprised a rather small fraction in most seed plant mt genomes. On the other hand, it was quite common that multipartite mt genomes could be generated through large repeat pairs with high frequency [35]. Indeed, 29 potential candidates of repeat pairs with more than 50 bp were found in Spirodela mtDNA by using REPuter [20] (Table S7). However, we could not detect repeat-specific contigs from the assembly that could be explained of isomeric and subgenomic molecules derived from a master circle after recombination. Probably, the high rate of non-coding sequence turnover in Spirodela mtDNA was mainly generated through the process of micro-homologous recombination or non-homologous end joining, later on of active rearrangement and continuous reshuffling. Still, the high proportion of enigmatic non-coding regions in mtDNA is quite extensive. To understand where all these enigmatic sequences might come from and why they appeared to be so common would require additional sequences from closely related species.
Phylogenetic Analysis and Gene Loss in Angiosperm Mitochondrial Genomes
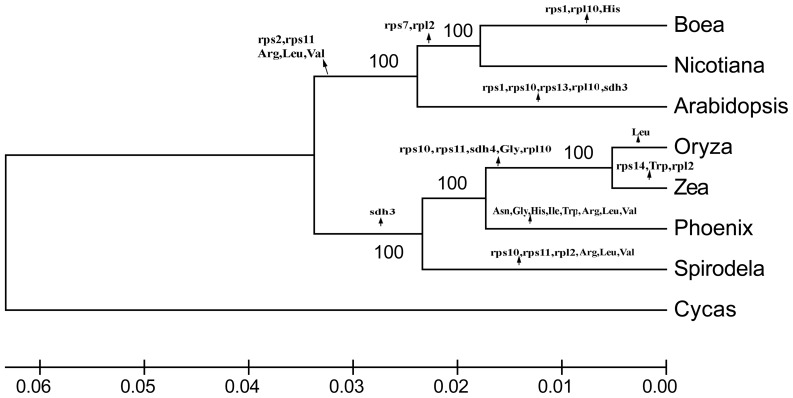
After re-examining mitochondrial genome annotations from seven species, a selection of 19 conserved genes (nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, cob, cox1, cox2, cox3, atp1, atp4, atp6, atp8, atp9 and rps3) was concatenated to permit alignment analysis of 19,824 sites in eight genomes, listed in Table S8 (dicot: Arabidopsis, Nicotiana and Boea; monocot: Spirodela, Phoenix, Oryza and Zea; outgroup: Cycas). The gene tree topology from multiple loci (Fig. 5) was largely congruent with the known phylogenetic relationships inferred from analysis of rbcL. There were two subclades of monocots and dicots within the angiosperm [42]. Previous studies of fossil records [43], morphology and molecular analysis [44] also supported that Alismatales (Spirodela) was a basal monocot followed by Arecales (Phoenix), whereas the Poales (rice and maize) resided in the most developed positions.
Figure 5. Phylogenetic tree based on 19 conserved genes in mitochondrial genomes.
The ML calculation was run by MEGA5 with 1,000 bootstrap replicates. All the gene losses were mapped on the tree branches. Cycas was included in the analysis as an outgroup. The signs of Amino Acid (Ala, Arg, Leu, Thr, His, Trp, Ile, Gly, Leu and Val) mean corresponding functional tRNA genes were absent in their mtDNAs.
