Abstract
Motivation: An increasing amount of evidence from experimental and computational analysis suggests that rare codon clusters are functionally important for protein activity. Most of the studies on rare codon clusters were performed on a limited number of proteins or protein families. In the present study, we present the Sherlocc program and how it can be used for large scale protein family analysis of evolutionarily conserved rare codon clusters and their relation to protein function and structure. This large-scale analysis was performed using the whole Pfam database covering over 70% of the known protein sequence universe. Our program Sherlocc, detects statistically relevant conserved rare codon clusters and produces a user-friendly HTML output.
Results: Statistically significant rare codon clusters were detected in a multitude of Pfam protein families. The most statistically significant rare codon clusters were predominantly identified in N-terminal Pfam families. Many of the longest rare codon clusters are found in membrane-related proteins which are required to interact with other proteins as part of their function, for example in targeting or insertion. We identified some cases where rare codon clusters can play a regulating role in the folding of catalytically important domains. Our results support the existence of a widespread functional role for rare codon clusters across species. Finally, we developed an online filter-based search interface that provides access to Sherlocc results for all Pfam families.
Availability: The Sherlocc program and search interface are open access and are available at http://bcb.med.usherbrooke.ca
Contact: rafael.najmanovich@usherbrooke.ca
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Recent studies suggest that beyond the amino acid sequence lies an additional layer of information, hidden within the codon sequence, able to mediate local kinetics of translation. In fact, synonymous codons that are used with low frequency, tend to have a depleted concentration of tRNAs (Duret, 2000; Ikemura, 1985; Moriyama and Powell, 1997; Percudani et al., 1997) thus causing ribosomes to pause at rare codons until the scarce activated tRNA brings the next amino acid to the growing polypeptide (Sørensen et al., 1989; Varenne et al., 1984). The observation that rare codons are not randomly distributed but rather organized in large clusters (Clarke and Clark, 2008) across species support the existence of a selective evolutionary pressure.
Many roles have been proposed to explain the heterogeneity of translational kinetics. For example, protein folding is a co-translational process during which the nascent polypeptide chain is in dynamic interaction with components of the cellular milieu such as the ribosome structure, chaperone proteins, solvent molecules and also with its own residues. The speed at which the polypeptide chain grows dictates the time scale within which the translated residues that have already exited the ribosomal channel undergo local folding events (α-helices are somehow an exception as they tend to start forming inside the tunnel). In this context, while not altering the sequence of the nascent protein, synonymous codon changes can lead to distinct folding pathways (Tsai et al., 2008).
Several studies focused on identifying rare codons in protein sequences and replacing them with frequent synonymous ones. The results were diverse: decrease of a protein's specific activity (Komar et al., 1999), a change in substrate specificity (Kimchi-Sarfaty et al., 2007) and a decrease in protein solubility and activation of a gene designed to detect misfolded proteins (Cortazzo et al., 2002). For these three studies, the results were suggested to be due to alterations in the folding pathway of the protein. It was also observed that slowly translated regions tend to preferentially code for β-strands and coils, whereas faster translated regions tend to code for α-helices (Thanaraj and Argos, 1996a). Recent work showed that translation speed decreases at the start of secondary structures in Escherichia coli (Saunders and Deane, 2010).
The ribosomal pauses caused by rare codons can in principle regulate-specific folding events but could also be involved in other mechanisms involving the nascent polypeptide chain such as protein targeting or co-translational molecular recognition events. A study that examined genes of Emericella nidulans observed a correlation between the position of hydrophobic stretches, predicted to span across a membrane or to be a cleavable signal sequence, and rare codon clusters suggesting a potential control over membrane protein targeting or membrane insertion (Dessen and Képès, 2000). It was observed that signal sequences of exported proteins of E. coli and Salmonella typhimurium were enriched with rare codons (Burns and Beacham, 1985; Power et al., 2004). One study suggested that rare codons favored the proper structural arrangement of an α-helix signal sequence, which ensured that the protein correctly followed its secretion pathway (Zalucki and Jennings, 2007). Moreover, a correlation between the position of rare codon clusters in mRNA and protein domain boundaries was observed (Komar and Jaenicke, 1995; Krasheninnikov et al., 1991; Purvis et al., 1987; Thanaraj and Argos, 1996b). All these results highlight the potential importance of rare codon clusters regarding various events of the early protein, may it be folding, membrane insertion or export/targeting. Also at a functional level, a bias in codon usage may be important to indirectly regulate the function of cell cycle-regulated genes (Frenkel-Morgenstern et al., 2012). Some algorithms have been implemented to detect rare codon clusters (Clarke and Clark, 2008; Makhoul and Trifonov, 2002; Widmann et al., 2008) and all were tested against a limited number of protein sequences or protein families.
In this study, we present a simple and efficient algorithm, Sherlocc, for the detection of statistically significant rare codon clusters within protein families and evolutionarily conserved across species. We performed a large-scale study of the distribution of conserved rare codon clusters across all protein families present in Pfam (over 11 000 families). We use the results of the large-scale analysis and integrate various sources of information to corroborate the different roles of rare codon clusters proposed in the literature. The program generates an HTML output that allows a user to visualize the position of the family-conserved rare codon clusters inside the Pfam protein family in which the organism-specific codon usage information has been integrated. The program as well as the filter-based search interface is available at bcb.med.usherbrooke.ca.
2 METHODS
2.1 Detection of rare codon clusters
Sherlocc, for SHERbrooke Locator Of Codon Clusters, is written in PERL. The protein family alignments on which the analysis was performed are from the Pfam-A Seed release 24.0 that contained 11 912 protein families (Finn et al., 2010) and are the primary input of the program. The Sherlocc algorithm passes through three stages, summarized in Supplementary Figure S1 and explained below.
