Abstract
The S41A mutant of riboflavin synthase from Escherichia coli catalyzes the formation of riboflavin from 6,7-dimethyl-8-ribityllumazine at a very low rate. Quenching of presteady-state reaction mixtures with trifluoroacetic acid afforded a compound with an absorption maximum at 412 nm (pH 1.0) that can be converted to a mixture of riboflavin and 6,7-dimethyl-8-ribityllumazine by treatment with wild-type riboflavin synthase. The compound was shown to qualify as a kinetically competent intermediate of the riboflavin synthase-catalyzed reaction. Multinuclear NMR spectroscopy, using various 13C- and 15N-labeled samples, revealed a pentacyclic structure arising by dimerization of 6,7-dimethyl-8-ribityllumazine. Enzyme-catalyzed fragmentation of this compound under formation of riboflavin can occur easily by a sequence of two elimination reactions.
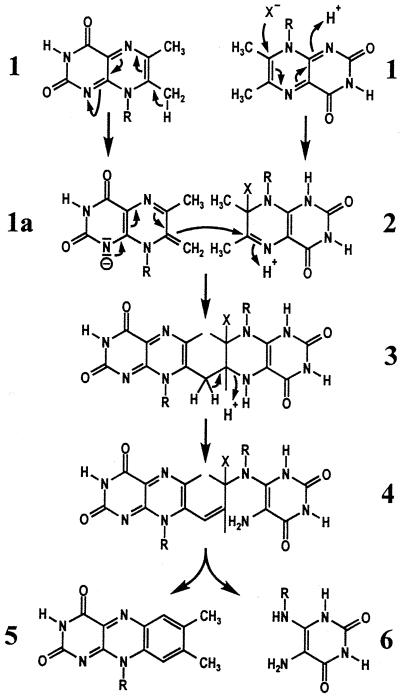
The final step in the biosynthesis of riboflavin is the dismutation of 6,7-dimethyl-8-ribityllumazine (1) affording riboflavin (5) and 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (6) in stoichiometric amounts (Fig. 1). The reaction involves the transfer of a 4-carbon moiety from a donor substrate molecule to an acceptor molecule.
Figure 1.
A shortened version of hypothetical reaction mechanisms proposed for the formation of riboflavin (5) from 6,7-dimethyl-8-ribityllumazine (1) by Plaut, Wood, and their coworkers (1–3). X, unknown nucleophile; R, ribityl.
Rowan and Wood (1) showed that the formation of riboflavin from 6,7-dimethyl-8-ribityllumazine can proceed in the absence of any catalyst under relatively mild experimental conditions. Thus, riboflavin is obtained on boiling of neutral or acidic aqueous solutions of 6,7-dimethyl-8-ribityllumazine (2, 3). The enzyme-catalyzed and the uncatalyzed reactions proceed with identical regiochemistry involving a head-to-tail arrangement of the two 4-carbon moieties from which the xylene ring of riboflavin is assembled (4–6).
The enzyme-catalyzed reaction requires the simultaneous presence of two substrate molecules with an essentially antiparallel orientation at the active site of the enzyme (4). Ligand-binding studies confirmed that each subunit of the homotrimeric riboflavin synthase can bind two lumazine molecules (7).
More recently, it was shown that each riboflavin synthase subunit comprises two tandem domains with similar sequence and folding topology. Each of these domains can accommodate one lumazine molecule, and the active sites are located at the interfaces of the N- and C-terminal domains, respectively (8, 9).
The position 7 methyl group of 6,7-dimethyl-8-ribityllumazine (1) is unusually acidic with a pK value of 8.5. The exomethylene type lumazine anion (1a) was proposed to attack the adduct of a second substrate molecule and a nucleophile (2) resulting in the formation of a lumazine dimer (3) as an initial reaction step (Fig. 1; refs. 4, 10, and 11). A ring-opening reaction was suggested to afford the intermediate 4.
The crystal structure of riboflavin synthase is still unknown despite considerable efforts. Recently, mutants of riboflavin synthase from Escherichia coli have been identified with reduced rates of riboflavin formation (12). Specifically, an S41A mutant catalyzed the formation of riboflavin at a rate that was lower by several orders of magnitude than the rate of the wild-type enzyme.
This paper describes a pentacyclic dimerization product of 6,7-dimethyl-8-ribityllumazine isolated after acid quenching of a presteady-state mixture containing 6,7-dimethyl-8-ribityllumazine and the S41A riboflavin synthase mutant of E. coli. The compound can be converted to riboflavin by riboflavin synthase and qualifies as a kinetically competent intermediate.
Experimental Procedures
Materials.
