Abstract
Enamel crystals are unique in shape, orientation and organization. They are hundreds of thousands times longer than they are wide, run parallel to each other, are oriented with respect to the ameloblast membrane at the mineralization front and are organized into rod or interrod enamel. The classical theory of amelogenesis postulates that extracellular matrix proteins shape crystallites by specifically inhibiting ion deposition on the crystal sides, orient them by binding multiple crystallites and establish higher levels of crystal organization. Elements of the classical theory are supported in principle by in vitro studies; however, the classical theory does not explain how enamel forms in vivo. In this review, we describe how amelogenesis is highly integrated with ameloblast cell activities and how the shape, orientation and organization of enamel mineral ribbons are established by a mineralization front apparatus along the secretory surface of the ameloblast cell membrane.
Keywords: ameloblastin, amelogenin, enamelin, mineralization front, tooth
Introduction
Dental enamel is comprised of highly oriented, thread-like crystallites of calcium hydroxyapatite (HAP) organized into rod and interrod structures.1 The rods are about 5 µm in cross-sectional diameter2 and comprised of bundles of parallel crystallites (about 26 nm×68 nm in cross-section) packed at a density of ∼550 crystallites·µm−2, or roughly 40 thousand crystallites per rod.3 Enamel crystallites appear to extend all the way from the dentino-enamel junction (DEJ) to the surface of the tooth.4
Understanding the mechanism of dental enamel formation requires reconciliation of ideas from scientists who grow crystals in vitro and scientists who study the biological process of amelogenesis in vivo. These two perspectives have never assimilated satisfactorily and seem to be on different courses. Better characterization of amelogenesis at the biochemical and genetic levels and in knockout mice with developmental enamel malformations has greatly advanced our understanding of events in vivo. Advances on the biological side are yielding new theories that are replacing the ‘classical' theory5,6 of dental enamel formation. This review states the case for a model of enamel formation postulating that the initial mineral in enamel is amorphous calcium phosphate (ACP) and that enamel mineral ribbons are the product of a specialized mineralization front: an apparatus along the secretory surface of the ameloblast plasma membrane that generates, shapes and elongates enamel mineral ribbons.
Basement membrane
Enamel crystallites initiate at the DEJ immediately following fenestration and removal of the basement membrane beneath fully differentiated preameloblasts.7 The basement membrane is replaced by a membrane-associated apparatus comprised of enamel proteins that coats the secretory surface of the ameloblast plasma membrane and is necessary for the initiation and elongation of enamel mineral ribbons. The genes encoding enamel proteins are themselves descended from a basement membrane gene: SPARC-like protein 1 (Sparcl1).8,9,10 The initiation of enamel formation requires successful replacement of the basement membrane with the mineralization front apparatus. This does not occur properly when proteins associated with the basement membrane, such as collagen 17,11,12,13 α6/β4 integrin14,15,16 or laminin-33217,18,19,20 or proteins associated with the mineralization front, such as enamelin21 or ameloblastin,22,23 are defective or missing.24,25,26 Amelogenin (the most abundant enamel matrix protein) appears to be sparse at the mineralization front23,27,28 and a thin layer of enamel (∼15 µm vs. ∼110 µm) is deposited in AmelX knockout mice.29
When the initial enamel starts to form, the distal surface of ameloblasts is folded, with cell processes extending to contact the irregular surface of the underlying predentin.30 The initial enamel has the same characteristic shape of mineral ribbons that is observed throughout the secretory stage of amelogenesis. At the ameloblast membrane (mineralization front), the mineral ribbons are thin slits (1.5 nm×15 nm) in cross-section.3 On the dentin surface, these ribbons are closely associated with collagen, which can be recognized by its characteristic cross-banding.30 From the onset, the initial enamel ribbons are tightly integrated (attached) to dentin, principally through connections to its organic matrix31,32,33 and extend from there to the mineralization front (Figure 1) where the ribbons are actively elongated by the addition of ions or ACP to the tips of the mineral ribbons. At first, irregular depressions on the dentin surface are filled, creating ‘islands of enamel'. The ameloblast cell processes retreat as the ribbons elongate. The cell membrane becomes smooth, forming an uninterrupted mineralization front, and the underlying enamel layer becomes continuous.30,34 As ribbon elongation proceeds, each ameloblast develops a Tomes' process. The mineralization front becomes discontinuous again, interrupted by the non-secretory portions of the Tomes' processes that do not support mineral ribbon elongation. The architecture of the ameloblast distal membrane and its division into secretory and non-secretory surfaces establishes the rod/interrod structural hierarchy characteristic of enamel.30,35,36 In rat incisors, ameloblasts move away from the dentin at a rate of 13.5 µm per day (0.56 µm·h−1 or about 9.3 nm·min−1).37 As the ribbons generally form at an angle to the line perpendicular to the DEJ, this represents a minimum rate for the lengthening of enamel ribbons at the mineralization front.