Stage 1. Sherlocc retrieves the nucleotide sequence of every protein in each Pfam protein family alignments from the European nucleotide archive (ENA) database (Leinonen et al., 2011) using cross-referencing from the Uniprot website (Magrane and Consortium, 2011). Using the appropriate translation table (also retrieved from the ENA database), the correspondence of the nucleotide sequence with the amino acid sequence provided in the Pfam alignment is verified.
Stage 2. Using the taxonomic identifier retrieved during Stage 1, the specie-specific codon usage frequencies are retrieved using the Kazusa codon usage frequency online database (Nakamura et al., 2000). The codon usage frequencies in this online database have been calculated using nucleotide sequences of individual organisms from the NCBI GenBank sequences (Benson et al., 2011). In our study, the proteins for which no codon usage frequency values could be assigned were discarded from the protein family alignments. We provide directly on our website the Stage 2 output files for each protein family so that a user can skip Stages 1 and 2 which are time-consuming due to multiple online queries.
Stage 3. To detect rare codon clusters, a seven codon-wide window (blue canvas in Fig. 1), centered at every position of the alignment, averages all codon usage frequencies inside the seven codon-wide window. This average calculated across all proteins of the alignment has subsequently the net effect of assuring that only positions that are rare across the majority of the members of the family are retained. The averages calculated by this window at all positions of all Pfam protein alignments were fitted into an extreme value distribution as described in Laskowski et al., (2005) and references therein. From this distribution (Fig. 2), a statistically significant threshold can be chosen. This threshold will allow us to discriminate positions of the alignment occupied by rare codons with a statistically significant low codon usage frequency average. In the example of Figure 1, this threshold is 13, and all codon usage frequency averages under this threshold are tagged as slow (orange; positions 166–169). To retain only the regions with a high density of slow positions (a rare codon cluster), a second seven position-wide window (purple canvas in Fig. 1) parses the alignments searching for windows with at least four pause positions out of seven. This method retains only the regions occupied by amino acids encoded by the ‘slowest’ codons among all existing positions in all protein families. This implies that even if mutations have led to a different codon and in some cases to a different amino acid, the low codon usage frequency was conserved.
Fig. 1.
Extract of an HTML output generated by Sherlocc. Each row represents a protein from the alignment and displays the amino acid, its corresponding codon and the corresponding codon usage frequency (bold). At the bottom (gray row), codon usage frequency averages calculated at each position by the first window (blue canvas) is displayed in bold (11.56 for position 168). Averages under the selected threshold are considered ‘slow’ and tagged in orange (positions 166–169). A second window (purple canvas) searches for 7 consecutive columns in which there is a minimum of 4 ‘slow’ positions: a rare codon cluster (in red: 166–169).
Fig. 2.
Distribution of codon usage frequency averages (mean: 23.754; SD: 4.868). The dataset was analyzed for rare codon clusters using thresholds 13–18.
2.2 Analysis of preferential positioning of rare codon clusters
A Pfam protein family represents a single domain, which can be part of a single or multi-domain protein. To investigate a preferential positioning of rare codon clusters in the protein as a whole, one must determine where the Pfam domain is positioned relative to the other domains of the protein (for a multi-domain protein). Each Pfam family of the dataset was classified as either strictly N-terminal (with respect to the entire protein) or not strictly N-terminal. To make such classification, the complete protein sequences (long sequences) of each member-protein in all Pfam families were retrieved from the Uniprot website (Magrane and Consortium, 2011). Every protein sequence of the Pfam family alignments (the short sequence) was aligned to its corresponding long sequence using Fasta (Pearson and Lipman, 1988). The number of residues in the long sequence before the start of the short sequence was calculated. The Pfam family was characterized as a strictly N-terminal domain only if the number of residues before the start of the short sequence was <50 residues for every member-protein; else it was classified as a not strictly N-terminal Pfam family. For simplicity of language, all not strictly N-terminal Pfam families are referred to as C-terminal Pfam families.
2.3 Comparison of translational pauses on structures of the same fold
To investigate if rare codon clusters regulate protein folding in a similar way for protein families of the same structural architecture, families containing rare codon clusters were grouped by structural topology. To do so, a representative PDB chain ID was assigned to each Pfam protein family using EBI SIFTS initiative cross-referencing (Velankar et al., 2005) and the PDB chains were assigned to a structural topology using the CATH database (Orengo et al., 1997). To find the position of rare codon clusters on the 3D structures, each member-protein sequence of the family was aligned with the PDB residue sequence using Fasta (Pearson and Lipman, 1988). The sequence with the highest similarity with the PDB residue sequence was used to infer rare codon cluster positions on the PDB structure. We compared among the similar folds, the region of the nascent chain predicted to extrude the ribosome tunnel at the start of the pause (when the ribosome is positioned at the start of a rare codon cluster). The ribosome tunnel can hold in average 30 residues of the nascent chain and sometimes more if the chain is arranged in the form of an α-helix (Etchells and Hartl, 2004). We marked the 10 residues positioned 30–40 residues away from the rare codon clusters toward the N-termini. This 10 residues interval accounts for the possibility that the polypeptide chain within the tunnel can be 30–40 residues long. Protein structures of the same structural topology were compared using PyMOL (Schrödinger, LLC).
3 RESULTS AND DISCUSSION
A total of 11 564 protein family alignments from the initial 11 912 were analyzed by Sherlocc. The 348 discarded families had for all members either no corresponding nucleotide sequence or codon usage frequency information. The whole dataset was analyzed for rare codon clusters using 6 different codon usage frequency thresholds (13–18). Smaller thresholds identify rare codon clusters occupied by codons with a lower codon usage frequency. The protein sequences from the families analyzed encompass 6439 different species, eukaryotes and prokaryotes. The 11 564 alignment files with mapped rare codon clusters (if any) for every threshold can be visualized on our website via a filter-based searchable interface.