6,7-Dimethyl-8-ribityllumazine was synthesized by published procedures (13). The preparation of [6α,(7α)-13C1]6,7-dimethyl-8-ribityllumazine (80% 13C abundance at C-6α and 20% 13C abundance at C-7α), [7α,(6α)-13C1]6,7-dimethyl-8-ribityllumazine (80% 13C abundance at C-7α and 20% 13C abundance at C-6α), and [6,(7)-13C1]6,7-dimethyl-8-ribityllumazine (80% 13C abundance at C-6 and 20% 13C abundance at C-7) has been reported (6). The preparation of [6α,6,7,7α-13C4]- and [U-13C13,U-15N4]6,7-dimethyl-8-ribityllumazine and [U-13C9,U-15N4]5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione will be described elsewhere. Concentrations of 6,7-dimethyl-8-ribityllumazine and riboflavin were determined photometrically (ɛ410 = 9,100 M−1 cm−1, pH 1.0, and ɛ470 = 10,300 M−1 cm−1, pH 7.0, respectively) (14).
Bacterial Strains.
The recombinant E. coli strain XL1-Blue[pERS] used for the hyperexpression of the E. coli riboflavin synthase gene has been described (15). The recombinant E. coli strain BL21 (DE3) expressing the E. coli riboflavin synthase S41A mutant is described elsewhere (12).
Bacterial Culture and Enzyme Purification.
Bacterial culture and enzyme purification were performed as described elsewhere (15).
Preparation of a Transient Molecular Species (Compound Q).
A reaction mixture containing 3.0 mM of 6,7-dimethyl-8-ribityllumazine and 141 mg of purified S41A mutant protein in a total volume of 3.2 ml of 30 mM K-phosphate (pH 6.9) was incubated at 37°C for 8 min. The solution was brought to pH 1 by the addition of 23 μl of 14 M trifluoroacetic acid. The mixture was centrifuged immediately at 17,000 × g and 10°C for 20 min. The supernatant was collected, and the pellet was washed twice with 0.5 ml of 0.1 M trifluoroacetic acid. Aliquots (0.3 ml) of the supernatant were applied to a Hypersil RP18 column (5 μm, 4 × 250 mm; Schambeck, Bad Honnef, Germany). The column was developed with 3 M formic acid at a flow rate of 1.5 ml min−1. The effluent was monitored spectrophotometrically by using a TIDAS diode array photometer (J & M Analytische Mess- und Regeltechnik GmbH, Aalen, Germany). The retention volumes of Compound Q and 6,7-dimethyl-8-ribityllumazine were 4.4 and 5.2 ml, respectively. Fractions were combined and lyophilized. Typically, 0.6–1.0 mg of Compound Q were obtained (yield, 20–30%). The concentration of Compound Q was determined photometrically (ɛ412 = 9,400 M−1 cm−1, pH 1.0).
Kinetic Studies.
A solution (600 μl) containing 60 μM riboflavin synthase in 0.3 M potassium phosphate buffer, pH 6.9, was mixed with a solution (600 μl) containing 360 μM 6,7-dimethyl-8-ribityllumazine or 180 μM Compound Q in the same buffer at 12°C. Aliquots (100 μl) were retrieved at intervals and the enzyme reaction was quenched by the addition of 5 μl of 2 M trifluoroacetic acid. The samples were centrifuged, and the supernatant was analyzed by HPLC.
HPLC.
Experiments were performed with a column of Hypersil RP18 (5 μm, 4 × 250 mm; Schambeck). An eluant containing methanol/formic acid/water (25:1:288, vol/vol) was used for determination of 6,7-dimethyl-8-ribityllumazine (retention volume, 6.5 ml) and Compound Q (retention volume, 7.4 ml). An eluant containing methanol/water (4:6, vol/vol) with 0.1 M ammonium formate was used for determination of riboflavin (retention volume, 2.2 ml). The flow rate was 1.5 ml min−1. Effluents were monitored photometrically (TIDAS diode array photometer, J & M Analytische Mess- und Regeltechnik).
NMR Spectroscopy.
1H, 13C, and 15N NMR spectra were recorded at 10°C with a DRX 500 spectrometer (Bruker, Karlsruhe, Germany) equipped with four channels and a pulsed gradient unit. The transmitter frequencies were 500.1, 125.6, and 50.7 MHz for 1H, 13C, and 15N, respectively. Two-dimensional inadequate, hmqc, and hmqc-tocsy experiments were performed according to standard Bruker software (xwinnmr). The duration for tocsy mixing was 60 ms in the hmqc-tocsy experiment. The delay for magnetization transfer was 6 ms in the inadequate experiment.
Measurements were performed in 50 mM phosphoric acid containing 10% D2O.
Results
Isolation of a Transient Molecular Species (Compound Q).