The loss of protein coding and tRNA genes in seven genomes relative to the outgroup was examined based on the phylogenetic tree. Generally, most losses were limited in their phylogenetic depth to a single family and must have occurred recently (Fig. 5). Three ribosomal protein genes rps10, rps11 and rpl2 were missing in Spirodela mtDNA. Frequent gene losses of ribosomal protein genes also occurred in other species. At a closer look, rps2 seemed to have been lost early in the evolution of dicots, whereas rps2 was present in Cycas, Marchantia, and other monocots [45]. The rps11 gene was missing in dicots (Arabidopsis, Nicotiana and Boea) and also in some monocots (Spirodela, Oryza and Zea). The corresponding mt rps2 and rps11 genes have been transferred to the nucleus in Arabidopsis, soybean, and tomato, suggesting that gene loss followed functional transfer to the nucleus [6], [45]. The unparallel loss of rps11 and rpl2 in Spirodela compared with other monocots suggested that the loss of many genes might have occurred independently in various lineages during speciation of angiosperms. The sdh3 gene was absent and the sdh4 gene was present in both Spirodela and Phoenix, whereas neither was retained in rice and maize (Fig. 5 and Table S8). A previous study showed that sdh4 losses were concentrated in the monocots and no losses were detected in basal angiosperms by Southern blot survey of 280 angiosperm genera, which further showed most of the losses were limited in phylogenetic depth to a single family [46].
Our data lend support to previous studies that most gene losses occurred with mt ribosomal protein genes and rarely with respiratory genes, which was well documented with a Southern blot survey of gene distribution in 281 diverse angiosperms [6]. When a gene was missing from mtDNA of a given species, it was generally assumed that the original copy had been transferred to the nucleus. Therefore, our results strongly suggested that intracellular gene transfer of ribosomal protein and tRNA genes from mitochondria to the nuclear genome was a frequent process, which in return allowed the nucleus to control the organelle by encoding organelle-destined proteins [33], [34]. Still, functional copies of these putative transferred genes will have to be confirmed after the whole nuclear genome sequence will be available. The finding of many intermediate stages of the cox2 gene transfer in legumes had shown that physical movement of mtDNA to the nuclear genome was an ongoing process [47].
Chloroplast DNA Insertions
The Spirodela mtDNA contained multiple cp-originated insertions, ranging in size from 69 to 1,048 bp. These sequences added up to 9,295 bp of the total amount of transferred cpDNA (Table S9), accounting for 4.07% of the mtDNA. A total of 4,436 bp was derived from the inverted repeats of the chloroplast genome, whereas 4,859 bp was transferred from single copy regions of cpDNA. The similarity level of each insertion to the chloroplast genome varied between 75% and 100%. Moreover, the migrated plastid fragments had 732 substitutions, 28 insertions, and 49 deletions within 9,295 bp. They also contained fragments of plastid genes, such as psbA, petB, psbC and ycf1 (Table S9). All of the protein-coding genes of plastid origin in Spirodela mtDNA were likely to be non-functional as a result of truncations and mutations, whereas four tRNAs of plastidal origin appeared to be intact. Indeed, chloroplast-derived sequences were very common in plant mt genomes, such as 6% in rice [25], 4% in maize [26] and 1% in Arabidopsis [28]. Surprisingly, 42.4% of the chloroplast genome of Vitis has been incorporated into its mt genome [48]. And a large segment of 113 Kb from chloroplast sequences was captured by the Cucurbita mt genome [5].
Integrated Nuclear DNA
It is believed that transposable elements in mitochondria are nuclear-derived and are therefore common in mt intergenic regions [38], [49]. For instance, 4% of Arabidopsis mtDNA was probably derived from transposons of nuclear origins [28]. Four fragments of transposable elements were found in maize mtDNA [26] and nineteen were identified in rice [25]. However, we could not find any transposons in the Spirodela mt genome when we searched against the Repbase repetitive element database [50]. This suggests that either very few nuclear sequences have migrated into Spirodela mtDNA or Spirodela mitochondria select against transposable elements.
Comparison of Genome Synteny
A significant degree of synteny was found within mitochondrial genomes of liverworts, mosses, and chlorophytes at the base of land plants, including a set of gene clusters (more than two genes together), such as the ribosomal protein cluster, ccm gene cluster, and two regions containing the nad and cox genes [51]. It was clear that the sequences of protein-coding genes were highly conserved, but the relative order of genes was greatly rearranged between Spirodela and rice (Fig. 2). Many ribosomal proteins were independently lost in both Spirodela and rice (Fig. 5); therefore, synteny between the remaining genes became harder to detect. The ancestral cob-nad1-cox3-cox2-nad6-atp6-rps7-rps12-nad2-nad4-nad5 gene order of basal land plants has been lost due to various recombination and rearrangement events in angiosperm mtDNA evolution. [4], [41], [52].