Supplementary Table S1 summarizes statistics calculated from families with clusters identified at thresholds 13–18. Lower thresholds decrease the number of Pfam families with rare codon clusters, going from 3360 to 154 for thresholds 18–13, respectively. Rare codon clusters are conserved in proteins families containing up to 606 and 36 protein sequences for thresholds 18 and 13, respectively. The average number of protein sequences in a Pfam family with rare codon clusters (9.8 and 2.6 for thresholds 18 and 13) is relatively low, revealing that the consensus low codon usage frequency region is generally shared among a limited number of protein sequences. Interestingly, there are some cases where the rare codon clusters are conserved in Pfam families containing up to 147 different species, prokaryotes and eukaryotes. The highest number of unique species per Pfam family with at least 1 cluster ranges from 147 to 20 (for thresholds 18–13). The size of the largest cluster is 114 and 23 (for thresholds 18–13). The Pfam families were sorted in descending order of each of the last three columns of Supplementary Table S1.
The purpose of the study is to identify evolutionarily conserved rare codon clusters. Pfam families consist of high-quality alignments of protein sequences constructed using Hidden Markov Model profiles which make an ideal dataset for this type of analysis. Although, it is important to note that protein sequences in the Pfam family alignments represent only a fraction of their open reading frames (ORFs). Supplementary Figure S2 shows distributions of the ratio of ORFs not represented by any Pfam family for each individual protein (284 929 proteins) in all analyzed families. The distribution reveals that for proteins longer or equal to 400 residues (Supplementary Figure S2C), a large proportion of the ORF was not analyzed. This implies that some observations still debated in the literature cannot be confirmed with the present study i.e. the propensity for rare codon clusters to be near domain boundaries observed by Thanaraj and Argos (1996b), Komar and Jaenicke (1995) and Komar (2009) but not observed by Saunders and Deane (2010). As seen in Supplementary Figure S2B, 50% of proteins <400 residues (63.4% of the dataset) have a small fraction (~24%) of ORF excluded from the analysis (not represented in any Pfam family).
3.1 Identifying statistically significant rare codon clusters in protein families
Plotting the distribution of all the codon usage frequency averages (blue canvas in Fig. 1), we can find a statistically significant threshold able to identify, among all amino acid positions of the 11 564 families, the ones occupied by the least used codons. We plotted such distribution (Fig. 2) of the calculated codon usage frequency averages (>1 700 000 values; mean: 23.754; and SD: 4.868).
This statistical approach helps us to deal with the fact that codon usage frequencies form a continuum and that no a priori threshold value exists that can discriminate a rare codon from a frequent one based on the codon usage value alone. Once the codon positions are tagged as ‘slow’ or ‘fast’ based on the threshold, the program can search for regions with a high density of these ‘slow’ positions (rare codon clusters) using the second mobile window (purple canvas in Fig. 1). For some families, the low codon usage frequency of a given position might not be conserved among all member-proteins of the family. The proteins for which this is the case can be identified using the HTML output.
3.2 Validation of identified clusters
We compared clusters identified by Sherlocc with cases found in the literature. For example, the impact of rare codons in the chloramphenicol acetyltransferase (CAT) protein has been previously studied experimentally in E. coli (Komar et al., 1999). The study showed that silent mutations of rare to frequent codons in the CAT protein accelerated the rate of synthesis and led to a 20% decrease in specific activity, which was suggested to be due to protein misfolding. Rare codon clusters have been identified computationally in a multi-organism sequence alignment of this protein (Widmann et al., 2008). Our algorithm, using a threshold of 18, also identified a rare codon cluster in the CAT protein family (PF00302). Another case involves the Salmonella phage P22 tailspike protein in which rare codons were previously identified using the MinMax algorithm (Clarke and Clark, 2008). Sherlocc identified rare codon clusters in the Salmonella phage P22 tailspike protein family (PF09251) down to a threshold of 15 (P-value: 1.72×10−8).
3.3 Filtering for the longest clusters
To investigate potential roles of rare codon clusters, the 673 Pfam families with rare codon clusters identified with a threshold of 15 (chosen to investigate the most statistically rare clusters while retaining sufficient data) were filtered to keep only families with at least 5 protein sequences and containing at least 1 long rare codon cluster of 12 residues in length or more. The protein families resulting from this filtering process are shown in Table 1. From the 673, 143 have at least 5 member-proteins and 72 have at least 1 rare codon cluster that spans a minimum of 12 residues. Combining the 2 filters leaves 13 Pfam families. From the 13 families, 4 have unknown localization (PF07227, PF05340, PF05831 and PF05265). From the remaining 9 families, 7 of them represent proteins that are inserted into membranes, mostly mitochondrial or thylakoïdal membranes (PF05115, PF00283, PF06444, PF00510, PF02326, PF01059 and PF06525). Another (PF04764) has an imprecise localization although it is known to be in chloroplast. The remaining family (PF05394) represents several avirulence proteins from Pseudomonas syringae and Xanthomonas campestris. For the 9 families with known localization, 7 (possibly 8, PF04764) of them are membrane proteins.
Table 1.
