Abstract
Objective
Circadian rhythms influence the timing of behavior, neurological diseases, and even death. Rare mutations in homologs of evolutionarily conserved clock genes are found in select pedigrees with extreme sleep timing, and there is suggestive evidence that certain common polymorphisms may be associated with self-reported day/night preference. However, no common polymorphism has been associated with the timing of directly observed human behavioral rhythms or other physiological markers of circadian timing at the population level.
Methods
We performed a candidate-gene association study with replication, evaluating associations between polymorphisms in homologs of evolutionarily conserved clock genes and the timing of behavioral rhythms measured by actigraphy. For validated polymorphisms, we evaluated associations with transcript expression and time of death in additional cohorts.
Results
rs7221412, a common polymorphism near period homolog 1 (PER1), was associated with the timing of activity rhythms in both the discovery and replication cohorts (joint p=2·1×10−7). Mean activity timing was delayed by 67 minutes in rs7221412GG vs. rs7221412AA homozygotes. rs7221412 also showed a suggestive time-dependent relationship with both cerebral cortex (p=0.05) and CD14+CD16− monocyte (p=0.02) PER1 expression and an interesting association with time of death (p=0.015) in which rs7221412GG individuals had a mean time of death nearly seven hours later than rs7221412AA/AG.
Interpretation
A common polymorphism near PER1 is associated with the timing of human behavioral rhythms, and shows evidence of association with time of death. This may be mediated by differential PER1 expression. These results may facilitate individualized scheduling of shift-work, medical treatments, or monitoring of vulnerable patient populations.
INTRODUCTION
Circadian rhythms modulate human behavior and physiology, including cognitive function1, as well as the timing and treatment of several neurological and medical disorders, including haemorrhagic2 and ischaemic3 stroke, amyloid-β secretion4, epilepsy5, myocardial infarction6, ventricular arrhythmias7, cancer8 and even time of death9. Moreover, circadian misalignment, as seen in shift-work, predisposes not only to accidents and lost productivity, but also to common diseases, such as diabetes10. Circadian traits are heritable11-14 and understanding their genetic architecture may facilitate individualization of the timing of school or work schedules, as well as medical investigations and therapies.
In animals, a conserved genetic network regulates circadian rhythms. At its core is a transcription-translation feedback loop formed by the genes PER1-3, CRY1-2, BMAL1, and CLOCK that generates near 24-hour rhythmicity10. In animals, mutations in these genes can lead to alterations in or loss of circadian rhythmicity10.
There is some genetic evidence for the role of homologs of these genes in the regulation of human circadian rhythms. Mutations in PER2 and CSNK1D, a PER2 kinase, have been implicated in select pedigrees with extreme circadian timing15, 16, but these are rare and their significance at the population level is unclear. More common variants in PER1, PER3, and CLOCK have evidence of association with self-reported day/night preference17-20, but these have not been shown to be associated with differences in the timing of directly observed human behavior. To our knowledge, no common genetic variant has been associated with the timing of directly observed human behavior or other any other physiological marker of circadian timing in a real-world setting at the population level.
To identify and characterize such variants, we performed a candidate gene association study, evaluating associations between polymorphisms in homologs of evolutionarily conserved clock genes and the timing of behavioral rhythms measured by actigraphy as a marker of circadian phase. For validated polymorphisms, we evaluated associations with intrinsic circadian period, transcript expression in cerebral cortex and peripheral blood mononuclear cells (PBMCs), and time of death.
METHODS
Participants
Characteristics of the study participants are shown in Table 1. Inclusion and exclusion criteria are described in the Supplementary Appendix. The discovery cohort consisted of 537 participants of European ancestry from the Rush Memory and Aging Project (“MAP” cohort), a community-based study of aging21-23. The replication cohort consisted of 38 individuals of European ancestry who had participated in detailed inpatient circadian period phenotyping studies at the Brigham and Women’s Hospital from 2001-2010 (“BWH Circadian” cohort). Transcript expression was examined in three datasets – one derived from port-mortem cerebral cortical samples from 193 individuals without dementia24 (“NIA” cohort), another derived from PBMCs from 228 individuals with multiple sclerosis25 (“BWH MS” cohort) and a third derived from CD14+CD16− monocytes from 69 healthy subjects without inflammatory or metabolic disease (“PhenoGenetic” cohort). Time of death was examined in 687 deceased participants of European ancestry from the MAP and the Religious Orders Study (“ROS/MAP” cohort”). The ROS/MAP cohort excluded any MAP participants who were part of the primary discovery cohort.
Table 1.