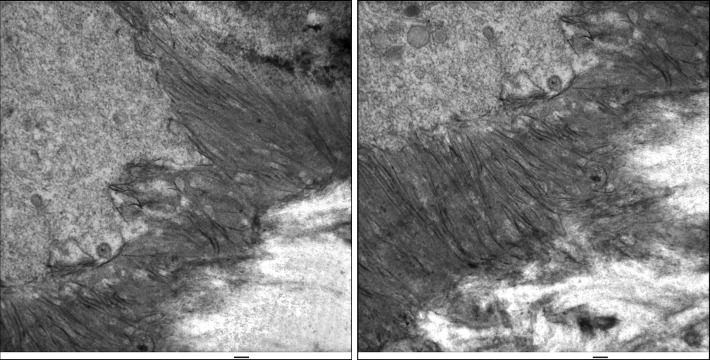
Figure 1.
Formation of initial enamel. Day 7 mouse mandibles were fixed with 2.5% glutaraldehyde in sodium cacodylate buffer and post-fixed with osmium tetroxide. Sections were stained with uranyl acetate, then lead citrate, and viewed by TEM. Ameloblasts are on the upper left. Banded collagen fibers are on the lower right. The enamel ribbons initiate on the dentin surface in close association with collagen and the mineralization front on the ameloblast membrane. Scale bars=100 nm. TEM, transmission electron microscopy.
Problems with the classical theory
The classical theory of enamel formation held that proteins bind specifically and selectively to the sides of crystalline ribbons, inhibiting their growth in width and thickness and permitting growth only in the C-axis direction.38 The purpose of proteolytic cleavages was to remove the protein inhibitors from the sides of the crystallites to allow them to mature (to grow in width and thickness). HAP's sixfold symmetry39 might have precluded the molecular recognition of selected crystal faces that was required to produce the observed slit-like cross-sections of enamel ribbons. This problem and others were solved by proposing the existence of an octacalcium phosphate precursor phase.40,41,42 Substantial evidence supports the classical theory in principle, but the classical theory is not consistent with observations of how enamel forms in vivo.
Enamel proteins bind HAP in vitro and inhibit their growth, and this growth inhibition can be reduced by the addition of enamel proteases such as matrix metalloproteinase 20 (MMP20)43 or kallikrein 4 (KLK4).44 In vivo, however, there is a high rate of mineralization on the sides of the enamel crystals in the transition/early maturation stage when enamel proteins are still abundant and should be inhibiting such growth.45 There is also a substantial amount of crystal maturation in Klk4 null mice, even though the enamel proteins are not effectively removed from the enamel layer during the maturation stage of amelogenesis.46
Electron diffraction and electron probe microanalysis of enamel ribbons near the mineralization front reveal a very poorly crystalline HAP with a low and variable Ca/P molar ratio and no evidence of octacalcium phosphate.47 Newly formed enamel mineral ribbons are ACP that transforms into HAP.32,48 The key finding is that the size, shape and spatial organization of the enamel ribbons are established prior to their crystallization. The characteristic enamel ribbons with slit-like cross-sections are not dictated by ‘stereochemical' interactions with selected crystal faces, as they are not yet crystalline. The mineral in the ribbons has no shape of its own. The ribbon shape must be due to an external influence, such as the shape of the space, or mold, within which it forms. Like the classical theory based upon an octacalcium phosphate precursor, the concept of an ACP precursor is also supported in principle by in vitro studies. The first synthetic HAP crystals suitable for X-ray diffraction were made by hydrolyzing solid CaHPO4 (monetite) into HAP.49 The solid–solid conversion of ACP into HAP occurs in vitro.50 Amelogenin stabilizes ACP and delays its conversion to HAP51 in vitro, although the ACP phase in enamel in vivo seems to be relatively short-lived.32 An ACP precursor phase is also thought to play a role in dentin and bone mineralization.52
Initial enamel ribbons
Enamel proteins, such as amelogenin, are secreted prior to the onset of enamel mineralization as the basement membrane is degraded and ameloblasts come into contact with the collagen-rich predentin matrix.53 Ameloblastin, enamelin, MMP20 and dentin sialophosphoprotein are also expressed by early ameloblasts54 and could be part of the organic component that helps fasten the incipient enamel ribbons to collagen. MMP20 activity seems to be critical for this attachment as a thin line of hypermineralized enamel at the DEJ in wild-type mice is missing in Mmp20 null mice55 and enamel delaminates at the DEJ (Figure 2). All forming enamel, including the initial enamel in contact with dentin, is directly associated with the ameloblast membrane and the ribbons are elongated by cycles of mineral deposition at the mineralization front throughout the secretory stage of amelogenesis.
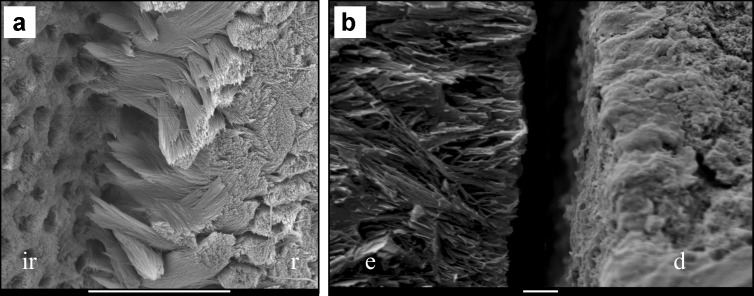
Figure 2.