Pfam families with the largest rare codon clusters
| Pfam ID | Process involved | Localization |
|---|---|---|
| PF05115 | Photosynthesis | Thyl. membrane |
| PF00283 | Photosynthesis | Thyl. membrane |
| PF06444 | Electron transfer | Thyl. membrane |
| PF00510 | Electron transfer | Thyl. membrane |
| PF02326 | ATP synthesis | Thyl. membrane |
| PF01059 | Electron transfer | Mith. membrane |
| PF06525 | Electron transfer in photosynthesis of plants and bacterias | Membrane |
| PF05394 | Plant infection | Extracellular |
| PF04764 | Unknown | Chloroplasta |
| PF05265 | Unknown | Unknown |
| PF05831 | Unknown | Unknown |
| PF05340 | Unknown | Unknown |
Families needed at least 5 protein sequences and a minimum of 1 rare codon cluster spanning at least 12 residues long in the alignment (see complete table in Supplementary Material).
Legend: Thyl., thylakoid; Mith., mitochondrion; A more detailed version of this table is available in Supplementary Material.
aPrecise localization is unknown.
A plausible explanation for finding long statistically significant rare codon clusters mostly in membrane proteins is the co-translational membrane insertion mechanism. Earlier studies on genes of Saccharomyces cerevisia, E. coli and E. nidulans reported that rare codon clusters could be involved in protein membrane insertion (Dessen and Képès, 2000; Képès, 1996). Other studies on E. coli emphasize the potential involvement of rare codon codons for protein export/secretion (Burns and Beacham, 1985; Power et al., 2004; Zalucki and Jennings, 2007; Zalucki et al., 2011). Despite the suggestions made in these studies, the role of rare codon clusters and their molecular mechanism involved in secretion or membrane translocation/insertion remain unclear. Although the current study does not fill a gap in this regard, our results from a large-scale analysis (11 564 protein families) that indicate a preferential N-terminal positioning of rare codon clusters (discussed further) and a high incidence of large evolutionarily conserved rare codon clusters in membrane related proteins further strengthens the evidence that rare codon clusters are involved in co-translational molecular recognition events involved either in targeting proteins for secretion or insertion into membranes. In many cases, molecular recognition events happen co-translationally. For example, the recognition of the SRP signal sequence in the nascent chain by the SRP protein happens co-translationally (Saraogi and Shan, 2011). The ribosome-nascent-chain-SRP complex traffics to a membrane-bound SRP receptor where the protein is translocated/inserted in the membrane (for review see Jha and Komar, 2011; Wang and Dalbey, 2011). All these mechanisms are dependent on molecular recognition of signal peptides (often positioned in the N-terminal) by a chaperone protein that will guide the protein to its precise localization. The recognition of the signal peptide is an early critical step during which the signal peptide sequence needs to be optimally exposed to the cellular milieu for recognition. While our evidence for co-translational molecular recognition is based on the restricted number of results obtained using parameters that select only the largest, most statistically-relevant evolutionarily conserved rare codon clusters, we believe that many more such cases exist, with perhaps smaller clusters or involving less rare codons.
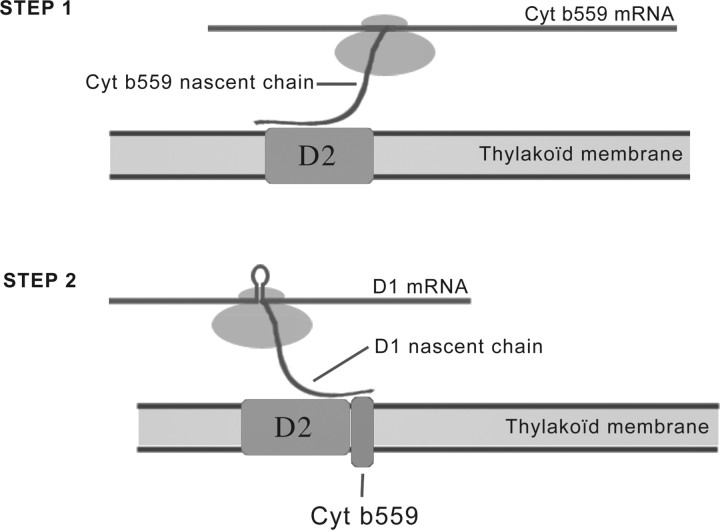
We investigated in more detail the potential co-translational rare-codon regulated membrane insertion using the Pfam family PF00283 (Table 1). This family represents the transmembrane segment of Cytochrome b559, which forms part of the reaction center of the multi-subunit protein-pigment complex PSII and that has been shown to be essential to the PSII assembly. The PSII complex is mainly constituted of D1 and D2 proteins, α and β subunits of Cytochrome b559, psbI and psbW gene products. One of the first steps of the PSII assembly is believed to be the formation of a D2-Cytochrome b559 complex (Müller and Eichacker, 1999). Translational slow down during synthesis of Cytochrome b559 could help its binding to the D2 protein already inserted in the membrane. Cytochrome b559 is composed of 3 segments: a stromal segment attached to a transmembrane segment of ~21 amino acids mainly encoded by rare codons and a final C-terminal lumenal segment. The translational pause caused by the rare codons of the transmembrane segment can give additional time for a signal sequence in the N-terminal stromal segment to co-translationally recognize the D2 protein (Fig. 3) either via a direct protein–protein interaction or a chaperone-based mechanism (like the previously mentioned example of the SRP protein). This would require the signal sequence to be extruded form the ribosome that is positioned at a rare codon cluster 30–40 residues downstream. This result complements the already known similar mechanism that has been observed for the next step of the assembly, which involves association of D1 to the D2-Cytochrome b559 complex. Experimental evidence indicates that the association of D1 to the D2-Cytochrome b559 complex happens co-translationally and via a direct interaction of the nascent D1 chain with the D2 protein (Kim et al., 1991; Zhang et al., 1999). mRNA secondary structures was suggested to cause a translational slowdown during D1 synthesis (Zama, 1995). In summary, we observe rare codon clusters in the first step of the insertion mechanism in addition to the already known second step (D1 insertion). Sherlocc, along with the filter-based searchable interface, can be a useful tool to study these mechanisms in more detail by providing location of putative translational pause sites.