Characteristics of Study Participants and Phenotyping
| Name | Memory and Aging Project (MAP Cohort) |
Brigham and Women’s Hospital Division of Sleep Medicine (BWH Circadian Cohort) |
National Institute of Aging Laboratory of Neuro-genetics (NIA Cohort) |
Brigham and Women’s Hospital Comprehensive Longitudinal Investigation of Multiple Sclerosis (BWH MS Cohort) |
PhenoGenetic Cohort |
Religious Orders Study and Memory and Aging Project Deceased Participants (ROS/MAP Cohort) |
|---|---|---|---|---|---|---|
|
| ||||||
| N | 537 | 38 | 193 | 228 | 69 | 687 |
| Age in Years - Mean (SD) [Range] |
82·9 (6·6) [59-100] |
32·9 (17·6) [18-70] |
80·9 (9·2) [65-100] |
40·9 (9·4) [21-68] |
32.4 (9.5) [19-51] |
88·3 (6·4) [67-109] |
| Number (%) Female |
412 (76%) | 14 (37%) | 86 (46%) | 183 (80%) | 41 (56%) | 426 (62%) |
| Female | ||||||
| Ethnic Ancestry (% European) |
100% | 100% | 100% | 100% | 100% | 100% |
| Number (%) with Dementia |
55 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 289 (42%) |
| Genotyping Platform |
Affymetrix 6.0 | Sequenom MassARRAY |
Affymetrix 500k |
Affymetrix 6.0 | Sequenom MassARRAY |
Affymetrix 6.0 |
| Actigraphy | 7 days with Phillips |
7 days with Phillips |
NA | NA | NA | NA |
| Respironics Actical |
Respironics Actiwatch-L |
|||||
| Expression Profiling Platform |
NA | NA | Illumina Human Refseq- 8 BeadChip |
Affymetrix Human Genome U133 Plus 2.0 |
Affymetrix Human Genome U133 Plus 2.0 |
NA |
| Comments | All without medical co morbidities |
All with relapsingremitting multiple sclerosis |
Healthy volunteers without inflammatory or metabolic disease |
No overlap with MAP participants used for primary association analysis |
||
All protocols were approved by the institutional review boards at each institution. All participants provided written informed consent.
Phenotyping
As described in the Supplementary Appendix, we recorded one week of actigraphy from participants in the MAP and BWH Circadian cohorts. Our primary actigraphic phenotype was the timing of the acrophase of activity, defined as the midpoint of the eight consecutive hours of each day with the greatest activity (Fig 1). The timing of the nadir and acrophase of activity calculated using cosinor analysis26 provided secondary actigraphic phenotypes.
Fig 1. Example actigraphic record.
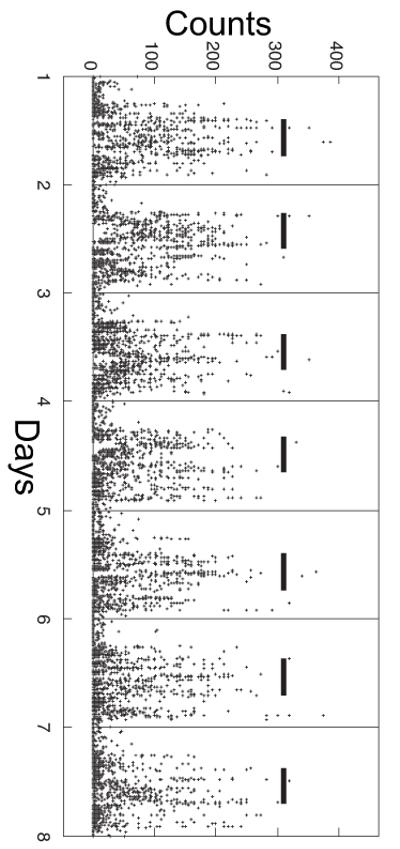
The activity acrophase for each day, defined as the midpoint of the eight consecutive hours of each day with the greatest activity, is indicated by a horizontal black bar. The vertical divisions indicate midnight.
To validate activity acrophase as a marker of circadian phase, we compared acrophase timing with that of dim-light melatonin onset (DLMO) and the core body temperature nadir under conditions of constant routine27 in a subset of 16 BWH Circadian participants closely following their actigraphy recordings.
To determine the intrinsic periods of the temperature and melatonin rhythms, each participant in the BWH Circadian cohort was studied in an inpatient forced desynchrony protocol as previously described28.
Time of death in the ROS/MAP cohort was determined from clinical records. Time of death is well captured because all participants are organ donors. Information about cause of death was not available on the majority of the ROS/MAP subjects.
Genotyping and Expression Data
A detailed description of genotyping procedures is found in the Supplemental Appendix.
Eighteen candidate genes were selected based on involvement in rare human pedigrees with extreme circadian phenotypes or functional importance in circadian rhythms in animals (Supplementary Table 1). After linkage-disequilibrium based pruning to reduce the multiple testing burden, and additional quality control, 135 tagging polymorphisms within these eighteen loci were selected for our primary analysis (Supplementary Table 2).
Genotypes at all 135 polymorphisms were determined in the MAP cohort. For the BWH Circadian cohort, polymorphisms significant in the MAP cohort along with flanking variants (Supplementary Table 3) were genotyped. For the PhenoGenetic, NIA, BWH MS, and ROS/MAP cohorts, genotypes for polymorphisms significant in both the MAP and BWH Circadian cohorts were extracted from pre-existing genome-wide datasets22, 24, 25 or genotyped as described in the Supplemental Appendix.
RNA data for the NIA and BWH MS cohorts were obtained from the NCBI GEO database (GSE8919 and GSE16412)24, 25, 29. RNA data for the PhenoGenetic cohort were obtained from CD14+CD16− monocytes as described in the Supplementary Appendix.
Analyses
A description of the statistical analyses, along with a comprehensive report on all methods, is available in the Supplementary Appendix.
RESULTS
We determined the average timing of the daily activity peak (the acrophase) in 537 participants from the Rush Memory and Aging Project (MAP) using one week of actigraphic data (Fig 1). Demographic and other characteristics of the participants are presented in Table 1. As expected, age and absence of dementia were associated with earlier timing of the activity peak27, 30, 31 (Supplementary Table 4).
