Abstract
The γ-carboxyglutamic acid (Gla) domain of blood coagulation factors is responsible for Ca2+-dependent phospholipid membrane binding. Factor X-binding protein (X-bp), an anticoagulant protein from snake venom, specifically binds to the Gla domain of factor X. The crystal structure of X-bp in complex with the Gla domain peptide of factor X at 2.3-Å resolution showed that the anticoagulation is based on the fact that two patches of the Gla domain essential for membrane binding are buried in the complex formation. The Gla domain thus is expected to be a new target of anticoagulant drugs, and X-bp provides a basis for designing them. This structure also provides a membrane-bound model of factor X.
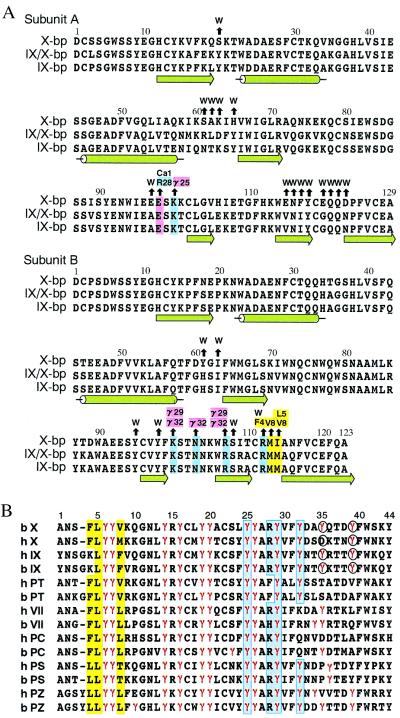
When the wall of a blood vessel is damaged, the activated factor VIIa-tissue factor complex initiates coagulation by activating factor X and factor IX, which leads to the formation of thrombin and ultimately a fibrin clot (1). Factors VII, IX, and X and prothrombin consist of a light chain and a heavy chain. The light chain contains the N-terminal γ-carboxyglutamic acid (Gla) domain and two epidermal growth factor (EGF)-like domains (the N-terminal Gla domain and two kringle domains in prothrombin). The heavy chain contains a trypsin-like serine protease domain. To find anticoagulant drugs, many inhibitors of thrombin and the trypsin-like serine protease domain active sites have been developed (2). As many other biologically important enzymes, however, belong to the members of the thrombin/trypsin family, a problem of selectivity exists when targeting one particular enzyme active site, and the inhibitor drugs cause the risk of producing the wrong drug profile or harmful side effects. Recently, Dennis et al. (3) found anticoagulant peptides by using phage display of naïve peptide libraries, which bind tightly to a site on factor VIIa that is distinct from the active site. Such exosite inhibitors are expected as a way of solving the above problem. Meanwhile, we found that naturally occurring anticoagulant proteins isolated from snake venoms, designated IX-bp and X-bp, form a complex with factors IX and X, respectively, whereas IX/X-bp forms a complex with either factor IX or X (4–6). These anticoagulant proteins are highly homologous with one another as shown in Fig. 1A, and belong to the C-type lectin superfamily (7), although lectin activity has not been reported. We already showed (6) that these proteins bind tightly to the Gla domain, which is responsible for phospholipid membrane binding in the presence of Ca2+ ions, and it is therefore likely that they block membrane interaction of the Gla domains that are essential for the amplification of the coagulation cascade (8). Fig. 1B shows amino acid sequence of the Gla domain containing 9–12 Gla residues within the first 40 residues of the N terminus of the mature protein. The Gla domain has a unique structure as described later, and is thus expected to be a new target of anticoagulant drugs, and the anticoagulant proteins from snake venom provide a basis for designing them. Structural studies of the present complex show how specifically the Gla domain binds to X-bp, or vice versa. The binding sites and residues on the Gla domain have an important correlation with those that have been proposed to interact with a membrane so far, suggesting a mechanism for membrane binding involving these interactions.
Figure 1.