Fig. 3.
Schematic model of the early steps of PSII assembly. Step 1 (hypothesized): rare codon cluster induced ribosomal pausing during synthesis of Cytochrome b559 provides additional time for its N-terminal residues to recognize the D2 protein located in the thylakoïd membrane. Step 2: mRNA secondary structure-induced ribosomal pausing during synthesis of D1 protein facilitates the co-translational recognition of the D1 nascent chain with the D2-Cytochrome b559 complex (Zhang et al., 1999).
3.4 Position of rare codon clusters relative to the N-terminal end of domains
We measured the distances in number of residues between the rare codon clusters and the N-terminal end of the Pfam domains (Fig. 4A). The majority of rare codon clusters are within the first 130 residues of the Pfam protein domains. There is a steep increase of rare codon clusters as we get close to the N-terminal. Figure 4B shows the distances normalized by the length of the respective Pfam domain. The Pfam lengths distribution is available in Supplementary Material (Supplementary Figure S3). From Figure 4B we see that the clusters have only a weak preference for N-terminal positions. When interpreting this result, it is important to keep in mind that Pfam protein families represent protein domains that can come from multi- or single-domain proteins. For this reason we categorized the protein families containing rare codon clusters as either strictly representing the first N-terminal domain (N-terminal Pfam family) or as representing the second, third or further non-N-terminal domain (what we refer for simplicity as a C-terminal Pfam family).
Fig. 4.
Distances between rare codon clusters and N-termini of Pfam domains. (A) The distances are measured in residues from the middle position of each rare codon cluster identified with a threshold of 15 to the N-termini. (B) The distances from A are normalized with the length of their respective Pfam domain.
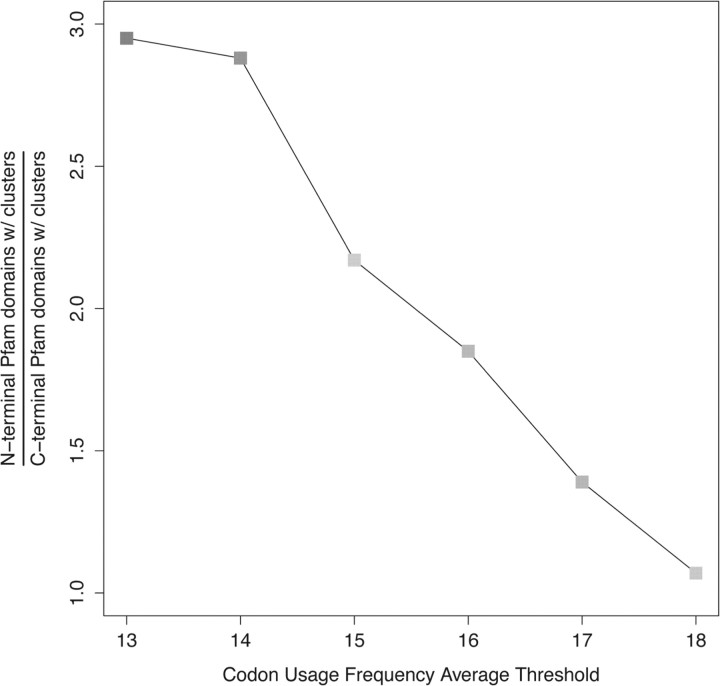
Figure 5 shows the fraction of the number of N-terminal families with clusters over the number of C-terminal families with clusters for 6 different codon usage frequency average thresholds (13–18). As we lower the threshold (lower frequency, increase rareness), we significantly raise the proportion of N-terminal Pfam families with clusters. When analyzing the dataset with a threshold of 13, there is a 3-fold increase of strictly N-terminal Pfam families with clusters. This value decreases as we raise the threshold ending at a ratio of 1:1 for threshold 18 (N-terminal/C-terminal). This result reveals that rare codon clusters identified with more stringent thresholds tend to be positioned closer to the N-terminal of the full protein sequences (i.e. the N-terminal of domains in single domain proteins or the first domain). Rare codon clusters with a higher codon usage have a more diffused positional preference.
Fig. 5.
Fraction of the number of N-terminal Pfam families with clusters over the number of C-terminal Pfam families with clusters for different thresholds. Lower thresholds identify rare codon clusters with the lowest codon usage in the whole dataset. As we reduce the threshold we notice clusters are mainly found in Pfam families that represent strictly N-terminal domains.
Clarke and Clark, 2010, studied ORFs of 26 prokaryotes and found an increased incidence of rare codon clusters at the N-terminal in 15 prokayotes and a smaller but significant increase of rare codons in the C-terminal for 11 of the prokaryotes studied. Our results, based on >6000 different species (eukaryotes and prokaryotes), do not clearly show an increase in the C-terminal end. As noted by Clarke and Clark (2010), the mechanism of translation is very different in eukaryotes versus prokaryotes which can explain the absence of signal in the C-terminal in eukaryotic proteins. The protein families analyzed here can be composed of proteins from prokaryotes and eukaryotes. In such cases, the contribution of the signals of rare codon clusters in the C-terminal in prokaryotic proteins can be dampened by the lack of signal in the eukaryotic sequences explaining why a clear peak is not observed in the C-terminal in the present study.