We identified 135 tagging polymorphisms in eighteen candidate genes known to be functionally important for circadian rhythms in animals (Supplementary Tables 1 and 2). These polymorphisms capture common genetic variation among our subjects of European ancestry. Each polymorphism was evaluated for association with acrophase timing in the MAP cohort. Of these, only rs7221412, 2·8kb downstream of PER1, was associated with acrophase timing after Bonferroni correction for 135 tests (Supplementary Table 5 and Fig 2A). Under our default additive model, each additional minor allele at rs7221412 was associated with a further delay in acrophase timing (p=6·0×10−7; Bonferroni corrected p=8·1×10−5), with 67 minutes separating the two homozygote allele classes. Genotype at rs7221412 was not associated with the amplitude of the activity rhythm (Supplementary Fig 2) and controlling for amplitude in the model relating genotype at rs7221412 to acrophase timing did not attenuate this relationship (unadjusted p=5·9×10−7). Similar results were seen in subjects with and without dementia (Supplementary Fig 3). Results were similar with the cosinor-determined activity nadir and acrophase as the phenotypes (Supplementary Figs 4 and 5), indicating that the results are not specific to the method used to analyse the activity data.
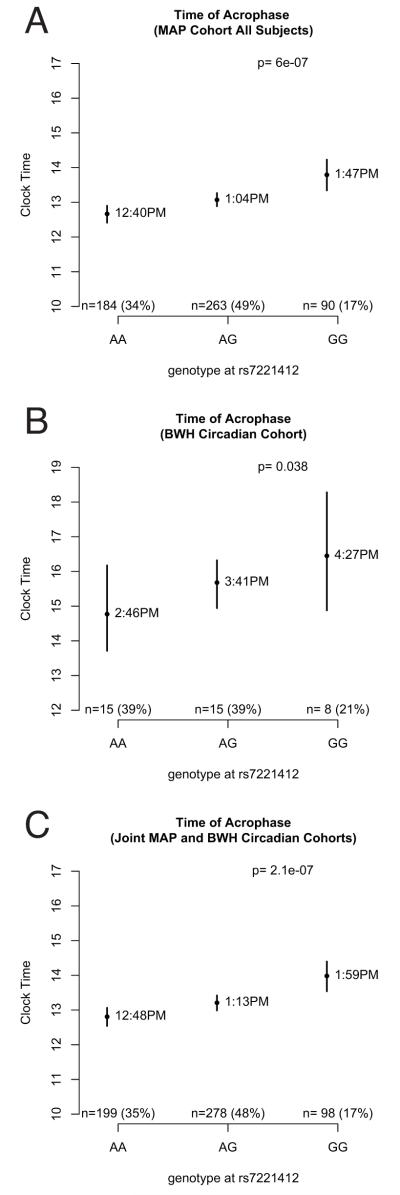
Fig 2. Association between the average timing of the acrophase of activity and genotype at rs7221412.
Dots indicate angular means, and bars indicate 95% confidence intervals of the mean. All significance values calculated using Fisher-Lee regression using an additive model, and adjusted for age, sex, and presence or absence of dementia. In the joint analysis, source cohort was added as an additional covariate. A: MAP cohort. B: BWH Circadian Cohort. C: Joint Analysis.
Characterization of the local association structure by testing all available polymorphisms (HapMap Release 22) within 20kb of rs7221412 for association with activity acrophase timing in the MAP cohort revealed a 14kb association peak overlapping with the 3′ end of the PER1 gene as well as downstream regions and containing rs7221412 at its centre (Fig 3).
Fig 3. Local association plot in the PER1 locus.
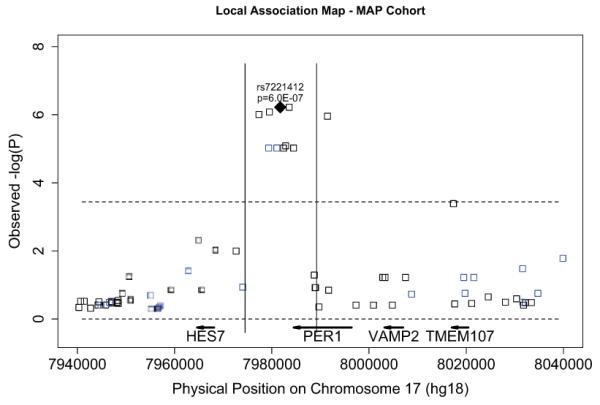
The plot displays the significance, as indicated by the −log10(p-value), of the associations between timing of the circadian acrophase, and genotype at polymorphisms flanking rs7221412. All significance values calculated with Fisher-Lee regression using an additive model, and adjusted for age, sex, and presence or absence of dementia. Blue squares indicate directly-genotyped polymorphisms; black square indicate polymorphisms whose genotypes were imputed (imputation quality score >0.90 for all imputed polymorphisms). Vertical black bars indicate the extent of the association peak. The black diamond indicates the index SNP rs7221412. The horizontal dotted line indicates the Bonferroni-corrected threshold for significance.
To confirm these observations, we genotyped rs7221412 and flanking polymorphisms in an independent cohort – the BWH Circadian cohort - and tested for associations with acrophase timing. rs7221412 remained significantly associated with acrophase timing (p=0·038, Fig 2B). In a combined analysis of both the MAP and BWH Circadian cohorts (n=575; Fig 2C) the strength of the association between rs7221412 and acrophase timing was enhanced (p=2·1×10−7).