Amino acid sequence alignments. (A) Anticoagulant proteins from snake venoms. Black arrows above the residues of X-bp point to amino acid residues of XGD1-44, Ca-1, or water molecules (W) with which they interact. Positively charged, negatively charged, and hydrophobic residues involved in those interactions are colored in blue, pink, and yellow, respectively. The numbering and secondary structure elements also are shown. (B) The Gla domains of vitamin K-dependent proteins (X, factor X; IX, factor IX; PT, prothrombin; VII, factor VII; PC, protein C; PS, protein S; PZ, protein Z) from human (h) and bovine (b). The numbering at the top refers to the bX sequence. Gla residues are denoted by γ and colored in red. Blue and yellow columns show hydrophilic and hydrophobic patches, respectively. Residues coordinating to the eighth Ca2+ ion are indicated in the circles.
Materials and Methods
Purification, Crystallization, and Data Collection.
X-bp was isolated and purified from the venom of Deinagkistrodon acutus (6). A Gla domain-containing peptide 1-44 (XGD1-44) was prepared and purified with factor X, which was isolated from bovine plasma (6). The mixture of X-bp and XGD1-44 was chromatographed on one column of Superdex 75 pg (1.6 × 60 cm) by using 50 mM Tris⋅HCl, pH 8.0, containing 0.1 M NaCl and 5 mM CaCl2 as the elution buffer to remove free XGD1-44. Formation of the stoichiometric complex was confirmed by gel filtration and reversed-phase HPLC analysis (Cosmosil 5C8 column, Nacalai Tesque, Kyoto). The complex was crystallized by the method of vapor diffusion from solution of 10 mM Tris⋅HCl, pH 7.9, 12% polyethylene glycol 8000, and 5 mM CaCl2. Crystals grew to maximal dimensions of 0.2 × 0.2 × 0.6 mm, belonged to space group P41212, with cell dimensions of a, b = 99.8 Å and c = 90.4 Å, and contained one complex in the asymmetric unit. Diffraction data were collected with the Weissenberg imaging-plate detector system (9) using synchrotron radiation (λ = 1.00 Å) at beamline 6A of the Photon Factory (Tsukuba, Japan). Data to 2.3-Å resolution were processed with DENZO/SCALEPACK (10), and statistics are given in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P41212 |
| Unit cell | a = b = 99.8 Å, c = 90.4 Å |
| Resolution range | 100–2.3 Å (2.44–2.30 Å) |
| No. of unique reflections | 20,411 (3,206) |
| Completeness | 96.4% (92.5%) |
| I/σ(I) > 3 | 82.8% (65.7%) |
| R-merge | 8.1% (33.0%) |
| Refinement statistics (30 to 2.3-Å resolution) | |
| Reflections | 18,805 |
| R-factor | 20.1% |
| Rfree-factor | 26.2% |
| No. of protein nonhydrogen atoms | 2,460 |
| No. of X-bp residues | 252 |
| No. of Gla-domain residues | 44 |
| No. of calcium ions | 12 |
| No. of water molecules | 181 |
| rms deviations from ideal geometry | |
| Bond lengths | 0.008 Å |
| Bond angles | 1.5° |
| Overall average B-factor | 24.3 Å2 |
| Average B-factor (protein) | 23.9 Å2 |
| Average B-factor (calcium) | 21.9 Å2 |
| Average B-factor (water) | 30.2 Å2 |
Structure Determination and Refinement.
The structure was determined by molecular replacement with program AMORE (11) by using the known structure of IX/X-bp (ref. 12; Protein Data Bank ID code 1IXX) as a search model. The analysis using diffraction data from 8 to 3.5 Å yielded a single independent solution with a final correlation coefficient of 47% and R value of 45.8%. The appropriately placed X-bp model was used for the calculation of the initial phases. The electron density map created with program QUANTA97 (Molecular Simulations, Waltham, MA) was sufficient to identify the main-chains and side-chains of X-bp. The structure was refined with program X-PLOR (13), using the parameters of Engh and Huber (14). Performing repeated cycles of refinement gradually extending to 2.6-Å resolution resulted in an R-factor of 31.6% and an Rfree-factor of 44.0%. The resultant 3Fo-2Fc map revealed an additional strong density corresponding to the bound Gla domain, which nearly fitted the known structure of the Gla domain of Ca-prothrombin fragment (15). Model building was followed by refinement. This procedure was repeated until the whole XGD1-44 sequence had been fitted to the density. Table 1 gives the statistics of the final model, which includes eight Ca2+ ions bound to XGD1-44, two Ca2+ ions bound to X-bp and two Ca2+ ions between symmetry-related X-bp molecules, and 181 water molecules.