The results point toward a differential usage of rare codon clusters based on the codon usage frequency. Clusters identified with lower thresholds (i.e. 13) theoretically cause longer ribosomal pausing. These clusters seem to be positioned closer to the N-termini where they can coordinate the insertion of membrane protein. Others have suggested that an N-terminal enrichment in rare codons could help to avoid ribosome collisions downstream (Lesnik et al., 2000). Clusters identified with higher thresholds i.e. 18 (causing shorter ribosomal pauses) are more dispersed along the Pfam domain. They could help fine-tune the multiple steps of the protein folding process. For instance, (Saunders and Deane, 2010) observed that rare codons are positioned preferably at the transition between secondary structures. We further discuss the involvement of rare codon clusters for protein structure below.
3.5 Implication of rare codon clusters in protein structure
Some studies focused on the role of rare codon clusters for protein structure and indirectly folding (Saunders and Deane, 2010; Thanaraj and Argos, 1996a; Widmann et al., 2008). However, the issue of how rare codons influence folding is still unclear (Deane and Saunders, 2011). We wished to revisit this potential role by comparing protein families of the same fold that have rare codon clusters. However, unlike previous studies, we extended our analysis to the entire Pfam database. Using a rather conservative threshold codon usage frequency average of 18 (P-value of 3.00×10−4), a total of 3360 Pfam families had rare codons clusters identified in them. We used a threshold of 18 here as opposed to 15 in all other analysis in this study because we wished to identify clusters with significantly low codon usage frequency averages but that gave us enough groups of protein families of the same structural fold to perform our comparative analysis. Grouping these protein families with rare codon clusters by structural topology led to 81 groups. For clarity of presentation we do not present this list here but it is accessible at our website.
We first investigate if the rare codon clusters partitions the nascent chain in corresponding structurally equivalent sections in different proteins of the same fold. Furthermore, we mapped on representative 3D structures of each family the position of the portion of the nascent chain that just extruded the ribosome tunnel at the moment the predicted pause occurred. This method allows us to visualize what nascent chain section of the corresponding structures would lie outside the ribosome tunnel at the time of the translational pauses. We found some similarities within the 81 structural groups. However, our results in this regard are not sufficiently clear or widespread to suggest a predominant role for rare codon clusters in inducing pauses that may be necessary for co-translational folding events necessary for correct folding in different proteins of the same structural fold. The few noteworthy exceptions are described further down in what follows. The actual rare codon clusters (for all thresholds) were mapped on PDB structures that have 100% identity with a member-protein sequence for visual inspection (Supplementary Material). The uncertainty regarding the length of the polypeptide chain held in the ribosome tunnel may affect our analysis. Experimental data confirms that α-helices can form in the ribosome tunnel (Bhushan et al., 2010), notably raising the number of residues inside the tunnel compared to a situation where the nascent chain would be unstructured. We already know if the residues in the tunnel form an α-helix in the final structure, it is uncertain if the helix is completely formed when inside the ribosome tunnel. We use a buffer window of 10 residues to account for this source uncertainty in our analysis.
Within the groups of structurally homologous proteins analyzed, most of the mapped regions were in different positions relative to the structural elements of the fold. Widmann et al., 2008 analyzed 16 different protein families of the α/β hydrolase fold and concluded the same regarding the position of rare codon clusters. What we observed from this large-scale analysis suggests that rare codons clusters that are conserved within a given Pfam family are not conserved across Pfam families of the same fold. From the 81 structural topology groups the 3 top groups that had the most protein family representatives with rare codon clusters are the Immunoglobulin-like fold, the Rossmann fold and the Jelly roll fold with 39, 29 and 27 representative protein families, respectively. Over the 81 topology groups, these 3 groups alone contained 23% of all protein families. These three topologies are constituted mainly of β-strands that are organized in large sandwich like architectures, β-barrels or parallel β-strands linked to α-helices. It was observed that β-strands are more stable when formed slowly and a previous study showed that rare codons preferentially code for β-strands (Thanaraj and Argos, 1996a).
Rare codon clusters can also ensure the proper formation of α-helices as well. Experimental evidence suggested that translational slowdown caused by rare codons were required for an N-terminal α-helical signal peptide to fold efficiently (Zalucki and Jennings, 2007). Recent studies show that interactions between certain amino acids of the nascent chain with the surface of the ribosome channel are possible (Lu and Deutsch, 2008; Seidelt et al., 2009). Interactions of the nascent chain in concert with precise translational slowdowns could guide the folding of the nascent chain helping it acquire its helical structure inside the ribosome tunnel. Doing so, once outside the tunnel, the helices can efficiently rearrange in more complex structures e.g. coiled-coils, helix-hairpins or helix–helix interfaces. These structures being formed rapidly, any non-favorable interactions of hydrophobic residues within the α-helices with the solvent could be reduced stabilizing the overall structure. This mechanism could occur for Chondroitin ABC lyase I. The protein is built from three structural domains: an N-terminal domain that binds a sodium or calcium ion (represented by PF09092), a central catalytic domain (PF09093) and a C-terminal domain (PF02278) (Huang et al., 2003). Sherlocc identified 11 rare codon clusters in the catalytic domain (PF09093) and 2 in the N-terminal ion-binding domain (PF09092). The majority of the rare codon clusters code for residues of the catalytic domain (Supplementary Fig. S4). This domain is formed by 10 α-helices in the shape of 5 hairpin-like pairs (Huang et al., 2003). One possible role for the detected rare codon clusters is to produce multiple translational pauses during the synthesis of its catalytic domain allowing a step-wise packing of α-helix pairs.
3.6 Codon usage frequency as a measure of translation speed
Secondary structures in the mRNA (Shpaer, 1985) as well as electrostatic interactions of the nascent chain with ribosome components (Lu and Deutsch, 2008; Seidelt et al., 2009) are other factors known to cause translational slowdowns. Predicting mRNA secondary structures in a large-scale context can be a very daunting task and some studies showed that translational pauses observed were not caused by mRNA secondary structures but rather by rare codons (Sørensen et al., 1989; Varenne et al., 1984). As for electrostatic interactions, they remain poorly documented and are therefore hard to analyze in a large-scale context.