To validate activity acrophase as a marker of circadian phase, we compared the timing of the activity acrophase to that of dim-light melatonin onset (DLMO) or of the temperature nadir under conditions of constant routine in a subset of the BWH Circadian cohort. In this validation subset, activity acrophase timing was strongly correlated with both the timing of DLMO and of the temperature nadir (r=0·7, p=0·0026 and r=0·75, p=0·009 respectively; Supplementary Fig 6), indicating that it was a reasonable marker of phase in this study. The small number of individuals in whom DLMO was measured (n=16) precluded meaningful direct assessment of the relationship between genotype at rs7221412 and DLMO.
Because differences in phase can reflect differences in the length of the period of individuals’ intrinsic biological rhythms31, 32, we tested for associations between rs7221412 genotype and intrinsic circadian period in the BWH Circadian cohort. rs7221412 genotype was not associated with the period of either the temperature or melatonin rhythms (Supplementary Fig 7), suggesting that it does not alter the length of the period of the intrinsic biological rhythm but rather displaces its entrained phase.
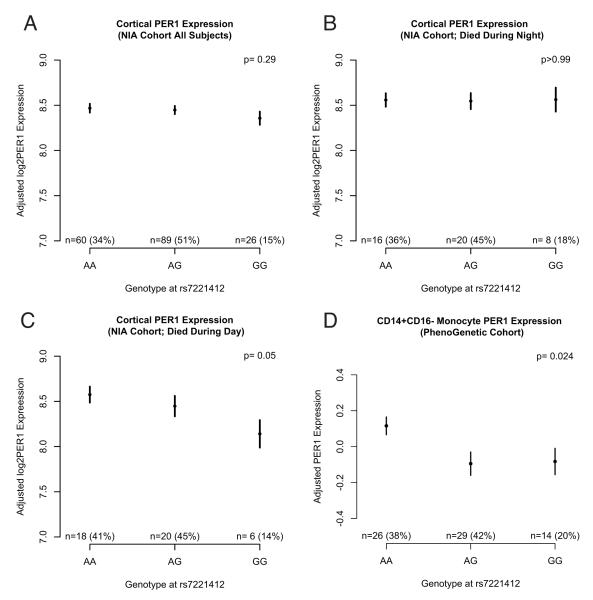
Alterations in PER1 expression can affect circadian phase33 and may be a mechanism mediating the association between rs7221412 and phase. Therefore, we assessed for associations between rs7221412 genotype and PER1 expression. In cerebral cortex expression data from the NIA cohort, rs7221412 genotype was not significantly associated with overall PER1 expression (p=0·29, Fig 4A). However, when the analysis was stratified by the inferred time of death (see Supplemental Appendix for details), a suggestive relationship emerged; rs7221412 genotype was associated with PER1 expression in samples from individuals inferred to have died during the day (p=0·05; Fig 4B), but not from those inferred to have died during the night (p>0.99; Fig 4C). A similar pattern was not seen for PER2 expression in cerebral cortex, suggesting that the association between rs7221412 and daytime PER1 expression was not merely attributable to overall phase changes in the molecular circadian clock (Supplementary Fig 8). In PBMCs from the PhenoGenetic and BWH MS Cohorts, all of which were collected during the day, similar associations were seen, (p=0·02 for CD14+CD16− monocytes in the PhenoGenetic cohort and p=0·20 for unselected PBMCs in the BWH MS cohort; Fig 4D and Supplementary Fig 9).
Fig 4. Adjusted cortical and CD14+CD16− monocyte PER1 transcript expression as a function of genotype at rs7221412.
A: cerebral cortex, NIA cohort, all participants. B: cerebral cortex, NIA cohort, participants inferred to have died during the night (see Supplementary Methods). C: cerebral cortex, NIA cohort, participants inferred to have died during the day (see Supplementary Methods). D: CD14+CD16− monocytes, PhenoGenetic cohort, all participants. Dots indicate means, and bars indicate 95% confidence intervals of the mean. Y-axis indicates log2 expression levels adjusted for methodological and clinical covariates as described in the Materials and Methods.
The incidence of death also varies by time of day9. We therefore assessed the association between rs7221412 genotype and time of death in the ROS/MAP cohort excluding subjects who were part of the primary association analysis. Time of death was obtained from autopsy reports. In both additive and recessive models, adjusting for age, sex, presence/absence of dementia, and source cohort, genotype at rs7221412 was associated with time of death (p=0·011 for the additive model and p=0·015 for the recessive model). Under the recessive model, the rs7221412GG group (n=134) had a mean time of death nearly seven hours later than the rs7221412AA/AG group (n=578) (5:50PM vs. 10:51AM; Supplementary Fig 10).
DISCUSSION
In this study, a polymorphism near PER1 was significantly associated with the timing of directly recorded human behavioral rhythms in two independent cohorts and with time of death in a third. The associated functional variant was mapped to a 14kb region straddling the 3′ end of the PER1 gene, and the region downstream of it. Moreover, there was suggestive evidence for a circadian time-dependent association between genotype at this variant and both cortical and PBMC PER1 expression, which could be a mechanism that mediates its effect. To our knowledge, this is the first genetic polymorphism shown to be associated with the timing of directly observed human behavioral rhythms at the population level, and the first showing evidence of association with time of death.