In summary, our data provides further evidence that SOLiD platforms can assemble both chloroplast and mitochondrial genomes with regular coverage without any organellar purification (Table 1) [11]. Our analysis of the mt genome of Spirodela, having the smallest size among sequenced monocots, elucidates the evolutionary change among monocot mt genomes. Although the critical genes for the electron transport chain in Spirodela mtDNA are well conserved, different types of ribosomal protein genes are missing in comparison to other monocots. The number of RNA editing in protein coding genes is within a typical range as other plants. Still, no known transposable elements can be found in its genome, suggesting a rather rare migration from the nucleus to the mitochondria. Sequence-based phylogenetic analysis clearly supports the hypothesis that Spirodela is at the very basal lineage of monocots. Comparative analyses of mitochondrial genes between Spirodela and rice have shown that the relative order of genes is greatly rearranged over a very short evolutionary time. In this regard, additional complete mitochondrial sequences from closely related species will be needed to fortify the distinct evolution of plant mitochondrial genomes.
Supporting Information
Primer pairs for gap closure and Sanger sequencing.
(XLS)
Primer pairs for RNA editing validation by RT-PCR.
(XLS)
Gene content for Spirodela mitochondrial genome. Gene content includes protein-coding genes, tRNA, rRNA and putative ORFs. “Ψ” means pseudo gene and “cp-” means chloroplast-derived gene.
(XLS)
Predicted RNA editing numbers in each protein-coding gene for Spirodela mtDNA. The cutoff value for each predicted site was the percentage of matches in alignment to the corresponding amino acid across species.
(XLS)
Type and number of codon modification in predicted RNA editing sites of Spirodela mtDNA.
(XLS)
Comparison of codon usage between Spirodela and Oryza. aResults for 35 protein coding genes in Spirodela with 30,790 bp. bResults for all CDS from Genbank in Oryza with 44,875 bp.
(XLS)
Predicted repeat pairs in Spirodela mtDNA by using REPuter. “F” and “P” means forward and palindromic matches.
(XLS)
Protein-coding and tRNA gene list in the 8 representative plant mtDNAs. “+” means present and “–“ means absent.
(XLS)
The regions in Spirodela mtDNA originated from cpDNA with corresponding coordinates and identity.
(XLS)
Acknowledgments
We thank David Sidote and Mark Diamond for SOLiD library construction and sequencing. We thank Gregory Thyssen and Qinghua Wang for their invaluable comments on the manuscript. We also thank Brian Schubert from Waksman Genomics Laboratory for program installation and server management.
Funding Statement
The authors have no support or funding to report.