Preferred fracture levels in Klk4 and Mmp20 null mice. (a) Klk4 null mouse enamel tends to fracture where the initial enamel forms the first interrod enamel (ir) and the base of the rods (r) just above the DEJ.46 Bar=10 µm. (b) Mmp20 null mouse enamel (e) separates from dentin (d) right at the DEJ. Bar=1 µm. DEJ, dentino-enamel junction; KLK4, kallikrein 4; MMP20, matrix metalloproteinase 20.
The mineralization front apparatus
The classical theory ignores the mineralization front apparatus; however, in vivo studies demonstrate that the mineralization front apparatus is literally the essence of enamel formation. When enamelin or ameloblastin are missing or defective, the mineralization front apparatus fails and the enamel layer is absent.25,26 No mineralization front apparatus equals no enamel. The mineralization front shapes the enamel ribbons before they are crystalline48 and orients them. In humans, the growing enamel ribbons are oriented perpendicular to the mineralization front.35 When ameloblasts develop Tomes' processes, the contour of the mineralization front changes so that different faces of the mineralization front are oriented in different directions. This is the basis for the hierarchical organization of enamel ribbons into rod and interrod structures (Figure 3).34,56 In rodents, the Tomes' processes in one row of ameloblasts are inclined in the same direction, while those of adjacent rows are inclined in the opposite direction,57 resulting in a decussating (X-shaped) pattern of enamel rods, each filled with enamel ribbons oriented along the long axis of the rod, but at an angle to the rod in the adjacent row. Near the end of the secretory stage of amelogenesis, ameloblasts retract their Tomes' processes and the final enamel, like the initial enamel deposited prior to formation of the Tomes' processes, lacks rod/interrod divisions and the ribbons run perpendicular to the enamel surface. The mineralization front determines the number, shape and orientation of the mineral ribbons and the topography of the mineralization front establishes rod/interrod organization.
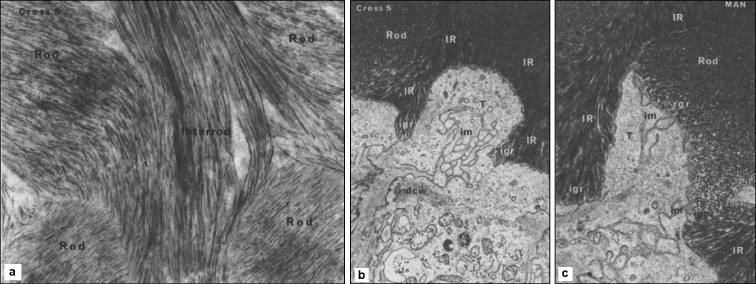
Figure 3.
Crystal shape, orientation, and organization are determined at the mineralization front. Three TEMs of an undecalcified sections of secretory stage inner enamel stained with uranyl acetate and lead citrate reproduced with permission from34 (a) Rat incisor section showing that within the rods, crystallites run parallel to the rod axis (×70 000). Between adjacent rods the interrod crystallites run at almost right angles. (b) Developing human tooth showing the relationship of the ameloblasts to the interrod and rod growth regions (×20 000). The Tomes' process (T) is surrounded by interrod enamel (IR) and is lined by a relatively smooth membrane. Interrod growth regions (igr) are seen at the prong tips on either side. The section has cut the rod growth region tangentially and it thus appears in the center of the process (im). dcw: distal cell web. (c) The rod growth region (rgr) and the forming rod are seen on one surface of Tomes' process (T). The opposite surface faces interrod enamel (lR). Interrod growth regions (igr) are seen at the prong tips on either side. Infolded membranes (im) are seen at interrod and rod growth regions. TEM, transmission electron microscopy.
There is virtually no evidence that describes in detail how enamel ribbons are extended at the mineralization front apparatus; however, the requirements of the process limit the possibilities and allow working hypotheses to be proposed. Perhaps calcium and phosphate channels in the plasma membrane above slits in the mineralization front apparatus concentrate these ions in a restricted extracellular space where ACP precipitates. Mineral precipitation may reduce the free calcium and phosphate ion concentrations within the space to allow more ions to be channeled into it to support continued ribbon elongation. The mechanism of calcium ion entry into the enamel matrix is unknown, but several intriguing observations have been reported. Ameloblasts do not concentrate calcium in anticipation of mineralization.58 Calcium flux into enamel is regulated by ameloblasts and is slower than it would be if it were not regulated.59 Calcium release-activated calcium modulator 1 acts as the pore-forming subunit for calcium release-activated calcium channels in the plasma membrane.60 Mutations in ORAI1 result in dental enamel malformations.61
Although the defining feature of the secretory stage of amelogenesis is the expansion of the enamel layer by the lengthening of mineral ribbons by the mineralization front apparatus, the mineral ribbons also grow in width and thickness,3 so that calcium deposition as a whole is relatively uniform throughout the matrix.62
Secretory stage enamel proteins
The major proteins in the secretory stage enamel extracellular matrix are amelogenin,63 ameloblastin64 and enamelin21 (which is not tuftelin). These proteins are cleaved by MMP2065 in the extracellular matrix. Amelx, Ambn, Enam and Mmp20 are genes specialized for dental enamel formation. Only dental enamel defects are observed in the corresponding four null mice and these genes degenerate in vertebrates that have stopped making teeth or enamel during evolution.66,67,68 In the pig, where tooth size is sufficient to isolate enamel protein cleavage products in quantity, the cleavage sites of enamel proteins have been characterized and correspond to the exact sites that are cleaved by MMP20 in vitro.69,70,71,72 Secretory stage enamel proteins in Mmp20 null mice are largely intact (uncleaved).73 Typical enamel ribbons are observed near the DEJ in Mmp20 null mice.74
Based upon the classical theory, one might expect that Mmp20 null mice would produce a normal enamel layer with crystallites that do not mature. One might also expect that the Amelx, Ambn or Enam null mice would make thicker hexagonal or plate-like crystals. None of these expectations are met. The Enam and Ambn null mice fail to make enamel and show significant ameloblast cell pathology. Tomes' processes do not form properly in Mmp20 null mouse ameloblasts, which produce a disorganized, thin enamel.