Modeling of Factor Xa Bound to X-bp.
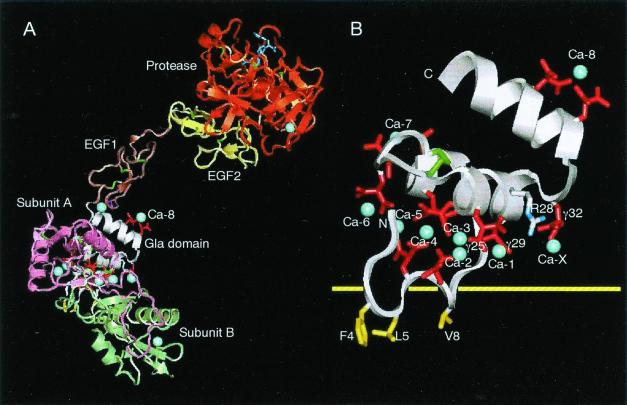
The model shown in Fig. 3A was constructed by employing the program QUANTA97, and by using the structure of factor IXa (16) as a template. The backbone chains of the protease, EGF1 and EGF2 domains of factor Xa (17), were superimposed on those of factor IXa. Similarly, the backbone chains of XGD1-44 bound to X-bp were superimposed on the part of the Gla domain of factor IXa. Clashes of the docking region between EGF1 and Gla domains were relieved by rearrangements of that region.
Figure 3.
Model of factor Xa bound to X-bp and XGD1-44 bound to membrane. (A) Factor Xa bound to X-bp. The Gla residues are in red, bound Ca2+ ions in blue (only labeled is Ca-8, which is identified in the present study), and disulfide bonds in green. The small molecule (dark blue) bound to the active site of the protease domain is the FX-2212a inhibitor (17). (B) Putative membrane-binding surface of XGD1-44. Same view as in A, but the scale of the figure is magnified for clarity. The hydrophobic patch includes Phe4, Leu5, and Val8, and hydrophilic patch includes Arg28, Gla25, Gla29, Gla32, and Ca-1 as a bridging Ca2+, which are on either side of the yellow horizontal line of the putative membrane surface. Ca-X is a putative Ca2+ ion that is taken in as another bridging Ca2+.
Results and Discussions
Structure of X-bp.
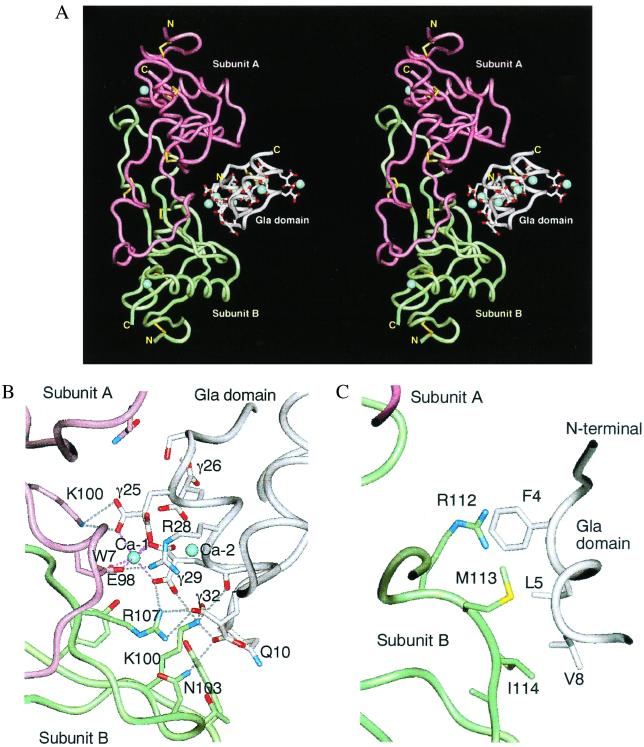
The homologous subunits A and B are related by a pseudodyad, which is perpendicular to the long axis of the molecule (Fig. 2A). The dimer is formed by domain swapping (18, 19), and the putative closed monomer of each subunit is a carbohydrate-recognition domain of a C-type lectin. The main-chain fold of X-bp is almost identical with those of known structures of the homologous proteins, IX/X-bp (12) and IX-bp (18), as might be expected by their high degree of sequence homology (Fig. 1A). A marked difference is found in the relative orientation of the two subunits A and B, as has been pointed out in a comparison between IX/X-bp and IX-bp (18). In the present case, when the B subunits of X-bp and IX-bp are superimposed, 5° rotation of the A subunit of X-bp around the axis, which is approximately perpendicular to the pseudotwofold axis and passing the position of the interchain disulfide bond, is required for the superposition of the A subunit of IX-bp. This finding suggests dynamic behavior between subunits A and B that is analogous to domain movements proposed in proteins (20). The rotational motion observed here is constrained severely by the presence of the interchain disulfide bond, which is concerned with the opening and closing of the concave surface that serves as the binding site of the Gla domain. Consequently, this motion might contribute to induced fit during Gla domain recognition.