References
- 1. Logan DC (2006) The mitochondrial compartment. J Exp Bot 57: 1225–1243. [DOI] [PubMed] [Google Scholar]
- 2. Mackenzie S, McIntosh L (1999) Higher plant mitochondria. Plant Cell 11: 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keeling PJ, Palmer JD (2008) Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9: 605–618. [DOI] [PubMed] [Google Scholar]
- 4. Sloan DB, Alverson AJ, Storchova H, Palmer JD, Taylor DR (2010) Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia . BMC Evol Biol 10: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, et al. (2010) Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol 27: 1436–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palmer JD, Adams KL, Cho Y, Parkinson CL, Qiu YL, et al. (2000) Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci U S A 97: 6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang Y, Wu H, Yang M (2012) A Complete Sequence and Transcriptomic Analysis of Date Palm (Phoenix dactylifera L.) Mitochondrial Genome. NCBI GenBank NC_0167401. [DOI] [PMC free article] [PubMed]
- 8. Cabrera LI, Salazar GA, Chase MW, Mayo SJ, Bogner J, et al. (2008) Phylogenetic relationships of aroids and duckweeds (Araceae) inferred from coding and noncoding plastid DNA. Am J Bot 95: 1153–1165. [DOI] [PubMed] [Google Scholar]
- 9. Stomp AM (2005) The duckweeds: A valuable plant for biomanufacturing. Biotechnology Annual Review 11: 69–99. [DOI] [PubMed] [Google Scholar]
- 10. Ljaz S (2010) Plant mitochondrial genome: “A sweet and safe home” for transgene. African Journal of Biotechnology 9: 4. [Google Scholar]
- 11. Wang W, Messing J (2011) High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS One 6: e24670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang T, Zhang X, Hu S, Yu J (2011) An efficient procedure for plant organellar genome assembly, based on whole genome data from the 454 GS FLX sequencing platform. Plant Methods 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lutz K, Wang W, Zdepski A, Michael T (2011) Isolation and analysis of high quality nuclear DNA with reduced organellar DNA for plant genome sequencing and resequencing. BMC Biotechnology 11: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lohse M, Drechsel O, Bock R (2007) OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52: 267–274. [DOI] [PubMed] [Google Scholar]
- 17.Stothard P (2000) The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28: 1102, 1104. [DOI] [PubMed]
- 18. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 19. Mower JP (2009) The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res 37: W253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, et al. (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29: 4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bergman CM, Quesneville H (2007) Discovering and detecting transposable elements in genome sequences. Brief Bioinform 8: 382–392. [DOI] [PubMed] [Google Scholar]
- 22. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 23. Revanna KV, Chiu CC, Bierschank E, Dong Q (2011) GSV: a web-based genome synteny viewer for customized data. BMC Bioinformatics 12: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Les DH, Crawford DJ, Landolt E, Gabel JD, Kimball RT (2002) Phylogeny and Systematics of Lemnaceae, the Duckweed Family. Systematic Botany 27: 221–240. [Google Scholar]
- 25. Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, et al. (2002) The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics 268: 434–445. [DOI] [PubMed] [Google Scholar]
- 26. Clifton SW, Minx P, Fauron CM, Gibson M, Allen JO, et al. (2004) Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol 136: 3486–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malek O, Knoop V (1998) Trans-splicing group II introns in plant mitochondria: the complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA 4: 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Unseld M, Marienfeld JR, Brandt P, Brennicke A (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet 15: 57–61. [DOI] [PubMed] [Google Scholar]
- 29. Chaw SM, Shih AC, Wang D, Wu YW, Liu SM, et al. (2008) The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol Biol Evol 25: 603–615. [DOI] [PubMed] [Google Scholar]
- 30. Zhang T, Fang Y, Wang X, Deng X, Zhang X, et al. (2012) The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: insights into the evolution of plant organellar genomes. PLoS One 7: e30531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Giege P, Brennicke A (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci 96: 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A (2008) The process of RNA editing in plant mitochondria. Mitochondrion 8: 35–46. [DOI] [PubMed] [Google Scholar]
- 33. Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schneider A (2011) Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem 80: 1033–1053. [DOI] [PubMed] [Google Scholar]
- 35.Mower JP, Sloan DB, Alverson AJ (2012) Plant Mitochondrial Genome Diversity: The Genomics Revolution In: Wendel JF, Greilhuber J, Dolezel J, Leitch IJ, editors. Plant Genome Diversity: Springer Vienna. 123–144.