Transition
As long as the mineralization front is sustained by ameloblasts, the enamel ribbons grow longer and the enamel layer thickens.75 After laying down a final layer of aprismatic enamel, secretory stage ameloblasts transition into modulating, maturation stage ameloblasts, with ∼25% of ameloblasts undergoing apoptosis.45,76 Ameloblast transition involves major changes in cell size and architecture,77 the onset of KLK4 secretion,78 and replacement of the mineralization front apparatus with a novel basement membrane containing amelotin79 and odontogenic, ameloblast-associated80 and other proteins. When the mineralization front apparatus is gone, the lengthening of enamel ribbons is over. All subsequent mineralization involves the widening and thickening of ribbon-like crystallites previously formed during the secretory stage.
Enamel maturation
During the maturation stage of amelogenesis, the enamel crystallites deposited during the secretory stage grow exclusively in width and thickness and the enamel proteins are removed.45 In humans, the maturation stage for the permanent teeth lasts about 4 or 5 years. Maturation stage ameloblasts modulate between ruffle-ended and smooth-ended morphologies.81 In rat mandibular incisors, maturation ameloblasts modulate three times a day with most of their time being spent in the ruffle-ended form.82 Calcium entry into the matrix appears to be mainly through ruffle-ended ameloblasts.83 The enamel beneath ruffle-ended ameloblasts is mildly acidic (pH ∼6) as a consequence of mineral deposition and is neutralized by bicarbonate to physiologic pH (∼7.2) under smooth-ended ameloblasts.84 As mineralization proceeds, enamel proteins are progressively degraded by KLK4 and the digestion products are reabsorbed into ameloblasts so that the crystallites can grow together and interlock.46 In developing teeth, KLK4 is specifically expressed by transition and maturation stage ameloblasts.85 There is trace expression of KLK4 in other tissues besides teeth,86 but the only phenotype in mice and humans that lack KLK4 is found in the enamel.87 In the absence of KLK4 expression, there is substantial retention of enamel proteins within the enamel matrix layer even as the teeth erupt. The enamel layer is increasingly hypomineralized from the enamel surface to the DEJ.55,88 Recently, there has been much interest in defining the mechanisms of ion transport (Ca2+, PO43−, H+, HCO32−)75,89,90,91 to improve our understanding of enamel maturation.
Conclusion
Dental enamel forms by the deposition of characteristic, non-crystalline, mineral ribbons by a mineralization front apparatus closely associated with the secretory surfaces of the ameloblast plasma membrane. The shape and orientation of enamel mineral ribbons is established at the mineralization front and is not due to stereospecific inhibition of mineral deposition on selected crystal faces by acidic enamel proteins. The hierarchical organization of enamel ribbons into rod and interrod enamel is established by the topographical re-configuration of the mineralization front that occurs with formation of the Tomes' process. The mineralization front apparatus is the key to enamel formation, and significant advances in our understanding of amelogenesis will be realized by gaining a better understanding of molecular events occurring at the enamel mineralization front.
Acknowledgments
We thank Dorothy Sorenson and Krystyna Pasyk of the U of M Microscopy and Image Analysis Laboratory for their help in producing the images in Figure 1. This study was supported by NIDCR/NIH grant projects DE011301 and DE061854.