Figure 2.
Overall structure of X-bp and XGD1-44 complex. (A) Stereoview of ribbon model viewed perpendicular to the pseudodyad of the molecule, showing the interface between X-bp and XGD1-44. The subunits A and B of X-bp are magenta and green. XGD1-44 is white. The side chain of Gla residues and disulfide bonds are displayed. The bound Ca2+ ions are denoted by blue spheres. (B) Same view as in A, but molecular detail of interaction between the hydrophilic patch of XGD1-44 and positively charged X-bp, and a bridging Ca2+. (C) Same view as in B, but between the N terminus hydrophobic patch of XGD1-44 and the C terminus of subunit B of X-bp.
Structure of XGD1-44 Fragment.
Crystal structures of the Gla domains are only available from the Ca-prothrombin fragment 1 (15) and factor VIIa/tissue factor complex (VIIa/TF; ref. 21). In the crystal structures of factor Xa (17, 22) and IXa (16), their Gla domains were absent or disordered. Therefore, the crystal structure of the Gla domain of factor X has been observed in this study. XGD1-44 in the complex adopts the same structural topology as the Gla domains of the Ca-prothrombin fragment 1 and VIIa/TF (the rms deviations for 44 common Cα-positions are 1.53 Å and 0.50 Å, respectively), in which the Gla domain is highly structured and seven Ca2+ ions are coordinated mainly by the Gla residues. In the present structure analysis, an additional Ca2+-binding site (the eighth site) was identified, formed by Gla35 and Gla39 at the C-terminal helix region through four Ca–O interactions. These two Gla residues are conserved only in factors IX and X, as shown in Fig. 1B (Gla35 of human factor X is replaced by Asp, but coordination to Ca2+ is possible by using two oxygen atoms of the carboxyl group of Asp). Factors IXa and Xa resemble each other in overall shape and bind to the homologous cofactors VIIIa and Va, forming intrinsic Xase and prothrombinase, respectively (1). The abovementioned eighth Ca2+-binding site of the Gla domain of factors IX and X points toward the concave surface corresponding to a putative VIIIa binding surface in the arched IXa molecule (16), suggesting that this site is involved in the formation of a complex with the cognate cofactors.
Interaction Between X-bp and XGD1-44.