Methodological Considerations
In this study, biological circadian phase was inferred from actigraphic data. Actigraphy is one of the few objective means to gather circadian data from large cohorts in their home environments. The relationship between activity and circadian rhythms can be complex, with social and other factors influencing activity patterns. However, in the subset of participants who underwent actigraphy followed by monitoring of melatonin and temperature rhythms, two standard markers of circadian phase, activity acrophase timing was strongly correlated with the timing of both, suggesting that activity acrophase timing is a reasonable marker of circadian phase under the conditions of this study. Others have found a similarly strong correlation between actigraphic timing and biochemical markers of phase34.
We used three markers of actigraphic phase in this study – non-parametrically determined acrophase, and parametrically determined nadir and acrophase by the cosinor method. Although the precise effect sizes for rs7221412 were slightly different for these three markers, there was a consistent direction of effect for all three, and the associations were highly statistically significant (joint p=2.0×10−7 for non-parametric acrophase, and p=7.1×10−6 for cosinor acrophase and nadir), indicating that our results are not specific to the precise actigraphic phase marker used, and are not merely due to changes in the shape of the activity profile.
This study was not a comprehensive screen for genetic variants influencing circadian phase. Because of our sample size, we limited the first stage to genes with biological plausibility, examined only common variants, and validated only the most significant variant. With this strategy, several polymorphisms previously associated with subjective diurnal preference17-20 were not assessed. Moreover, of the 138 SNPs assessed, only 1 was found to be statistically significantly associated with the timing of activity rhythms below the Bonferroni-corrected threshold of 3.7×10−4, although a further 8 SNPs had an unadjusted p<0.05. This does not preclude a role for human homologs of animal clock genes other than PER1 in the generation of human circadian rhythms. Rather, the design and sample size of our study meant that it would have had power to detect only relatively common variants with large effect sizes, and less frequent variants with smaller effect sizes would have been missed. Indeed, post-hoc power estimations by simulation (see Supplementary Methods) indicate that we would have had 80% power to detect only those variants with a minor allele frequency of 0.45 and a difference in timing between the minor and major allele homozygote genotypes of at least 64 minutes. At a minor allele frequency of 0.10, the required effect size rises to a minimum difference in timing of 150 minutes or greater. It is likely that with larger sample sizes in future studies, additional variants in other genes will be shown to be associated with human circadian phase.
Genetic Determinants of Circadian Phase
Rare mutations in homologs of conserved circadian genes have been associated with extreme sleep timing in select human pedigrees15, 16. While these mutations corroborate the role, in humans, of clock genes identified from animal models, their population-level relevance is unclear. Meanwhile, several common variants have suggestive evidence of association with self-reported day/night preference17-20 or usual bedtime12. However, not all of those results have been replicated, and none of these variants has been associated with the timing of directly observed human behavior or another physiological marker of circadian timing in a real-world setting – an important consideration given that self-reported day/night preference is prone to misperception and poor recall, and that its relationship to physiological markers of circadian timing in real-world settings can be incomplete35.
This previous work established the importance, in humans, of evolutionarily conserved clock genes and provided evidence that variants in these genes can in principle influence human sleep and circadian traits. However, the variant rs7221412 differs from these previously identified variants in a number of important respects. First, unlike the highly penetrant variants identified in rare pedigrees15, 16, rs7221412 is a much more common variant (MAF 0·43) meaning that it is likely to play a much more important role at the population level. Second, genotype at rs7221412 was shown to be associated with the timing of directly observed human behavior – an objective physiological marker of circadian timing – as opposed to self-reported subjective day/night preference as has been the case for many of the previously reported variants17-20. This is important because while subjective diurnal preference is likely to be influenced in part by underlying biological circadian phase, these are not identical. For example, a PER3 variable length tandem repeat (VNTR) polymorphism has been reported to be associated with subjective diurnal preference17; however, it was subsequently shown not to be associated with underlying circadian phase as measured using actigraphy or serial serum cortisol, and indeed it appears that this PER3 VNTR influences subjective diurnal preference by affecting sleep homeostatic function rather than circadian phase35. Third, our result was highly statistically significant, with an unadjusted p-value approaching genome-wide significance, retained significance following rigorous correction for multiple comparisons, and was replicated in an independent cohort. By contrast, many of the previously reported associations between common variants and subjective diurnal preference had nominal p-values with borderline statistical significance (0.01<p<0.05)17-19 that would not have been statistically significant after correction for multiple comparisons18, 19. Moreover, most of these associations have not been replicated, and where replication was attempted, discordant results have been found20, 36. Fourth, genotype at rs7221412 showed evidence of association not only with the timing of human behavioral rhythms, but also with PER1 expression in two additional cohorts, and with time of death in another, providing several lines of converging evidence of the population-level physiological and clinical significance of this variant. Examination of rs7221412 and other common variants near PER1 in association with objective measures of circadian timing in additional large cohorts will be needed to further establish the association of this and other PER1 variants with circadian timing in different human populations.
Remarkably for so common a variant, the effect size was large, with over one hour separating the daily activity peak in homozygotes for one or the other allele. This result was significant even after correction for multiple comparisons. Several other polymorphisms near rs7221412 are in linkage disequilibrium with it and exhibited a similar level of association, so identification of the precise causal variant within the PER1 locus will require further studies, likely involving sequencing of the 3′ end of PER1 as well as the PER1 downstream region in a large number of people with objective characterization of circadian timing.