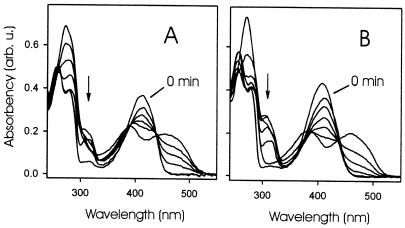
Presteady-state kinetic experiments were performed with wild-type riboflavin synthase of E. coli at 20°C. A transient species with an absorption maximum around 312 nm was detected around 1.5 s in stopped-flow single-turnover experiments (Fig. 2A); the formation of riboflavin was virtually complete after 1 min.
Figure 2.
Optical spectra obtained in single-turnover experiment. The reaction mixture contained 30 mM potassium phosphate buffer, pH 6.9, 25 μM 6,7-dimethyl-8-ribityllumazine, and 4 μM riboflavin synthase. (A) Wild type. (B) S41A mutant. Spectra were obtained at 0, 0.025, 0.08, 0.17, 0.3, and 5 min (A) or at 0, 0.08, 3, 6, 15, 30, and 240 min (B). Arrows indicate the optical transient at 312 nm.
The S41A mutant of riboflavin synthase catalyzes the formation of riboflavin from 6,7-dimethyl-8-ribityllumazine at a much lower rate than the wild-type enzyme; a turnover number of 1.0 h−1 per subunit at 37°C can be estimated from steady-state experiments. In presteady-state experiments with this mutant, the transient species characterized by absorbance at 312 nm reached a substantially higher level as compared with the wild-type enzyme (Fig. 2B).
To isolate the transient molecular species, we incubated a mixture containing 6,7-dimethyl-8-ribityllumazine and S41A mutant protein at a molar ratio of 6:1 (i.e., two substrate molecules per protein subunit). When the absorbance at 312 nm reached a maximum value after 8 min, the reaction mixture was acidified to pH 1, and the intermediate (designated as Compound Q) was isolated chromatographically as described under Experimental Procedures.
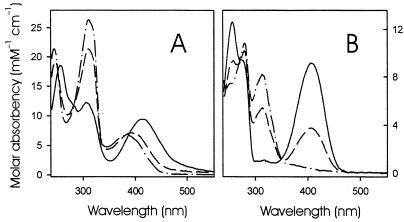
Compound Q shows absorption maxima at 256, 306, and 412 nm at pH 3.4 (Fig. 3A). The spectrum is similar to that of 6,7-dimethyl-8-ribityllumazine (Fig. 3B), but the ratio of ɛ256/ɛ412 is 2.0 for Compound Q and 1.4 for 6,7-dimethyl-8-ribityllumazine.
Figure 3.
Optical spectra of Compound Q (A) and 6,7-dimethyl-8-ribityllumazine (B) at pH 3.4 (solid lines), pH 8.8 (dashed lines), and pH 11.1 (dotted lines).
At pH 8.8, the spectrum of Compound Q shows maxima at 247, 310, and 392 nm, whereas 6,7-dimethyl-8-ribityllumazine shows maxima at 258, 279, 313, and 406 nm. Photometric titrations afforded pK values of 8.4 and 8.2 for 6,7-dimethyl-8-ribityllumazine and Compound Q, respectively.
Catalytic Competence of Compound Q.
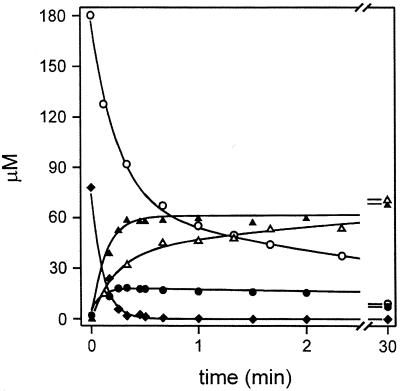
The compound is converted into a mixture of 6,7-dimethyl-8-ribityllumazine and riboflavin by treatment with wild-type riboflavin synthase. In presteady-state experiments, it was consumed more rapidly by riboflavin synthase than 6,7-dimethyl-8-ribityllumazine (Fig. 4). Although the consumption of Compound Q was virtually complete after 20 s, the consumption of the 6,7-dimethyl-8-ribityllumazine required more than 30 min in the control experiment.
Figure 4.
Single-turnover experiments using wild-type riboflavin synthase and 6,7-dimethyl-8-ribityllumazine, respectively, with Compound Q as substrate. 6,7-Dimethyl-8-ribityllumazine as substrate (○) afforded riboflavin (▵). Compound Q as substrate (♦) afforded 6,7-dimethyl-8-ribityllumazine (●) and riboflavin (▴).