Codon usage frequencies have been shown to correlate with tRNA concentrations for prokaryotes as well as eukaryotes (Duret, 2000; Ikemura, 1985; Moriyama and Powell, 1997; Percudani et al., 1997). However, there are exceptions (Parmley and Huynen, 2009; Saunders and Deane, 2010) as tRNA concentrations are tissue-specific and can vary depending on cell condition/cycle or growth rate (Dong et al., 1996; Kanduc, 1997). Considering that species-specific tRNA concentrations have been tabulated for a limited number of species, codon usage frequencies [tabulated for 8792 different species (Nakamura et al., 2000] is the only measure that can be used as a surrogate for translation speed in a large-scale context. Based on these frequencies, one can measure the codon adaptation index (CAI; Sharp and Li, 1987) or the adapted version of by Carbone et al. (2003) which could boost the codon usage bias signal. Although as noted by Clarke and Clark (2010), the CAI is useful to predict highly expressed genes, but not suited to study local translation rates. The smoothing technique combined to the exhaustive probabilistic approach presented in this study, allows the use of a single threshold able to increase the statistical significance of the low codon usage frequency signal we observe.
4 CONCLUSIONS
The primary objective of this study is to perform a large scale survey of the occurrence of evolutionarily conserved rare codon clusters in the almost entirety of Pfam protein families and present Sherlocc, a program able to identify the regions of protein families that are occupied by the lowest codon usage frequencies. As we are interested in the large-scale analysis of evolutionarily conserved rare codons we made use of the curated seed sequence alignments of Pfam domains.
We identified cases where rare codon clusters are conserved in a large number of organisms. We observed that more stringent thresholds identify rare codon clusters mainly in protein N-terminal Pfam domains suggesting that domains closer to the N-termini of proteins require longer pauses. We cannot say strictly if such pauses are required for folding or molecular recognition but based on the involvement of families with rarer clusters with membrane insertion or the recognition of large complexes, we suggest that rare codon cluster are important in co-translational molecular recognition events. We also identified specific cases where the ribosomal pausing caused by rare codon clusters could regulate the folding of functionally important domains.
Proteins are synthesized in a non-linear kinetic landscape and their mRNA sequence seems to convey more information than that which is necessary to encode protein sequences, information that can be used not only to regulate folding events but perhaps more importantly, to regulate co-translational molecular recognition events such as the recognition of signal peptides, the formation of complexes or membrane insertion. The Sherlocc program and the online Sherlocc Finder Interface are efficient tools that can be used to study the widespread translational pauses in protein families.
Supplementary Material
ACKNOWLEDGEMENTS
RJN is part of Centre de Recherche Clinique Étienne-Le Bel as well as a member of the Institut de Pharmacologie de Sherbrooke and Proteo, the Québec network for research on protein function, structure and engineering.
Funding: MC is funded through a grant from the Québec Consortium for Drug Development (CQDM). RJN holds a Junior 1 fellowship from the Fonds de Recherche du Québec — Santé (FRQ-S).
Conflict of Interest: none declared.
REFERENCES
- Benson D.A., et al. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S., et al. alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2010;17:313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- Burns D.M., Beacham I.R. Rare codons in E. coli and S. typhimurium signal sequences. FEBS Lett. 1985;189:318–324. doi: 10.1016/0014-5793(85)81048-6. [DOI] [PubMed] [Google Scholar]
- Carbone A., et al. Codon adaptation index as a measure of dominating codon bias. Bioinformatics. 2003;19:2005–2015. doi: 10.1093/bioinformatics/btg272. [DOI] [PubMed] [Google Scholar]
- Clarke T.F., Clark P.L. Rare codons cluster. PLoS ONE. 2008;3:e3412. doi: 10.1371/journal.pone.0003412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T.F., Clark P.L. Increased incidence of rare codon clusters at 5′and 3′gene termini: implications for function. BMC genomics. 2010;11:118. doi: 10.1186/1471-2164-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazzo P., et al. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem. Biophys. Res. Commun. 2002;293:537–541. doi: 10.1016/S0006-291X(02)00226-7. [DOI] [PubMed] [Google Scholar]
- Deane C.M., Saunders R. The imprint of codons on protein structure. Biotechnol. J. 2011;6:641–649. doi: 10.1002/biot.201000329. [DOI] [PubMed] [Google Scholar]
- Dessen P., Képès F. The PAUSE software for analysis of translational control over protein targeting: application to E. nidulans membrane proteins. Gene. 2000;244:89–96. doi: 10.1016/s0378-1119(00)00002-0. [DOI] [PubMed] [Google Scholar]
- Dong H., et al. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–289. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- Etchells S.A., Hartl F.U. The dynamic tunnel. Nat. Struct. Mol. Biol. 2004;11:391–392. doi: 10.1038/nsmb0504-391. [DOI] [PubMed] [Google Scholar]
- Finn R.D., et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel-Morgenstern M., et al. Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels. Mol. Syst. Biol. 2012;8:572. doi: 10.1038/msb.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., et al. Crystal structure of Proteus vulgaris chondroitin sulfate ABC lyase I at 1.9A resolution. J. Mol. Biol. 2003;328:623–634. doi: 10.1016/s0022-2836(03)00345-0. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Jha S., Komar A.A. Birth, life and death of nascent polypeptide chains. Biotechnol. J. 2011;6:623–640. doi: 10.1002/biot.201000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduc D. Changes of tRNA population during compensatory cell proliferation: differential expression of methionine-tRNA species. Arch. Biochem. Biophys. 1997;342:1–5. doi: 10.1006/abbi.1996.9869. [DOI] [PubMed] [Google Scholar]