Functional Correlates
The Per locus was first identified in Drosophila melanogaster37 and shortly thereafter three orthologs were identified in the mouse10. PER1-3 play a critical role in a negative-feedback transcriptional cycle at the core of the molecular circadian clock10. Transcription of PER1-3 is activated by a complex of CLOCK and BMAL110. Once translated, PER1-3 heterodimerize with CRY1-2 and translocate into the nucleus to inhibit their own transcriptional activation by the CLOCK:BMAL1 complex, forming a 24-hour negative feedback loop10.
Another PER1 polymorphism, rs2735611 4·5kb upstream of rs7221412 (r2=0·049), has been associated with self-reported day/night preference, although phase was not measured directly19. In our study, rs2735611 was ascertained in the MAP but not BWH Circadian cohort, and was not associated with activity acrophase timing (p=0·24). As day/night preference was not assessed in the MAP cohort, we cannot comment on whether rs2735611 would have been associated with this phenotype.
Coding mutations in PER215 and CKSKNID16, a PER2 kinase, are associated with autosomal dominant familial advanced sleep phase syndrome (ADFASPS) in rare pedigrees. In these pedigrees, altered phosphorylation of PER2 is hypothesized to lead to a shortened intrinsic period and hence advanced phase. By contrast, rs7221412 was not associated with significant differences in the length of the intrinsic circadian period. While it remains possible that our study did not have sufficient power to detect a subtle effect, an alternative is that the association of rs7221412 with phase may be mediated by factors independent of period, such as a differential phase-response to circadian synchronizing signals. We hypothesize that these changes in turn may be mediated by altered PER1 expression. In this regard, rs7221412A was suggestively associated with higher cortical PER1 expression during the presumptive day, but not night. This effect was also seen in PBMCs collected during the day, suggesting that rs7221412A may be associated with a differential increase PER1 expression during the day, leading to an earlier peak in the activity acrophase. This putative association between genotype at rs7221412 and PER1 expression will need to be replicated in other human tissues obtained from other cohorts but is consistent with the observation that the association peak containing rs7221412 overlaps not only with several exons and introns at the 3′ end of PER1 but also with the 3′ untranslated region and a phylogenetically conserved region of open chromatin immediately downstream of PER1 as revealed by examination of existing maps of DNase hypersensitivity 38 and phylogenetic conservation39 (Supplementary Fig 11). These conserved downstream regions in particular are suggestive of a potential cis-regulatory region. Identification of the precise functional variant mediating the rs7221412 association will allow dissection of its molecular and physiological effects in animal model systems.
Time of Death
It has been shown that the probability of non-traumatic death9 as well as stroke3, ST-elevation myocardial infarction6, and ventricular arrhythmia7, varies by time of day, with a peak in the late morning or early afternoon. Consistent with this, in the ROS/MAP cohort, death was most likely to occur in the late morning / early afternoon. However, this varied with rs7221412 genotype, with the rs7221412GG group having a mean time of death nearly seven hours later than the rs7221412AA/AG group. There are several possible interpretations for this disparity. First, it may reflect primarily genotype-related group differences in circadian phase in the days leading up to death. Whereas when individuals are relatively well, their biological circadian tendencies are constrained by social commitments, proximate to death, these social constraints are no longer relevant and the rs7221412GG and rs7221412AA/AG groups may have drifted apart by seven hours in the phase of their underlying biological rhythms, accounting for the large difference in mean time of death. A second possibility is that rs7221412 genotype confers differential susceptibility to causes of death that peak at different times of the day. Cause of death in the ROS/MAP cohort was not available for the vast majority of participants. Moreover, circadian phase was not measured by actigraphy near the time of death (and probably would not have been valid if it had been measured given the nature of terminal illness in many people). Therefore, it was not possible to distinguish these two possibilities with the available data. Replication of these results in additional cohorts in whom time of death and cause of death are carefully recorded will allow differentiation of the two potential interpretations noted above.
Conclusions and Future Directions
This work identifies a 14kb region overlapping with the 3′ end of PER1 containing a polymorphism associated with a greater than one hour difference in the timing of directly observed human behavioral rhythms and a greater than six hour difference in average time of death. It suggests that there may exist additional common variants that influence circadian traits. Studies with larger sample sizes will be helpful to replicate the current findings and facilitate identification and validation of additional variants influencing circadian phase. Meanwhile, further work is needed to delineate the effect of rs7221412 on other clinical traits and diseases with circadian modulation, such as cognitive function, stroke, epilepsy, tumor growth, or cardiac arrhythmia. One might hypothesize, for instance, that genotype at rs7221412 might influence the optimal timing of cognitive testing in dementia, or drug therapy in epilepsy, stroke, or cardiac disease. Moreover, recognition and prediction of individual biological rhythms may provide important dividends not just in the clinical arena but also in identifying individuals suited to early or late work, school, or other schedules.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Mr. Dong Tran for programming assistance, Mr. Gregory Klein and Mr. John Gibbons for assistance with data distribution, Drs. Mirjam Y Münch, Daniel A. Cohen, and Elizabeth B. Klerman for their assistance in obtaining the phenotyping data from the BWH Circadian cohort, Patrice Vamivakas and Lisa-Marie Farwell for assistance with genotyping, Michelle H. Lee for assistance with data from the PhenoGenetic cohort, the participants in the Rush Memory and Aging Project and their families, and the participants in the Religious Orders Study.
We thank the Brigham and Women’s Hospital PhenoGenetic Project for providing CD14+CD16− peripheral blood mononuclear cells from healthy subjects that were used to examine PER1 expression in this study.