The partitioning factor for enzyme-catalyzed formation of riboflavin and 6,7-dimethyl-8-ribityllumazine from Compound Q was 5:1. The newly formed 6,7-dimethyl-8-ribityllumazine was converted subsequently to riboflavin at a low rate similar to that for consumption of proffered 6,7-dimethyl-8-ribityllumazine. Thus, Compound Q fulfills the requirements for a kinetically competent intermediate of the enzyme reaction.
Structure of Compound Q.
Structure determination of the intermediate was hampered by the poor stability of Compound Q, by relatively broad NMR signals, and by the paucity of hydrogen atoms in the heterocyclic chromophore. Preliminary experiments revealed that stability and NMR linewidths of Compound Q were acceptable in 50 mM phosphoric acid at 10°C. To enhance the sensitivity of heteronuclear NMR analysis and to facilitate structure determination by analysis of heteroatom connectivity, we prepared samples of Compound Q from various 13C- and 15N-labeled samples of 6,7-dimethyl-8-ribityllumazine as substrates.
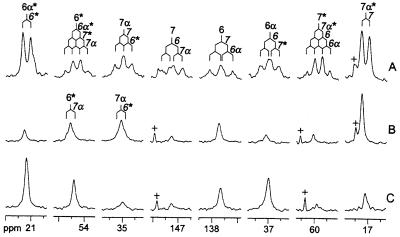
Proton-decoupled 13C NMR signals of Compound Q prepared from [6α,6,7,7α-13C4]6,7-dimethyl-8-ribityllumazine are shown in Fig. 5A. The spectrum of labeled Compound Q showed 8 multiplets caused by 13C–13C coupling. More specifically, two doublets, four double-doublets, and two pseudoquartets indicated two carbon atoms with one, four atoms with two, and two atoms with three directly adjacent 13C atoms, respectively. Two signals had chemical shifts typical for alkene carbon atoms, and six signals had chemical shifts typical for carbon atoms with sp3 hybridization.
Figure 5.
13C NMR signals of Compound Q obtained from [6α,6,7,7α-13C4]6,7-dimethyl-8-ribityllumazine (A) from a mixture of [7α,(6α)-13C1]6,7-dimethyl-8-ribityllumazine (13C abundance: C-7α, 80%; C-6α, 20%) and [6,(7)-13C1]6,7-dimethyl-8-ribityllumazine (13C abundance: C-6, 80%; C-7, 20%) (B), and from a mixture of [6α,(7α)-13C1]6,7-dimethyl-8-ribityllumazine (13C abundance: C-6α, 80%; C-7α, 20%) and [6,(7)-13C1]6,7-dimethyl-8-ribityllumazine (13C abundance: C-6, 80%; C-7, 20%) (C). 13C coupling patterns are indicated. +, impurities.
hmqc experiments revealed two 13C-labeled methyl groups and two methylene 13C atoms with pairs of diasterotopic hydrogens. Four 13C atoms including both alkene carbon atoms gave no correlations in the hmqc experiments. In the inadequate spectrum, the two methyl signals gave correlations to the signals at 54.4 and 59.9 ppm, respectively, exhibiting pseudoquartet coupling signatures and reflecting quaternary carbon atoms (Fig. 5A). In conjunction with chemical-shift topology, the spin network is in line with a [13C8]dimethylcyclohexene motif.
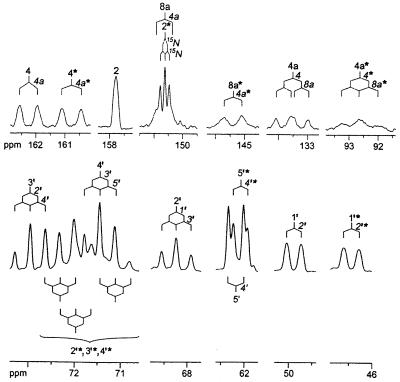
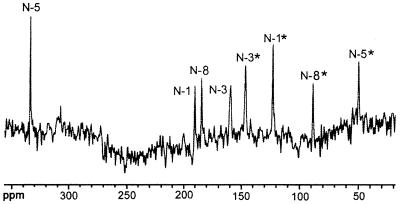
Compound Q prepared from [U-13C13,U-15N4]6,7-dimethyl-8-ribityllumazine afforded the 8 13C NMR signals described above (data not shown), as well as 18 additional 13C NMR signals shown in Fig. 6. The 15N NMR spectrum of the labeled compound showed 8 signals (Fig. 7). These results confirmed that a [13C2615N8] dimer without inherent symmetry was formed from the 13C1315N4-labeled substrate. 13C–13C and 13C–15N coupling constants; 15N NMR signal modulation under 1H decoupling conditions (Table 1); and correlation patterns in hmqc, hmqc-tocsy, and inadequate spectra (Fig. 8 and Table 2) established Compound Q as the dimethyl-bis-(ribityl)-octaaza-pentacene-tetraone derivative shown in Fig. 9.