- 36. Handa H (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana . Nucleic Acids Res 31: 5907–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Satoh M, Kubo T, Nishizawa S, Estiati A, Itchoda N, et al. (2004) The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol Genet Genomics 272: 247–256. [DOI] [PubMed] [Google Scholar]
- 38. Satoh M, Kubo T, Mikami T (2006) The Owen mitochondrial genome in sugar beet (Beta vulgaris L.): possible mechanisms of extensive rearrangements and the origin of the mitotype-unique regions. Theor Appl Genet 113: 477–484. [DOI] [PubMed] [Google Scholar]
- 39. Lilly JW, Havey MJ (2001) Small, repetitive DNAs contribute significantly to the expanded mitochondrial genome of cucumber. Genetics 159: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McDermott P, Connolly V, Kavanagh TA (2008) The mitochondrial genome of a cytoplasmic male sterile line of perennial ryegrass (Lolium perenne L.) contains an integrated linear plasmid-like element. Theor Appl Genet 117: 459–470. [DOI] [PubMed] [Google Scholar]
- 41. Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD (2011) The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS One 6: e16404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janssen T, Bremer K (2004) The age of major monocot groups inferred from 800+ rbcL sequences. Botanical Journal of the Linnean Society: 4.
- 43. Stockey RA (2006) The fossil record of basal monocots. 22: 16. [Google Scholar]
- 44. Borsch T, Hilu KW, Quandt D, Wilde V, Neinhuis C, et al. (2003) Noncoding plastid trnT-trnF sequences reveal a well resolved phylogeny of basal angiosperms. J Evol Biol 16: 558–576. [DOI] [PubMed] [Google Scholar]
- 45. Perrotta G, Grienenberger JM, Gualberto JM (2002) Plant mitochondrial rps2 genes code for proteins with a C-terminal extension that is processed. Plant Mol Biol 50: 523–533. [DOI] [PubMed] [Google Scholar]
- 46. Adams KL, Rosenblueth M, Qiu YL, Palmer JD (2001) Multiple losses and transfers to the nucleus of two mitochondrial succinate dehydrogenase genes during angiosperm evolution. Genetics 158: 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adams KL, Song K, Roessler PG, Nugent JM, Doyle JL, et al. (1999) Intracellular gene transfer in action: Dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc Natl Acad Sci 96: 13863–13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goremykin VV, Salamini F, Velasco R, Viola R (2009) Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol 26: 99–110. [DOI] [PubMed] [Google Scholar]
- 49. Knoop V, Unseld M, Marienfeld J, Brandt P, Sunkel S, et al. (1996) copia-, gypsy- and LINE-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana . Genetics 142: 579–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smit A, Hubley R, Green P (1996–2010) RepeatMasker Open-3.0. Available: http://www.repeatmasker.org. Accessed 2012 Sep 11.
- 51. Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, et al. (2007) The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol 24: 699–709. [DOI] [PubMed] [Google Scholar]
- 52. Chang S, Yang T, Du T, Huang Y, Chen J, et al. (2011) Mitochondrial genome sequencing helps show the evolutionary mechanism of mitochondrial genome formation in Brassica . BMC Genomics 12: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer pairs for gap closure and Sanger sequencing.
(XLS)
Primer pairs for RNA editing validation by RT-PCR.
(XLS)
Gene content for Spirodela mitochondrial genome. Gene content includes protein-coding genes, tRNA, rRNA and putative ORFs. “Ψ” means pseudo gene and “cp-” means chloroplast-derived gene.
(XLS)
Predicted RNA editing numbers in each protein-coding gene for Spirodela mtDNA. The cutoff value for each predicted site was the percentage of matches in alignment to the corresponding amino acid across species.
(XLS)
Type and number of codon modification in predicted RNA editing sites of Spirodela mtDNA.
(XLS)
Comparison of codon usage between Spirodela and Oryza. aResults for 35 protein coding genes in Spirodela with 30,790 bp. bResults for all CDS from Genbank in Oryza with 44,875 bp.
(XLS)
Predicted repeat pairs in Spirodela mtDNA by using REPuter. “F” and “P” means forward and palindromic matches.
(XLS)
Protein-coding and tRNA gene list in the 8 representative plant mtDNAs. “+” means present and “–“ means absent.
(XLS)
The regions in Spirodela mtDNA originated from cpDNA with corresponding coordinates and identity.
(XLS)