For the NIA cohort, many data and biomaterials were collected from several National Institute on Aging (NIA) and National Alzheimer’s Coordinating Centre (NACC, grant #U01 AG016976) funded sites. Amanda J. Myers, PhD (University of Miami, Department of Psychiatry) and John A. Hardy, PhD (Reta Lila Weston Institute, University College London) collected and prepared the series. Marcelle Morrison-Bogorad, PhD., Tony Phelps, PhD and Walter Kukull PhD are thanked for helping to co-ordinate this collection.The directors, pathologist and technicians involved include: National Institute on Aging: Ruth Seemann, John Hopkins Alzheimer’s Disease Research Centre (NIA grant # AG05146): Juan C. Troncoso, MD, Dr. Olga Pletnikova, University of California, Los Angeles (NIA grant # P50 AG16570):Harry Vinters, MD, Justine Pomakian, The Kathleen Price Bryan Brain Bank, Duke University Medical Centre (NIA grant #AG05128, NINDS grant # NS39764, NIMH MH60451 also funded by Glaxo Smith Kline): Christine Hulette, MD, Director, John F. Ervin, Stanford University: Dikran Horoupian, MD, Ahmad Salehi, MD, PhD, New York Brain Bank, Taub Institute, Columbia University (NYBB): Jean Paul Vonsattel, MD, Katerina Mancevska, Massachusetts General Hospital: E. Tessa Hedley-Whyte, MD, Karlotta Fitch, University of Michigan (NIH grant P50-AG08671): Dr. Roger Albin, Lisa Bain, Eszter Gombosi, University of Kentucky (NIH #AG05144): William Markesbery, MD, Sonya Anderson, Mayo Clinic, Jacksonville: Dennis W. Dickson, MD, Natalie Thomas, University Southern California: Caroll A. Miller, MD, Jenny Tang, M.S., Dimitri Diaz, Washington University, St Louis Alzheimer’s Disease Research Center (NIH #P50AG05681): Dan McKeel, MD, John C. Morris, MD, Eugene Johnson, Jr., PhD, Virginia Buckles, PhD, Deborah Carter, University of Washington, Seattle (NIH #P50 AG05136):Thomas Montine, MD, PhD, Aimee Schantz, MEd., University of Pennsylvania School of Medicine, Alzheimer’s Disease Research Center: John Q Trojanowski, MD, Virginia M Lee, MD, Vivianna Van Deerlin, MD, Terry Schuck, Boston University Alzheimer’s Disease Research Center (NIH grant P30-AG13846): Ann C. McKee, Carol Kubilus Sun Health Research Institute, Arizona (NIA #P30 AG19610): Joseph Rogers, PhD, Thomas G. Beach, MD, PhD, Lucia I. Sue Emory University: Bruce H. Wainer, MD, PhD, Marla Gearing, PhD, University of Texas, Southwestern Medical School: Charles L. White, III, M.D., Roger Rosenberg, Marilyn Howell, Joan Reisch, University of California, Davis: William Ellis, MD, Mary Ann Jarvis, Rush University Medical Center, Rush Alzheimer’s Disease Center (NIH #AG10161): David A. Bennett, M.D. Julie A. Schneider, MD, MS, Karen Skish, MS, PA (ASCP)MT, Wayne T Longman, University of Miami/NPF Brain Endowment Bank: Deborah C. Mash, MD, Margaret J Basile, Mitsuko Tanaka The authors also wish to thank the following funding sources: Canadian Institutes of Health Research Bisby Fellowship; American Academy of Neurology Clinical Research Training Fellowship; Dana Foundation Clinical Neuroscience Grant; NIH grants R01 NS072337, R01 AG24480, R01 AG17917, R01 AG151819, R01 AG30146, P30 AG10161, R01 HL080978, P01 AG09975, R21 AT02571, ARRA RC2 GM093080, AFOSR FA 9550-06-0080/O5NL132, AG034504, and the Translational Genomics Research Institute.