Figure 6.
13C NMR signals of Compound Q obtained from [U-13C13,U-15N4]6,7-dimethyl-8-ribityllumazine. 13C- and 15N-coupling patterns are indicated.
Figure 7.
15N NMR spectrum of Compound Q obtained from [U-13C13,U-15N4]6,7-dimethyl-8-ribityllumazine.
Table 1.
NMR-data of Compound Q
| Position | δ, ppm†
|
J, Hz‡
|
|||
|---|---|---|---|---|---|
| δ13C | δ15N | δ1H | JHH | Jcc | |
| C-7α* | 17.1 | 1.12 | 37 | ||
| C-6α* | 21.2 | 1.22 | 40 | ||
| C-7α | 35.0 | 3.00§, 3.30§ | 24 | 40 | |
| C-6α | 37.0 | 3.07, 3.36 | 25 | 43 | |
| C-1′* | 46.4 | 3.09, 3.53 | 42 | ||
| C-1′ | 49.9 | 4.58, 4.01 | 39 | ||
| C-6* | 54.4 | 36 | |||
| C-7* | 59.9 | 38 | |||
| C-5′ or 5′* | 62.1 | 3.45–3.20 | 40 | ||
| C-5′ or 5′* | 62.2 | 3.45–3.20 | 41 | ||
| C-2′ | 68.2 | 4.01 | 40 | ||
| C-4′ | 71.4 | 3.52 | 42 | ||
| C-2′*, C-3′*, | 71.6 | 3.50–3.25 | nd | ||
| C-4′* | 72.0 | ||||
| 72.6 | |||||
| C-3′ | 73.0 | 3.50 | 42 | ||
| C-4a* | 92.6 | 75 | |||
| C-4a | 133.5 | 75, 61 | |||
| C-6 | 137.6 | 56 | |||
| C-8a* | 145.4 | 80 | |||
| C-7 | 147.3 | 55 | |||
| C-8a | 150.6 | nd | |||
| C-2 | 150.6 | ||||
| C-2 | 157.8 | ||||
| C-4* | 160.8 | 80.5 | |||
| C-4 | 162.3 | 77.5 | |||
| N-5 | 334.6 | ||||
| N-1 | 190.7 | ||||
| N-8 | 184.9 | ||||
| N-3 | 159.8¶ | ||||
| N-3* | 147.0¶ | ||||
| N-1* | 123.4¶ | ||||
| N-8* | 88.8 | ||||
| N-5* | 49.9¶ | ||||
The ribityl signals are tentatively assigned by comparison with chemical shifts of 6,7-dimethyl-8-ribityllumazine (see also Table 3). For atom numbering, see Fig. 9. nd, not determined because of signal overlapping.
Chemical shifts, referenced to external trimethylsilyl propane sulfonate.
HH coupling constants are extracted from hmoc experiments and CC couplings are extracted from 13C spectra of 13C-enriched Compound Q.
Exchangeable with D2O.
Signal phase is modulated by 1H decoupling.
Figure 8.
Part of a two-dimensional INADEQUATE spectrum of Compound Q obtained from [U-13C13,U-15N4]6,7-dimethyl-8-ribityllumazine.
Table 2.
Correlation patterns observed in spectra of [U-13C26] Compound Q
| Position | inadequate | hmqc | hmqc-tocsy |
|---|---|---|---|
| C-7α* | 7* | 7a* | 7a* |
| C-6α* | 6* | 6a* | 6a* |
| C-7α | 7a | 7a | |
| C-6α | 6a | 6a | |
| C-1′* | 2′* | 1′* | 1′* |
| C-1′ | 2′ | 1′ | 1′, 3′ |
| C-6* | 6α* | ||
| C-7* | 7α* | ||
| C-5′ or 5′* | 4′, 4′* | 5′ or 5′* | 5′ or 5′*, 4′ or 4′* |
| C-5′ or 5′* | 5′ or 5′* | 5′ or 5′*, 4′ or 4′* | |
| C-2′ | 1′ | 2′ | 1′, 2′, 3′ |
| C-4′ | 5′ | 4′ | 4′, 5′, 3′, 2′ |
| C-2′*, C-3′*, C-4′* | 1′*, 5′* | 2′*, 3′*, 4′* | 2′*, 3′*, 4′*, 5′*, 1′* |
| C-3′ | 3′ | 3′, 2′ | |
| C-4a | 4 | ||
| C-8a | 4a | ||
| C-4 | 4a |
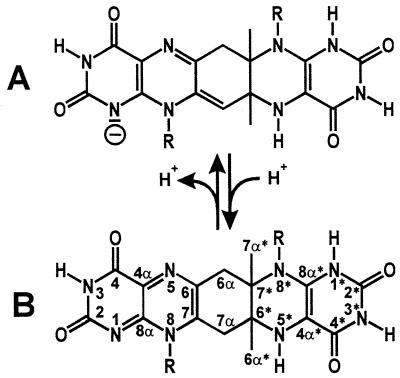
Figure 9.