References
- Selvig KA. The crystal structure of hydroxyapatite in dental enamel as seen with the electron microscope. J Ultrastruct Res. 1972;41 3:369–375. doi: 10.1016/s0022-5320(72)90076-7. [DOI] [PubMed] [Google Scholar]
- Meckel AH, Griebstein WJ, Neal RJ. Structure of mature human dental enamel as observed by electron microscopy. Arch Oral Biol. 1965;10 5:775–783. doi: 10.1016/0003-9969(65)90131-7. [DOI] [PubMed] [Google Scholar]
- Kerebel B, Daculsi G, Kerebel LM. Ultrastructural studies of enamel crystallites. J Dent Res. 1979;58 Spec Issue B:844–851. doi: 10.1177/00220345790580023701. [DOI] [PubMed] [Google Scholar]
- Daculsi G, Menanteau J, Kerebel LM, et al. Length and shape of enamel crystals. Calcif Tissue Int. 1984;36 5:550–555. doi: 10.1007/BF02405364. [DOI] [PubMed] [Google Scholar]
- Aoba T. Recent observations on enamel crystal formation during mammalian amelogenesis. Anat Rec. 1996;245 2:208–218. doi: 10.1002/(SICI)1097-0185(199606)245:2<208::AID-AR8>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Fincham AG. Molecular mechanisms of dental enamel formation. Crit Rev Oral Biol Med. 1995;6 2:84–108. doi: 10.1177/10454411950060020701. [DOI] [PubMed] [Google Scholar]
- Nanci A.Enamel: composition, formation, and structureIn: Nanci A ed. Ten cate's oral histology development, structure, and function St Louis; Mosby; 2008141–190. [Google Scholar]
- Delgado S, Casane D, Bonnaud L, et al. Molecular evidence for precambrian origin of amelogenin, the major protein of vertebrate enamel. Mol Biol Evol. 2001;18 12:2146–2153. doi: 10.1093/oxfordjournals.molbev.a003760. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Weiss KM. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci U S A. 2003;100 7:4060–4065. doi: 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Suzuki T, Weiss KM. Genetic basis for the evolution of vertebrate mineralized tissue. Proc Natl Acad Sci U S A. 2004;101 31:11356–11361. doi: 10.1073/pnas.0404279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer JW, Lanschuetzer C. Type XVII collagen gene mutations in junctional epidermolysis bullosa and prospects for gene therapy. Clin Exp Dermatol. 2003;28 1:53–60. doi: 10.1046/j.1365-2230.2003.01192.x. [DOI] [PubMed] [Google Scholar]
- Pasmooij AM, Pas HH, Jansen GH, et al. Localized and generalized forms of blistering in junctional epidermolysis bullosa due to COL17A1 mutations in the Netherlands. Br J Dermatol. 2007;156 5:861–870. doi: 10.1111/j.1365-2133.2006.07730.x. [DOI] [PubMed] [Google Scholar]
- Asaka T, Akiyama M, Domon T, et al. Type XVII collagen is a key player in tooth enamel formation. Am J Pathol. 2009;174 1:91–100. doi: 10.2353/ajpath.2009.080573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerio JE, Pulkkinen L, McMillan JR, et al. Pyloric atresia-junctional epidermolysis bullosa syndrome: mutations in the integrin beta4 gene (ITGB4) in two unrelated patients with mild disease. Br J Dermatol. 1998;139 5:862–871. doi: 10.1046/j.1365-2133.1998.02515.x. [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Rouan F, Bruckner-Tuderman L, et al. Novel ITGB4 mutations in lethal and nonlethal variants of epidermolysis bullosa with pyloric atresia: missense versus nonsense. Am J Hum Genet. 1998;63 5:1376–1387. doi: 10.1086/302116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A, Pulkkinen L, Murrell D, et al. Epidermolysis bullosa with congenital pyloric atresia: novel mutations in the beta 4 integrin gene (ITGB4) and genotype/phenotype correlations. Pediatr Res. 2001;49 5:618–626. doi: 10.1203/00006450-200105000-00003. [DOI] [PubMed] [Google Scholar]
- Buchroithner B, Klausegger A, Ebschner U, et al. Analysis of the LAMB3 gene in a junctional epidermolysis bullosa patient reveals exonic splicing and allele-specific nonsense-mediated mRNA decay. Lab Invest. 2004;84 10:1279–1288. doi: 10.1038/labinvest.3700164. [DOI] [PubMed] [Google Scholar]
- McLean WH, Irvine AD, Hamill KJ, et al. An unusual N-terminal deletion of the laminin alpha3α isoform leads to the chronic granulation tissue disorder laryngo-onycho-cutaneous syndrome. Hum Mol Genet. 2003;12 18:2395–2409. doi: 10.1093/hmg/ddg234. [DOI] [PubMed] [Google Scholar]
- Pfendner EG, Lucky AW.Junctional epidermolysis bullosaIn: Pagon RA, Bird TD, Dolan CR et al. eds. SourceGeneReviews™. Seattle; University of Washington; 2008. Available at http://www.ncbi.nlm.nih.gov/books/NBK1125 (accessed 14 September 2012). [PubMed] [Google Scholar]
- Ryan MC, Lee K, Miyashita Y, et al. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 1999;145 6:1309–1323. doi: 10.1083/jcb.145.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CC, Fukae M, Uchida T, et al. Cloning and characterization of porcine enamelin mRNAs. J Dent Res. 1997;76 11:1720–1729. doi: 10.1177/00220345970760110201. [DOI] [PubMed] [Google Scholar]