About 1,660 Å2 of solvent accessible surface is buried between XGD1-44 and X-bp. Of the 750 Å2 buried on the side of XGD1-44, about 47% is caused by two patches that contribute to the direct interaction with X-bp. The remaining 53% is occupied by ordered water molecules, which form extensive water-mediated hydrogen bonds and may contribute to the stability of the two direct contacts, as observed in antigen–antibody interfaces (23). The hydrophilic patch is formed by a cluster of negatively charged Gla residues 25, 29, and 32, Arg28, and Ca-1, as shown in Fig. 2B. Glu98(A) (A designates subunit A of X-bp) is bound to Ca-1, probably by displacing one of two water molecules, which would normally coordinate to Ca-1 before complex formation taking place, forming a Ca2+-bridging interaction. The negatively charged surface of XGD1-44 is in intimate contact with a cluster of positively charged residues of X-bp through nine salt bridges as illustrated in Fig. 2B. Residues involved in this interaction are strictly conserved in X-bp, and also conserved in XGD1-44, except for Arg28 and Gla32. Arg28 is replaced by Phe, His, and Lys in prothrombin, bovine factor VII, and human protein C, respectively, whereas Gla32 is replaced by Lys and Arg in human and bovine factor VII, respectively, and by Gln in both human and bovine protein C. Therefore, it is reasonable to speculate that interactions similar to those observed here occur in the complex between IX/X-bp and the Gla domains of factors IX and X, and between IX-bp and factor IX, with residues 28 and 32 being important for conferring specificity. This finding is consistent with the finding that X-bp, IX/X-bp, and IX-bp have no affinity for prothrombin, factor VII, and protein C.
Three hydrophobic residues in the N-terminal loop, Phe4, Leu5, and Val8 form the second patch that interacts with the hydrophobic residues in the loop of subunit B of X-bp (Fig. 2C). The aromatic ring of Phe4 has a parallel stacking arrangement over the Arg112(B) guanidino group, as observed frequently in proteins (24). Leu5 participates in a hydrophobic interaction with I114(B) and, similarly, Val8 interacts with Met113(B) and I114(B). These three hydrophobic residues are conserved among Gla domain-containing proteins except in the case of factor IX where Phe4 is replaced by Lys (Fig. 1B). This amino acid difference may account for the difference in binding affinity to X-bp inasmuch as that of factor IX was 10-fold lower than that of factor X (6). In both proteins S and Z, Phe4 is replaced by Leu, which is hydrophobic but unfavorable for the stacking interaction with Arg112(B).
Thus, interactions similar to those observed here may be present in the complex between homologous coagulation factor-binding proteins (IX/X-bp and IX-bp) and cognate Gla domains, with Gla domain residues 4, 28, and 32 acting as likely discriminators in complex formation. On the other hand, it is uncertain which are the discriminator residue(s) on the coagulation factor-binding proteins, because the residues directly involved in the interactions are strictly conserved among these proteins (Fig. 1A). Amino acid sequences are highly homologous, so that variable residues are concentrated only in the loop between the second α-helix and the second β-strand in subunit A (Fig. 1A), suggesting that this is a discriminator site (6). This region and the C-terminal region of subunit A interact with XGD1-44 through water molecules. Water-mediated hydrogen bonds may play an important role in determining specificity as well as stability, as suggested in situations involving protein–protein and protein–DNA recognition (25). It seems that strict selection by coagulation factor binding is not biologically important for snake venom. IX/X-bp binds not only to factor IX but also to factor X, and X-bp binds also to factor IX with affinity 10 times lower than to factor X (6). Such a selectivity of recognition of factor IX and/or factor X is ascribed to the interaction of water-mediated hydrogen bonds.
A Model for the Membrane-Binding of Factor X.