Compound Q. (A) Monoanion. (B) Neutral molecular form, which structure was determined by NMR spectroscopy. R, ribityl.
The methylene group 7α of Compound Q is structurally equivalent to the acidic 7α methyl group of 6,7-dimethyl-8-ribityllumazine (16). In keeping with this, the hydrogen atoms bound to 7α were exchanged rapidly with deuterium atoms from the solvent D2O. The pH-dependent equilibrium between neutral and acidic forms of Compound Q is shown in Fig. 9.
The 13C NMR chemical shifts prediction, using the SpecInfo archive and interpretation system, agreed well with the experimental data (Table 3). Moreover, 13C and 15N chemical shifts of the lumazine, respectively the diaminopyrimidine moiety of Compound Q, were in almost perfect agreement with the chemical shifts of 6,7-dimethyl-8-ribityllumazine or FMN, respectively with those of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione or FMNH2 (Table 3).
Table 3.
Comparison of selected 13C and 15N NMR chemical shifts of Compound Q
| Position | A
|
B
|
C
|
D
|
E†
|
F†
|
|||
|---|---|---|---|---|---|---|---|---|---|
| δ13C | δ15N | δ13C | δ13C | δ13C | δ13C | δ15N | δ13C | δ15N | |
| C-1′* | 46.4 | 53.1 | 47.4 | ||||||
| C-1′ | 49.9 | 55.0 | 51.2 | ||||||
| C-2′ | 68.2 | 70.2 | 69.1 | ||||||
| C-4a* | 92.6 | 108.2 | 82.5 | 102.8 | |||||
| C-4a | 133.5 | 136.7 | 130.9 | 136.2 | |||||
| C-8a | 150.6 | 150.3 | 150.4 | 152.1 | |||||
| C-2* | 150.6 | 152.0 | 150.7 | 151.1 | |||||
| C-2 | 157.8 | 154.9 | 157.9 | 159.8 | |||||
| C-4* | 160.8 | 155.4 | 160.9 | 158.3 | |||||
| C-4 | 162.3 | 158.8 | 163.1 | 163.7 | |||||
| N-5 | 334.6 | 334.7 | |||||||
| N-1 | 190.7 | 190.8 | |||||||
| N-8 | 184.9 | 163.5 | |||||||
| N-3 | 159.8 | 160.5 | |||||||
| N-3* | 147.0 | 149.7 | |||||||
| N-1* | 123.4 | 128.0 | |||||||
| N-8* | 88.8 | 87.2 | |||||||
| N-5* | 49.9 | 58.0 | |||||||
A, selected 13C and 15N NMR chemical shifts of Compound Q; B, predicted values by specinfo; C, chemical shifts of corresponding positions in 6,7-dimethyl-8-ribityllumazine; D, 5-amino-6-ribitylamino-4(3H)-pyrimidinedion; E, FMN; F, FMNH2.
From ref. 17.
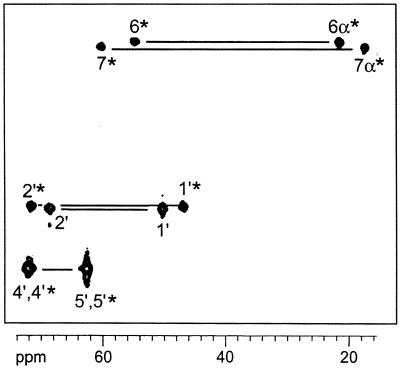
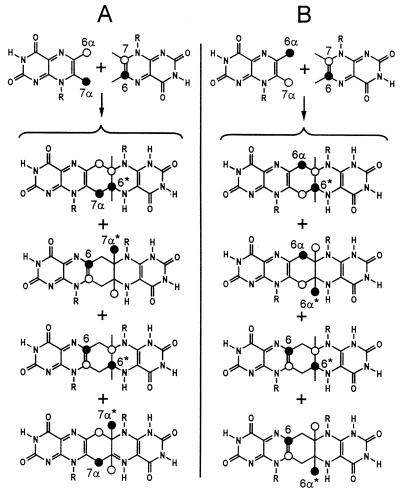
To analyze the regiochemistry of Compound Q formation, mixtures of [7α,(6α)-13C1]- and [6,(7)-13C1]6,7-dimethyl-8-ribityllumazine or [6α,(7α)-13C1]- and [6,(7)-13C1]6,7-dimethyl-8-ribityllumazine were converted into Compound Q. The 13C NMR signals of Compound Q prepared from these experiments are shown in Fig. 5 B and C. Analysis of signal intensities is conducive to isotopomer mixtures shown in Fig. 10. The regiochemistry of the reaction is obvious from the 13C NMR signal intensities of the resulting isotopomer mixtures.