- Uchida T, Murakami C, Dohi N, et al. Synthesis, secretion, degradation and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies. J Histochem Cytochem. 1997;45 10:1329–1340. doi: 10.1177/002215549704501002. [DOI] [PubMed] [Google Scholar]
- Nanci A, Zalzal S, Lavoie P, et al. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J Histochem Cytochem. 1998;46 8:911–934. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- Hu JC, Yamakoshi Y. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit Rev Oral Biol Med. 2003;14 6:387–398. doi: 10.1177/154411130301400602. [DOI] [PubMed] [Google Scholar]
- Hu JC, Hu Y, Smith CE, et al. Enamel defects and ameloblast-specific expression in Enam knock-out/lacz knock-in mice. J Biol Chem. 2008;283 16:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, et al. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167 5:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini G, Lavoie P, Smith C, et al. Immunochemical characterization of a chicken egg yolk antibody to secretory forms of rat incisor amelogenin. J Histochem Cytochem. 2001;49 3:285–292. doi: 10.1177/002215540104900302. [DOI] [PubMed] [Google Scholar]
- Nanci A, Hashimoto J, Zalzal S, et al. Transient accumulation of proteins at interrod and rod enamel growth sites. Adv Dent Res. 1996;10 2:135–149. doi: 10.1177/08959374960100020501. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Hall B, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276 34:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Ronnholm E. An electron microscopic study of the amelogenesis in human teeth. I. The fine structure of the ameloblasts. J Ultrastruct Res. 1962;6:229–248. doi: 10.1016/s0022-5320(62)90055-2. [DOI] [PubMed] [Google Scholar]
- Diekwisch TG, Berman BJ, Gentner S, et al. Initial enamel crystals are not spatially associated with mineralized dentine. Cell Tissue Res. 1995;279 1:149–167. doi: 10.1007/BF00300701. [DOI] [PubMed] [Google Scholar]
- Bodier-Houlle P, Steuer P, Meyer JM, et al. High-resolution electron-microscopic study of the relationship between human enamel and dentin crystals at the dentinoenamel junction. Cell Tissue Res. 2000;301 3:389–395. doi: 10.1007/s004410000241. [DOI] [PubMed] [Google Scholar]
- Fang PA, Lam RS, Beniash E. Relationships between dentin and enamel mineral at the dentino-enamel boundary: electron tomography and high-resolution transmission electron microscopy study. Eur J Oral Sci. 2011;119 Suppl 1:120–124. doi: 10.1111/j.1600-0722.2011.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshawsky H, Josephsen K, Thylstrup A, et al. The development of enamel structure in rat incisors as compared to the teeth of monkey and man. Anat Rec. 1981;200 4:371–399. doi: 10.1002/ar.1092000402. [DOI] [PubMed] [Google Scholar]
- Ronnholm E. The amelogenesis of human teeth as revealed by electron microscopy. II. The development of the enamel crystallites. J Ultrastruct Res. 1962;6:249–303. doi: 10.1016/s0022-5320(62)80036-7. [DOI] [PubMed] [Google Scholar]
- Ronnholm E., III The structure of the organic stroma of human enamel during amelogenesis. J Ultrastruct Res. 1962;6:368–389. doi: 10.1016/s0022-5320(62)80041-0. [DOI] [PubMed] [Google Scholar]
- Smith CE, Nanci A. Protein dynamics of amelogenesis. Anat Rec. 1996;245 2:186–207. doi: 10.1002/(SICI)1097-0185(199606)245:2<186::AID-AR7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Addadi L, Weiner S.Stereochemical and structural relations between macromolecules and crystals in biomineralizationIn: Mann S, Webb J, Williams RJP eds. Biomineralization, chemical and biochemical perspectives Weinheim/New York; VCH; 1989133–152. [Google Scholar]
- Hughes J, Rakovan J. The crystal structure of apatite, Ca5(PO4)3(F,OH,Cl) Rev Mineral Geochem. 2002;1:1–12. [Google Scholar]
- Brown W, Lehr J, Smith J, et al. Crystallography of octacalcium phosphate. J Am Chem Soc. 1957;79 19:5318–5319. [Google Scholar]
- Brown WE, Eidelman N, Tomazic B. Octacalcium phosphate as a precursor in biomineral formation. Adv Dent Res. 1987;1 2:306–313. doi: 10.1177/08959374870010022201. [DOI] [PubMed] [Google Scholar]
- Dowker S, Anderson P, Elliott J, et al. Crystal chemistry and dissolution of calcium phosphate in dental enamel. Mineral Mag. 1999;63 6:791–800. [Google Scholar]
- Uskokovic V, Khan F, Liu H, et al. Hydrolysis of amelogenin by matrix metalloprotease-20 accelerates mineralization in vitro. . Arch Oral Biol. 2011;56 12:1548–1559. doi: 10.1016/j.archoralbio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Fan D, Fan Y, et al. Enamel proteases reduce amelogenin–apatite binding. J Dent Res. 2008;87 12:1133–1137. doi: 10.1177/154405910808701212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9 2:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Hu Y, Lertlam R, et al. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284 28:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis WJ, Burke GY, Neuringer JR, et al. Earliest enamel deposits of the rat incisor examined by electron microscopy, electron diffraction, and electron probe microanalysis. Anat Rec. 1988;220 3:233–238. doi: 10.1002/ar.1092200303. [DOI] [PubMed] [Google Scholar]