X-bp is bound to the Gla domain at the bottom of the predicted composite model of factor Xa in Fig. 3A. This position corresponds to the plane of the membrane surface if a horizontal membrane orientation is assumed as in Fig. 3B. The 65-Å distance from this plane to the Xa active site is consistent with the 61 Å expected from fluorescence studies (26). It is probable that X-bp inhibits the binding of the Gla domain to the membrane by effectively blocking its binding interface. In this context, it is of particular interest to note that both ionic and hydrophobic interaction sites observed between XGD1-44 and X-bp correspond to those which have been proposed from x-ray (15, 21) and NMR (27, 28) studies to account for the phospholipid binding mechanism of the Gla domain (for a recent review, see ref. 29). First, we propose that hydrophilic patch 1 of the Gla domain is the major contributor toward the ionic interaction with phospholipid and in the Ca2+ ion bridging (30), as follows. (i) Ca-1 is used in a binding interaction with the negative head groups on the membrane surface. (ii) Gla32, with potential for Ca2+ ion binding and the only Gla residue not involved in binding Ca2+ ions within the Gla domain of the present structure and Ca-prothrombin fragment 1 (15), forms another Ca2+ ion bridging to a phospholipid surface. There is also evidence from site-directed mutagenesis studies that the corresponding residue Gla33 in prothrombin is of some importance in membrane binding (31). Further evidence is that a point mutation of Gla32 to Gln has been found in a family diagnosed with factor X deficiency (32). (iii) Positively charged Arg28 is used in a salt bridge interaction with membrane phosphate groups. Thus, all three interactions may participate in concert in binding to membrane. Second, we understand that the hydrophobic residues Phe4, Leu5, and Val8 of hydrophobic patch 2 of the Gla domain of factor X are another contributor to membrane binding. Solution study of the Gla-EGF domain of factor X showed the Ca2+-induced exposure of those hydrophobic amino acid side chains into the solvent (27). Those hydrophobic residues are thus readily accessible to interaction with the phospholipid membrane when Ca2+ ions are bound. The above two patches can make simultaneous contact with the membrane surface, but the total contact area is still relatively small and not extensive enough to be irreversible. The binding mode of the factor Xa interaction with membrane surface may represent a general feature of the homologous Gla domains in this family of proteins.
This crystallographic study demonstrated that the anticoagulation caused by X-bp is based on the fact that two patches of the Gla domain essential for membrane binding are buried in the complex formation with X-bp. The formation of the complex could block efficiently membrane interaction of the Gla domain that is essential for the amplification of the coagulation cascade. The Gla domain is expected to be a new target of anticoagulant drugs because of its unique structure. X-bp (also IX-bp and IX/X-bp), which specifically interacts with the Gla domain, may provide a new framework for the drug design. This specific interaction also provides a membrane-bound model of factor X.
Acknowledgments
We thank Drs. N. Sakabe, N. Watanabe, M. Suzuki, and N. Igarashi for use of the facilities at Photon Factory, Tsukuba, Japan. This study was supported in part by a fund for Promoting Science and Technology of Science and Technology Agency (to H.M.), and by a Research Grant for Cardiovascular Diseases from Ministry of Health and Welfare to (T.M.).
Abbreviations
- X-bp
factor X-binding protein
- IX-bp
factor IX-binding protein
- IX/X-bp
factor IX/factor X-binding protein
- EGF
epidermal growth factor
- Gla
γ-carboxyglutamic acid
- XGD1-44
Gla domain-containing peptide 1-44 of factor X
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1IOD).
References