Figure 10.
Regiochemistry of 6,7-dimethyl-8-ribityllumazine conversion into Compound Q. (A) Isotopomer mixture of Compound Q obtained from [7α,(6α)-13C1]- and [6,(7)-13C1]6,7-dimethyl-8-ribityllumazine. (B) Isotopomer mixture of Compound Q obtained from [6α,(7α)-13C1]- and [6,(7)-13C1]6,7-dimethyl-8-ribityllumazine. 13C enrichments ≈80% 13C (●) or 20% 13C (○) are indicated. R, ribityl.
Discussion
The structure of Compound Q was difficult to elucidate by NMR spectroscopy because of the paucity of protons coupled to each other. This problem could be circumvented by the use of totally respectively partially 13C/15N-labeled 6,7-dimethyl-8-ribityllumazine as enzyme substrate, which enabled the deduction of the structure from the 1H/13C/15N spin network.
The results leave no doubt that the mutant protein catalyzes the formation of a pentacyclic compound carrying two methyl and two ribitol substituents. All NMR spectroscopic data summarized in Tables 1 and 2 are consistent with the structure shown in Fig. 9.
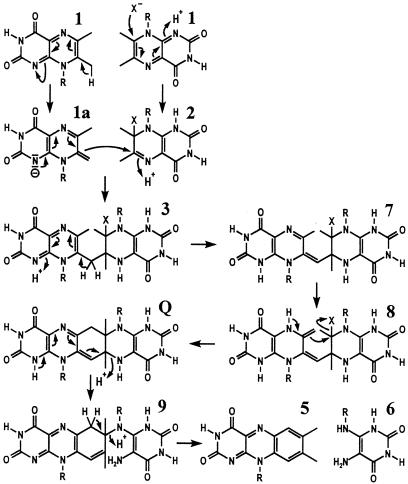
Conversion of Compound Q to riboflavin is explained easily by a series of two elimination steps as shown in Fig. 11. On the other hand, the formation of 6,7-dimethyl-8-ribityllumazine from Compound Q is explained less easily.
Figure 11.
Hypothetical mechanism of riboflavin formation by riboflavin synthase by means of the pentacyclic intermediate, Compound Q. X, unknown nucleophile; R, ribityl.
Plaut, Wood, and their coworkers (5, 18) had implicated the unusual CH acidity of the position 7 methyl group of 6,7-dimethyl-8-ribityllumazine (pK = 8.5) in the initial steps of the enzyme-catalyzed reaction. As shown by NMR experiments, deprotonation of 6,7-dimethyl-8-ribityllumazine results in the formation of a complex equilibrium mixture comprising a minor exomethylene species (19, 20). The exomethylene-type anion was supposed to attack a molecular species resulting from the addition of a nucleophile to a second lumazine molecule (10, 11, 21).
Fig. 11 shows a hypothetical reaction sequence conducive to the formation of Compound Q instead of the dimerization product (4; Fig. 1) that had been proposed earlier. A nucleophilic attack of one molecule of 6,7-dimethyl-8-ribityllumazine (exomethylene form, 1a) at C-6 of a second molecule of 6,7-dimethyl-8-ribityllumazine covalently bound at the acceptor site of the protein (2) could yield intermediate 3 (4, 10, 11). Deprotonation of 3 at C-7a and protonation at N-1 could afford 7. Tautomerization of 7 into compound 8 and subsequent nucleophilic substitution of the acceptor residue (X−) could yield Compound Q. A vinylogous elimination of N-5* could afford intermediate 9. A subsequent vinylogous elimination of N-8* under aromatization could yield riboflavin (5) and 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (6). It must be emphasized that this reaction sequence remains speculative at the present level of experimental evidence.
In single-turnover experiments, riboflavin is formed more rapidly from Compound Q than from the natural substrate, 6,7-dimethyl-8-ribityllumazine. Moreover, Compound Q is consumed rapidly and completely under formation of 6,7-dimethyl-8-ribityllumazine and riboflavin, whereas 6,7-dimethyl-8-ribityllumazine disappears rather slowly from the reaction mixture. Hence, Compound Q fulfills the formal requirements for a kinetically competent intermediate.