- Beniash E, Metzler RA, Lam RS, et al. Transient amorphous calcium phosphate in forming enamel. J Struct Biol. 2009;166 2:133–143. doi: 10.1016/j.jsb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perloff A, Posner AS. Preparation of pure hydroxyapatite crystals. Science. 1956;124 3222:583–584. doi: 10.1126/science.124.3222.583-a. [DOI] [PubMed] [Google Scholar]
- Boskey A, Posner AS. Conversion of amorphous calcium phosphate to microcrystalline hydroxyapatite. J Phys Chem. 1973;77 79:2313–2317. [Google Scholar]
- Kwak SY, Wiedemann-Bidlack FB, Beniash E, et al. Role of 20-kDa amelogenin (P148) phosphorylation in calcium phosphate formation in vitro. . J Biol Chem. 2009;284 28:18972–18979. doi: 10.1074/jbc.M109.020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker GL. Calcification mechanisms: roles for cells and mineral. J Oral Pathol. 1977;6 5:307–315. doi: 10.1111/j.1600-0714.1977.tb01653.x. [DOI] [PubMed] [Google Scholar]
- Inai T, Kukita T, Ohsaki Y, et al. Immunohistochemical demonstration of amelogenin penetration toward the dental pulp in the early stages of ameloblast development in rat molar tooth germs. Anat Rec. 1991;229:259–270. doi: 10.1002/ar.1092290213. [DOI] [PubMed] [Google Scholar]
- Begue-Kirn C, Krebsbach PH, Bartlett JD, et al. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sci. 1998;106 5:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- Hu Y, Hu JC, Smith CE, et al. Kallikrein-related peptidase 4, matrix metalloproteinase 20, and the maturation of murine and porcine enamel. Eur J Oral Sci. 2011;119 Suppl 1:217–225. doi: 10.1111/j.1600-0722.2011.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risnes S, Septier D, Deville de Periere D, et al. TEM observations on the ameloblast/enamel interface in the rat incisor. Connect Tissue Res. 2002;43 2/3:496–504. doi: 10.1080/03008200290000899. [DOI] [PubMed] [Google Scholar]
- Kallenbach E. The fine structure of Tomes' process of rat incisor ameloblasts and its relationship to the elaboration of enamel. Tissue Cell. 1973;5 3:501–524. doi: 10.1016/s0040-8166(73)80041-2. [DOI] [PubMed] [Google Scholar]
- Reith EJ, Boyde A. Histochemical and electron probe analysis of secretory ameloblasts of developing rat molar teeth. Histochemistry. 1978;55:17–26. doi: 10.1007/BF00496690. [DOI] [PubMed] [Google Scholar]
- Reith EJ, Boyde A. The enamel organ, a control gate for calcium influx into the enamel. J Dent Res. 1979;58 Spec Issue B:980. doi: 10.1177/00220345790580025001. [DOI] [PubMed] [Google Scholar]
- Prakriya M, Feske S, Gwack Y, et al. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443 7108:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- McCarl CA, Picard C, Khalil S, et al. ORAI1 deficiency and lack of store-operated Ca2+ entry cause immunodeficiency, myopathy, and ectodermal dysplasia. J Allergy Clin Immunol. 2009;124 6:1311–1318.e7. doi: 10.1016/j.jaci.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munhoz CO, Leblond CP. Deposition of calcium phosphate into dentin and enamel as shown by radioautography of sections of incisor teeth following injection of 45Ca into rats. Calcif Tissue Res. 1974;15 3:221–235. doi: 10.1007/BF02059059. [DOI] [PubMed] [Google Scholar]
- Snead ML, Zeichner-David M, Chandra T, et al. Construction and identification of mouse amelogenin cDNA clones. Proc Natl Acad Sci U S A. 1983;80:7254–7258. doi: 10.1073/pnas.80.23.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebsbach PH, Lee SK, Matsuki Y, et al. Full-length sequence, localization, and chromosomal mapping of ameloblastin: a novel tooth-specific gene. J Biol Chem. 1996;271 8:4431–4435. doi: 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Simmer JP, Xue J, et al. Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene. 1996;183:123–128. doi: 10.1016/s0378-1119(96)00525-2. [DOI] [PubMed] [Google Scholar]
- Sire JY, Delgado SC, Girondot M. Hen's teeth with enamel cap: from dream to impossibility. BMC Evol Biol. 2008;8:246. doi: 10.1186/1471-2148-8-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Gatesy J, Murphy WJ, et al. Molecular decay of the tooth gene Enamelin (ENAM) mirrors the loss of enamel in the fossil record of placental mammals. PLoS Genet. 2009;5 9:e1000634. doi: 10.1371/journal.pgen.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Gatesy J, Cheng J, et al. Pseudogenization of the tooth gene enamelysin (MMP20) in the common ancestor of extant baleen whales. Proc Biol Sci. 2011;278 1708:993–1002. doi: 10.1098/rspb.2010.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu OH, Fincham AG, Hu CC, et al. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78 3:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- Iwata T, Yamakoshi Y, Hu JC, et al. Processing of ameloblastin by MMP-20. J Dent Res. 2007;86 2:153–157. doi: 10.1177/154405910708600209. [DOI] [PubMed] [Google Scholar]