- 1.Mann K G. Thromb Haemostasis. 1999;82:165–174. [PubMed] [Google Scholar]
- 2.Leung D, Abbenante G, Fairlie D P. J Med Chem. 2000;43:305–341. doi: 10.1021/jm990412m. [DOI] [PubMed] [Google Scholar]
- 3.Dennis M S, Elgenbrot C, Skelton N J, Ultsch M H, Santell L, Dwyer M A, O'Connell M P, Lazarus R A. Nature (London) 2000;404:465–470. doi: 10.1038/35006574. [DOI] [PubMed] [Google Scholar]
- 4.Atoda H, Hyuga M, Morita T. J Biol Chem. 1991;266:14903–14911. [PubMed] [Google Scholar]
- 5.Atoda H, Ishikawa M, Yoshihara E, Sekiya F, Morita T. J Biochem. 1995;118:965–973. doi: 10.1093/jb/118.5.965. [DOI] [PubMed] [Google Scholar]
- 6.Atoda H, Ishikawa M, Mizuno H, Morita T. Biochemistry. 1998;37:17361–17370. doi: 10.1021/bi981177x. [DOI] [PubMed] [Google Scholar]
- 7.Drickamer K. Prog Nucleic Acid Res Mol Biol. 1993;45:207–232. [PubMed] [Google Scholar]
- 8.Furie B, Bouchard B A, Furie B C. Blood. 1999;93:1798–1808. [PubMed] [Google Scholar]
- 9.Sakabe N. Nucl Instrum Methods Phys Res Sect A. 1991;303:448–463. [Google Scholar]
- 10.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 11.Collaborative Computational Project number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 12.Mizuno H, Fujimoto Z, Koizumi M, Kano H, Atoda H, Morita T. Nat Struct Biol. 1997;4:438–411. doi: 10.1038/nsb0697-438. [DOI] [PubMed] [Google Scholar]
- 13.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 14.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 15.Soriano-Garcia M, Padmanabhan K, de Vos A M, Tulinsky A. Biochemistry. 1992;31:2554–2566. doi: 10.1021/bi00124a016. [DOI] [PubMed] [Google Scholar]
- 16.Brandstetter H, Bauer M, Huber R, Lollar P, Bode W. Proc Natl Acad Sci USA. 1995;92:9796–9800. doi: 10.1073/pnas.92.21.9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamata K, Kawamoto H, Honma T, Iwana T, Kim S H. Proc Natl Acad Sci USA. 1998;95:6630–6635. doi: 10.1073/pnas.95.12.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuno H, Fujimoto Z, Koizumi M, Kano H, Atoda H, Morita T. J Mol Biol. 1999;289:103–112. doi: 10.1006/jmbi.1999.2756. [DOI] [PubMed] [Google Scholar]
- 19.Benett M J, Schlunegger M P, Eisenberg D. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gernstein M, Lesk A M, Chotia C. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 21.Banner D W, D'Arcy A, Chene C, Winkler F K, Guha A, Konigsberg W H, Nemerson Y, Kirchhofer D. Nature (London) 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 22.Padmanabhan K, Padmanabhan K P, Tulinsky A. J Mol Biol. 1993;232:947–966. doi: 10.1006/jmbi.1993.1441. [DOI] [PubMed] [Google Scholar]
- 23.Covell D G, Wallqvist A. J Mol Biol. 1997;269:281–297. doi: 10.1006/jmbi.1997.1028. [DOI] [PubMed] [Google Scholar]
- 24.Flocco M M, Mowbray S L. J Mol Biol. 1994;235:709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- 25.Janin J. Structure. 1999;7:R277–R279. doi: 10.1016/s0969-2126(00)88333-1. [DOI] [PubMed] [Google Scholar]
- 26.Husten E J, Esmon C T, Johnson A E. J Biol Chem. 1987;262:12953–12961. [PubMed] [Google Scholar]
- 27.Sunnerhagen M, Forsén S, Hoffrén A-M, Drakenberg T, Teleman O, Stenflo J. Nat Struct Biol. 1995;2:504–509. doi: 10.1038/nsb0695-504. [DOI] [PubMed] [Google Scholar]
- 28.Freedman S J, Furie B C, Furie B, Baleja J D. Biochemistry. 1995;34:12126–12137. doi: 10.1021/bi00038a005. [DOI] [PubMed] [Google Scholar]
- 29.Nelsestuen G L, Shah A M, Harvey S B. Vitam Horm (San Francisco) 2000;58:355–389. doi: 10.1016/s0083-6729(00)58031-5. [DOI] [PubMed] [Google Scholar]
- 30.Schwalbe R A, Ryans J, Stern D M, Kisiel W, Dahlback B, Nelsestuen G L. J Biol Chem. 1989;264:20288–20296. [PubMed] [Google Scholar]
- 31.Ratcliffe J V, Furie B, Furie B C. J Biol Chem. 1993;268:24339–24345. [PubMed] [Google Scholar]
- 32.Zama T, Murata M, Watanabe R, Yokoyama K, Moriki T, Ambo H, Murakami H, Kikuchi M, Ikeda Y. Br J Haematol. 1999;106:809–811. doi: 10.1046/j.1365-2141.1999.01614.x. [DOI] [PubMed] [Google Scholar]