Compound Q is quite bulky as compared with the natural enzyme substrate, 6,7-dimethyl-8-ribityllumazine. It is noteworthy that this rigid and bulky molecular species can access the active site of the enzyme.
Riboflavin synthase is an essential enzyme in certain microorganisms (e.g., enterobacteria and yeasts) as a consequence of the apparent absence of a flavin uptake system. The enzyme could therefore represent a antiinfective target. The detailed analysis of the reaction mechanism could provide clues for the development of mechanism-based enzyme inhibitors designed for therapeutic application.
Acknowledgments
We thank M. Goese (Technische Universität München) for a sample of [U-13C5]ribulose 5-phosphate. This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie and the Hans Fischer-Gesellschaft, and by a fellowship from the Alexander von Humboldt–Stiftung (to B.I.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rowan, T. & Wood, H. C. S. (1963) Proc. Chem. Soc. London, 21–22.
- 2.Rowan, T. & Wood, H. C. S. (1968) J. Chem. Soc. C, 452–458. [DOI] [PubMed]
- 3.Beach, R. L. & Plaut, G. W. E. (1969) Tetrahedron Lett., 3489–3492. [DOI] [PubMed]
- 4.Beach R L, Plaut G W E. J Am Chem Soc. 1970;92:2913–2916. doi: 10.1021/ja00712a052. [DOI] [PubMed] [Google Scholar]
- 5.Paterson, T. & Wood, H. C. (1969) J. Chem. Soc. Chem. Commun., 290–291.
- 6.Sedlmaier H, Müller F, Keller P, Bacher A. Z Naturforsch. 1986;42:425–429. doi: 10.1515/znc-1987-0416. [DOI] [PubMed] [Google Scholar]
- 7.Otto M K, Bacher A. Eur J Biochem. 1981;115:511–517. doi: 10.1111/j.1432-1033.1981.tb06232.x. [DOI] [PubMed] [Google Scholar]
- 8.Schott K, Kellermann J, Lottspeich F, Bacher A. J Biol Chem. 1990a;265:4204–4209. [PubMed] [Google Scholar]
- 9.Meining W, Tibbelin G, Ladenstein R, Eberhardt S, Fischer M, Bacher A. J Struct Biol. 1998;121:53–60. doi: 10.1006/jsbi.1997.3935. [DOI] [PubMed] [Google Scholar]
- 10.Paterson, T. & Wood, H. C. S. (1972) J. Chem. Soc. Perkin Trans. 1, 1051–1056. [DOI] [PubMed]
- 11.Plaut G W E, Beach R. In: Flavins and Flavoproteins. Singer T P, editor. Amsterdam: Elsevier; 1976. pp. 737–746. [Google Scholar]
- 12.Illarionov B, Kemter K, Eberhardt S, Richter G, Cushman M, Bacher A. J Biol Chem. 2001;276:11524–11530. doi: 10.1074/jbc.M008931200. . (First Published January 18, 2001; 10.1074/jbc.M008931200) [DOI] [PubMed] [Google Scholar]
- 13.Bacher A, Eberhardt S, Fischer M, Mörtl S, Kis K, Kugelbrey K, Scheuring J, Schott K. Methods Enzymol. 1997;280:389–399. doi: 10.1016/s0076-6879(97)80130-9. [DOI] [PubMed] [Google Scholar]
- 14.Plaut G W E, Harvey R A. Methods Enzymol. 1971;18:515–538. [Google Scholar]
- 15.Eberhardt S, Richter S, Gimbell W, Werner T, Bacher A. Eur J Biochem. 1996;242:712–719. doi: 10.1111/j.1432-1033.1996.0712r.x. [DOI] [PubMed] [Google Scholar]
- 16.Beach R L, Plaut G W E. J Org Chem. 1971;36:3937–3943. [Google Scholar]
- 17.Moonen C T W, Vervoort J, Müller F. Biochemistry. 1984;23:4859–4867. doi: 10.1021/bi00316a007. [DOI] [PubMed] [Google Scholar]
- 18.Plaut G W E, Beach R, Aogaichi T. Biochemistry. 1970;9:771–785. doi: 10.1021/bi00806a010. [DOI] [PubMed] [Google Scholar]
- 19.Bown D H, Keller P J, Floss H G, Sedlmaier H, Bacher A. J Org Chem. 1986;51:2461–2467. [Google Scholar]
- 20.Beach R L, Plaut G W E. Biochemistry. 1970;9:760–770. doi: 10.1021/bi00806a009. [DOI] [PubMed] [Google Scholar]
- 21.Plaut G W E, Beach R. In: Chemistry and Biology of Pteridines. Pfleiderer W, editor. Berlin: de Gruyter; 1975. pp. 101–123. [Google Scholar]