- Képès F. The “+70 pause”: hypothesis of a translational control of membrane protein assembly. J.Mol.Biol. 1996;262:77–86. doi: 10.1006/jmbi.1996.0500. [DOI] [PubMed] [Google Scholar]
- Kim J., et al. Ribosomes pause at specific sites during synthesis of membrane-bound chloroplast reaction center protein D1. J. Biol. Chem. 1991;266:14931–14938. [PubMed] [Google Scholar]
- Kimchi-Sarfaty C., et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Komar A.A. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Komar A.A., Jaenicke R. Kinetics of translation of gamma B crystallin and its circularly permutated variant in an in vitro cell-free system: possible relations to codon distribution and protein folding. FEBS Lett. 1995;376:195–198. doi: 10.1016/0014-5793(95)01275-0. [DOI] [PubMed] [Google Scholar]
- Komar A.A., et al. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462:387–391. doi: 10.1016/s0014-5793(99)01566-5. [DOI] [PubMed] [Google Scholar]
- Krasheninnikov I.A., et al. Nonuniform size distribution of nascent globin peptides, evidence for pause localization sites, and a contranslational protein-folding model. J. Protein Chem. 1991;10:445–453. doi: 10.1007/BF01025472. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., et al. Protein function prediction using local 3D templates. J. Mol. Biol. 2005;351:614–626. doi: 10.1016/j.jmb.2005.05.067. [DOI] [PubMed] [Google Scholar]
- Leinonen R., et al. The European nucleotide archive. Nucleic Acids Res. 2011;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnik T., et al. Ribosome traffic in E. coli and regulation of gene expression. J. Theor. Biol. 2000;202:175–185. doi: 10.1006/jtbi.1999.1047. [DOI] [PubMed] [Google Scholar]
- Lu J., Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M., Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhoul C.H., Trifonov E.N. Distribution of rare triplets along mRNA and their relation to protein folding. J. Biomol. Struct. Dyn. 2002;20:413–420. doi: 10.1080/07391102.2002.10506859. [DOI] [PubMed] [Google Scholar]
- Moriyama E.N., Powell J.R. Codon usage bias and tRNA abundance in Drosophila. J. Mol. Evol. 1997;45:514–523. doi: 10.1007/pl00006256. [DOI] [PubMed] [Google Scholar]
- Müller B., Eichacker L.A. Assembly of the D1 precursor in monomeric photosystem II reaction center precomplexes precedes chlorophyll a-triggered accumulation of reaction center II in barley etioplasts. Plant Cell. 1999;11:2365–2377. [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., et al. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo C.A., et al. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Parmley J.L., Huynen M.A. Clustering of codons with rare cognate tRNAs in human genes suggests an extra level of expression regulation. PLoS Genetics. 2009;5:e1000548. doi: 10.1371/journal.pgen.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R., et al. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- Power P.M., et al. Whole genome analysis reveals a high incidence of non-optimal codons in secretory signal sequences of Escherichia coli. Biochem. Biophys. Res. Commun. 2004;322:1038–1044. doi: 10.1016/j.bbrc.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Purvis I.J., et al. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J. Mol. Biol. 1987;193:413–417. doi: 10.1016/0022-2836(87)90230-0. [DOI] [PubMed] [Google Scholar]
- Saraogi I., Shan S.-O. Molecular mechanism of co-translational protein targeting by the signal recognition particle. Traffic. 2011;12:535–542. doi: 10.1111/j.1600-0854.2011.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders R., Deane C.M. Synonymous codon usage influences the local protein structure observed. Nucleic Acids Res. 2010;38:6719–6728. doi: 10.1093/nar/gkq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger, LLC. The PyMol Molecular Graphics System. Version 1.3. [Google Scholar]
- Seidelt B., et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpaer E.G. The secondary structure of mRNAs from Escherichia coli: its possible role in increasing the accuracy of translation. Nucleic Acids Res. 1985;13:275–288. doi: 10.1093/nar/13.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M.A., et al. Codon usage determines translation rate in Escherichia coli. J. Mol. Biol. 1989;207:365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- Thanaraj T.A., Argos P. Protein secondary structural types are differentially coded on messenger RNA. Protein Sci. 1996a;5:1973–1983. doi: 10.1002/pro.5560051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj T.A., Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996b;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-J., et al. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J. Mol. Biol. 2008;383:281–291. doi: 10.1016/j.jmb.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V.N., et al. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Varenne S., et al. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Velankar S., et al. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33:D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Dalbey R.E. Inserting membrane proteins: the YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. Biophys. Acta. 2011;1808:866–875. doi: 10.1016/j.bbamem.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Widmann M., et al. Analysis of the distribution of functionally relevant rare codons. BMC Genomics. 2008;9:207. doi: 10.1186/1471-2164-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalucki Y.M., Jennings M.P. Experimental confirmation of a key role for non-optimal codons in protein export. Biochem. Biophys. Res. Commun. 2007;355:143–148. doi: 10.1016/j.bbrc.2007.01.126. [DOI] [PubMed] [Google Scholar]
- Zalucki Y.M., et al. Coupling between codon usage, translation and protein export in Escherichia coli. Biotechnol. J. 2011;6:660–667. doi: 10.1002/biot.201000334. [DOI] [PubMed] [Google Scholar]
- Zama M. Discontinuous translation and mRNA secondary structure. Nucleic Acids Symp. Ser. 1995;34:97–98. [PubMed] [Google Scholar]
- Zhang L., et al. Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 1999;274:16062–16067. doi: 10.1074/jbc.274.23.16062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.