- Nagano T, Kakegawa A, Yamakoshi Y, et al. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88 9:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun YH, Yamakoshi Y, Yamakoshi F, et al. Cleavage site specificity of MMP-20 for secretory-stage ameloblastin. J Dent Res. 2010;89 8:785–790. doi: 10.1177/0022034510366903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakoshi Y, Richardson AS, Nunez SM, et al. Enamel proteins and proteases in Mmp20 and Klk4 null and double-null mice. Eur J Oral Sci. 2011;119 Suppl 1:206–216. doi: 10.1111/j.1600-0722.2011.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniash E, Skobe Z, Bartlett JD. Formation of the dentino-enamel interface in enamelysin (MMP-20)-deficient mouse incisors. Eur J Oral Sci. 2006;114 Suppl 1:24–29. doi: 10.1111/j.1600-0722.2006.00293.x. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Papagerakis P, Smith CE, et al. Regulation of dental enamel shape and hardness. J Dent Res. 2010;89 10:1024–1038. doi: 10.1177/0022034510375829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Warshawsky H. Quantitative analysis of cell turnover in the enamel organ of the rat incisor. Evidence for ameloblast death immediately after enamel matrix secretion. Anat Rec. 1977;187 1:63–98. doi: 10.1002/ar.1091870106. [DOI] [PubMed] [Google Scholar]
- Kallenbach E. Fine structure of rat incisor ameloblasts in transition between enamel secretion and maturation stages. Tissue Cell. 1974;6 1:173–190. doi: 10.1016/0040-8166(74)90030-5. [DOI] [PubMed] [Google Scholar]
- Hu JC, Zhang C, Sun X, et al. Characterization of the mouse and human PRSS17 genes, their relationship to other serine proteases, and the expression of PRSS17 in developing mouse incisors. Gene. 2000;251 1:1–8. doi: 10.1016/s0378-1119(00)00203-1. [DOI] [PubMed] [Google Scholar]
- Moffatt P, Smith CE, St-Arnaud R, et al. Cloning of rat amelotin and localization of the protein to the basal lamina of maturation stage ameloblasts and junctional epithelium. Biochem J. 2006;399 1:37–46. doi: 10.1042/BJ20060662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt P, Smith CE, St-Arnaud R, et al. Characterization of Apin, a secreted protein highly expressed in tooth-associated epithelia. J Cell Biochem. 2008;103 3:941–956. doi: 10.1002/jcb.21465. [DOI] [PubMed] [Google Scholar]
- Josephsen K, Fejerskov O. Ameloblast modulation in the maturation zone of the rat incisor enamel organ. A light and electron microscopic study. J Anat. 1977;124 1:45–70. [PMC free article] [PubMed] [Google Scholar]
- Smith CE, McKee MD, Nanci A. Cyclic induction and rapid movement of sequential waves of new smooth-ended ameloblast modulation bands in rat incisors as visualized by polychrome fluorescent labeling and GBHA-staining of maturing enamel. Adv Dent Res. 1987;1 2:162–175. doi: 10.1177/08959374870010020401. [DOI] [PubMed] [Google Scholar]
- Reith EJ, Boyde A. Autoradiographic evidence of cyclical entry of calcium into maturing enamel of the rat incisor tooth. Arch Oral Biol. 1981;26 12:983–987. doi: 10.1016/0003-9969(81)90107-2. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Takagi T, Suzuki M. Cyclical changes in pH in bovine developing enamel as sequential bands. Arch Oral Biol. 1991;36 3:227–231. doi: 10.1016/0003-9969(91)90090-h. [DOI] [PubMed] [Google Scholar]
- Simmer J, Hu Y, Richardson A, et al. Why does enamel in Klk4 null mice break above the dentino-enamel junction. Cells Tissues Organs. 2011;194 2/3/4:211–215. doi: 10.1159/000324260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer JP, Richardson AS, Smith CE, et al. Expression of kallikrein-related peptidase 4 in dental and non-dental tissues. Eur J Oral Sci. 2011;119 Suppl 1:226–233. doi: 10.1111/j.1600-0722.2011.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PS, Hart TC, Michalec MD, et al. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41 7:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Richardson AS, Hu Y, et al. Effect of Kallikrein 4 loss on enamel mineralization: comparison with mice lacking matrix metalloproteinase 20. J Biol Chem. 2011;286 20:18149–18160. doi: 10.1074/jbc.M110.194258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Nanci A, Kurtz I, et al. Regulation of pH during amelogenesis. Calcif Tissue Int. 2009;86 2:91–103. doi: 10.1007/s00223-009-9326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephsen K, Takano Y, Frische S, et al. Ion transporters in secretory and cyclically modulating ameloblasts: a new hypothesis for cellular control of preeruptive enamel maturation. Am J Physiol Cell Physiol. 2010;299 6:C1299–C1307. doi: 10.1152/ajpcell.00218.2010. [DOI] [PubMed] [Google Scholar]
- Lacruz RS, Smith CE, Moffatt P, et al. Requirements for ion and solute transport, and pH regulation during enamel maturation. J Cell Physiol. 2012;227 4:1776–1785. doi: 10.1002/jcp.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]