REFERENCES
- 1.Wyatt JK, Ritz-De Cecco A, Czeisler CA, Dijk DJ. Circadian temperature and melatonin rhythms, sleep, and neurobehavioral function in humans living on a 20-h day. Am J Physiol. 1999;277:R1152–1163. doi: 10.1152/ajpregu.1999.277.4.r1152. [DOI] [PubMed] [Google Scholar]
- 2.Casetta I, Granieri E, Portaluppi F, Manfredini R. Circadian variability in hemorrhagic stroke. JAMA. 2002;287:1266–1267. doi: 10.1001/jama.287.10.1266. [DOI] [PubMed] [Google Scholar]
- 3.Turin TC, Kita Y, Rumana N, et al. Morning surge in circadian periodicity of ischaemic stroke is independent of conventional risk factor status: findings from the Takashima Stroke Registry 1990-2003. Eur J Neurol. 2009;16:843–851. doi: 10.1111/j.1468-1331.2009.02605.x. [DOI] [PubMed] [Google Scholar]
- 4.Kang JE, Lim MM, Bateman RJ, et al. Amyloid-beta dynamics are regulated by orexin and the sleep-wake cycle. Science. 2009;326:1005–1007. doi: 10.1126/science.1180962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loddenkemper T, Vendrame M, Zarowski M, et al. Circadian patterns of pediatric seizures. Neurology. 2011;76:145–153. doi: 10.1212/WNL.0b013e318206ca46. [DOI] [PubMed] [Google Scholar]
- 6.Holmes DR, Jr., Aguirre FV, Aplin R, et al. Circadian rhythms in patients with ST-elevation myocardial infarction. Circ Cardiovasc Qual Outcomes. 2010;3:382–389. doi: 10.1161/CIRCOUTCOMES.109.913343. [DOI] [PubMed] [Google Scholar]
- 7.Englund A, Behrens S, Wegscheider K, Rowland E, European 7219 Jewel Investigators Circadian variation of malignant ventricular arrhythmias in patients with ischemic and nonischemic heart disease after cardioverter defibrillator implantation. J Am Coll Cardiol. 1999;34:1560–1568. doi: 10.1016/s0735-1097(99)00369-1. [DOI] [PubMed] [Google Scholar]
- 8.Eriguchi M, Levi F, Hisa T, et al. Chronotherapy for cancer. Biomed Pharmacother. 2003;57(Suppl 1):92s–95s. doi: 10.1016/j.biopha.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Mitler MM, Hajdukovic RM, Shafor R, et al. When people die. Cause of death versus time of death. Am J Med. 1987;82:266–274. doi: 10.1016/0002-9343(87)90067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans DS, Snitker S, Wu SH, et al. Habitual sleep/wake patterns in the Old Order Amish: heritability and association with non-genetic factors. Sleep. 2011;34:661–669. doi: 10.1093/sleep/34.5.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottlieb DJ, O’Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barclay NL, Eley TC, Buysse DJ, et al. Diurnal preference and sleep quality: same genes? A study of young adult twins. Chronobiol Int. 2010;27:278–296. doi: 10.3109/07420521003663801. [DOI] [PubMed] [Google Scholar]
- 14.Koskenvuo M, Hublin C, Partinen M, et al. Heritability of diurnal type: a nationwide study of 8753 adult twin pairs. J Sleep Res. 2007;16:156–162. doi: 10.1111/j.1365-2869.2007.00580.x. [DOI] [PubMed] [Google Scholar]
- 15.Toh KL, Jones CR, He Y, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Padiath QS, Shapiro RE, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 17.Archer SN, Robilliard DL, Skene DJ, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–415. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 18.Carpen JD, Archer SN, Skene DJ, et al. A single-nucleotide polymorphism in the 5′-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14:293–297. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- 19.Carpen JD, von Schantz M, Smits M, et al. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J Hum Genet. 2006;51:1122–1125. doi: 10.1007/s10038-006-0060-y. [DOI] [PubMed] [Google Scholar]
- 20.Katzenberg D, Young T, Finn L, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–576. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 21.Bennett DA, Schneider JA, Buchman AS, et al. The Rush Memory and Aging Project: study design and baseline characteristics of the study cohort. Neuroepidemiology. 2005;25:163–175. doi: 10.1159/000087446. [DOI] [PubMed] [Google Scholar]
- 22.Chibnik LB, Shulman JM, Leurgans SE, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69:560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchman AS, Wilson RS, Bennett DA. Total daily activity is associated with cognition in older persons. Am J Geriatr Psychiatry. 2008;16:697–701. doi: 10.1097/JGP.0b013e31817945f6. [DOI] [PubMed] [Google Scholar]
- 24.Myers AJ, Gibbs JR, Webster JA, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 25.De Jager PL, Jia X, Wang J, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson W, Tong YL, Lee JK, Halberg F. Methods for cosinor-rhythmometry. Chronobiologia. 1979;6:305–323. [PubMed] [Google Scholar]
- 27.Duffy JF, Dijk DJ, Klerman EB, Czeisler CA. Later endogenous circadian temperature nadir relative to an earlier wake time in older people. Am J Physiol. 1998;275:R1478–1487. doi: 10.1152/ajpregu.1998.275.5.r1478. [DOI] [PubMed] [Google Scholar]
- 28.Czeisler CA, Duffy JF, Shanahan TL, et al. Stability, precision, and near-24-hour period of the human circadian pacemaker. Science. 1999;284:2177–2181. doi: 10.1126/science.284.5423.2177. [DOI] [PubMed] [Google Scholar]
- 29.Webster JA, Gibbs JR, Clarke J, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harper DG, Stopa EG, McKee AC, et al. Dementia severity and Lewy bodies affect circadian rhythms in Alzheimer disease. Neurobiol Aging. 2004;25:771–781. doi: 10.1016/j.neurobiolaging.2003.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Duffy JF, Cain SW, Chang AM, et al. Quantification of Behavior Sackler Colloquium: Sex difference in the near-24-hour intrinsic period of the human circadian timing system. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1010666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gronfier C, Wright KP, Jr., Kronauer RE, Czeisler CA. Entrainment of the human circadian pacemaker to longer-than-24-h days. Proc Natl Acad Sci U S A. 2007;104:9081–9086. doi: 10.1073/pnas.0702835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 2009;276:5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- 34.Archer SN, Viola AU, Kyriakopoulou V, et al. Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. Sleep. 2008;31:608–617. doi: 10.1093/sleep/31.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viola AU, Archer SN, James LM, et al. PER3 polymorphism predicts sleep structure and waking performance. Curr Biol. 2007;17:613–618. doi: 10.1016/j.cub.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 36.Robilliard DL, Archer SN, Arendt J, et al. The 3111 Clock gene polymorphism is not associated with sleep and circadian rhythmicity in phenotypically characterized human subjects. J Sleep Res. 2002;11:305–312. doi: 10.1046/j.1365-2869.2002.00320.x. [DOI] [PubMed] [Google Scholar]
- 37.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyle AP, Davis S, Shulha HP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.