Abstract
The Krüppel-like zinc finger transcription factor, Glis3, has been associated with neonatal diabetes in humans and mice, and implicated in the regulation of pancreatic β-cell generation. However, its precise function in the development of pancreatic β-cells has not yet been elucidated. In this study, we provide evidence that Glis3 regulates Neurogenin 3 (Ngn3) through its distal promoter region. Previous studies showed that the distal region and proximal region of Ngn3 promoter contains various transcription binding sites, including binding sites for pancreatic and duodenal homeobox 1 (Pdx1), Hnf1β and Hnf6. Interestingly, putative Glis3 binding sites (Glis3BS) were found in the distal region of Ngn3 promoter close to the Hnf6 binding sites. This suggested that along with Hnf6, Glis3 may also be involved in the regulation of Ngn3 expression. This hypothesis is supported by data showing that Glis3 can bind to the Ngn3 promoter directly and activate Ngn3 transcriptional activity. Additionally, Glis3 can interact directly with Hnf6 in vitro and in vivo. The amino-terminus in Glis3 and the homeodomain of Hnf6 are critical for this interaction. These data suggest that crosstalk between Glis3 and Hnf6 may play an important role in the regulation of Ngn3 during pancreatic endocrine progenitor cell specification and development.
Keywords: Glis3, Glis3 binding site, Hnf6, Ngn3
INTRODUCTION
During pancreas development various gene regulatory networks (GRN) involving various transcription factors and morphogens govern the development of the endocrine compartment (Gittes, 2009). Pancreas development is initiated in the dorsal and ventral cells of the early endodermal primodium at E8.5 that subsequently fuse to create a single organ (Gittes, 2009; Jensen, 2004). The precursors of the ductal, endocrine, and exocrine cells are located in the pluripotent epithelium surrounded by mesenchymal cells and become committed at mid-gestation (Gu et al., 2002). The pancreatic progenitor marker, Pdx1, is expressed in the endodermal region during pancreas development and throughout endocrine cell formation in both the dorsal and ventral buds. The induction of the transcription factor Neurogenin3 (Ngn3) in Pdx1 positive cells is essential for the generation of proendocrine progenitors at the secondary transition initiated at E13.5. These progenitor cells give rise to all pancreatic endocrine cell types (Gittes, 2009; Rukstalis and Habener, 2009). The branched ductal compartment and mature exocrine acini develop during the secondary transition at E15.5–E16.5 in the mouse (Gittes, 2009; Pierreux et al., 2006; Rukstalis and Habener, 2009). Following endocrine aggregation, the islets of Langerhans also take shape by the end of this time frame.
The expression of the basic helix-loop-helix transcription factor, Ngn3, is highly dynamic during pancreas development (Apelqvist et al., 1999; Gradwohl et al., 2000; Jensen et al., 2000; Oliver-Krasinski et al., 2009; Rukstalis and Habener, 2009). Although Ngn3 first appears in the early pancreatic epithelium at E9 (Jensen et al., 2000; Rukstalis and Habener, 2009; Schwitzgebel et al., 2000), it reaches its highest level of expression during the second transition between E13.5–E15.5, and then decreases substantially thereafter (Gradwohl et al., 2000; Gu et al., 2002; Rukstalis and Habener, 2009). Ngn3 is not only critical during normal embryonic endocrine cell differentiation, but has also been found to be critical for endocrine cell generation in the adult (Xu et al., 2008). Moreover, Ngn3 is critical in viral-mediated somatic exocrine-to-endocrine cell conversion in the adult organ (Zhou et al., 2008). Although Ngn3 is known to be an endocrine progenitor marker, its gene regulation is not fully understood. Ngn3 expression is repressed by notch-mediated intracellular signaling (Rukstalis and Habener, 2009) and is directly and negatively regulated by Hes1. Current knowledge of positive control of the Ngn3 promoter activation is limited to the involvement of Hnf6 also known as Onecut1 (Oc1), which is expressed in pancreatic progenitor cells, but not adult endocrine cells (Jacquemin et al., 2000; Lemaigre et al., 1996; Poll et al., 2006). Two individual Hnf6 binding sites have been identified within the distal promoter of Ngn3 spaced approximately 250 bp apart (Jacquemin et al., 2000; Lee et al., 2001). Furthermore, another Onecut member, Onecut2 (Oc2), can also positively regulate Ngn3 (Jacquemin et al., 2000; 2003; Lee et al., 2001). The role of Oc2 appears to be partially redundant to Hnf6 (Jacquemin et al., 1999).
During pancreas development Hnf6 and Oc2 function as activators of Ngn3 gene transcription (Gu et al., 2002; Jacquemin et al., 1999; 2000; Poll et al., 2006). Deficiency of Hnf6 in mice was shown to lead to ventral pancreatic hypoplasia and reduced Pdx1 expression (Jacquemin et al., 2003). Ngn3 expression is almost abolished in the absence of Hnf6, causing a significant reduction in endocrine cell formation. In addition, Hnf6 deficient mice develop biliary cysts in the liver and similar abnormalities in pancreatic duct that have been linked to defective cilia formation (Jacquemin et al., 2003).
The Krüppel-like zinc finger transcription factor Gli-similar (Glis) family consists of three members, Glis1, Glis2 and Glis3 (Kang et al., 2010; Kim et al., 2002; 2003; Zhang et al., 2002). Among them, Glis3, is highly expressed in kidney and pancreas and has critical function in the development and maintenance of these tissues (Kang et al., 2009a; 2009b). Mutations in the Glis3 gene in human and mouse result in neonatal diabetes and hypothyroidism (Dupuis et al., 2010; Kang et al., 2009b; Senee et al., 2006). Interestingly, Glis3 regulates the preproinsulin gene by binding to its proximal promoter region (Kang et al., 2009b). More recently, Glis3 single polynucleotide polymorphisms have been associated with increased risk for type 1 and type 2 diabetes (Barrett et al., 2009; Dupuis et al., 2010).
In this study, we characterize the role of Glis3 in the regulation Ngn3 and the Ngn3 promoter. The Ngn3 distal regulatory cluster is positioned at −3.9 kb of the Ngn3 promoter (Jacquemin et al., 2000; Lee et al., 2001). We found that putative Glis3BS are located near the Ngn3 distal regulatory cluster region. Binding of Glis3 to these response elements and regulation of Ngn3 by Glis3 were supported by EMSA and reporter gene analysis. Furthermore, we also demonstrated that Glis3 can interact with Hnf6. The significance of the Glis3/Hnf6 interaction in endocrine progenitor and Ngn3 regulation requires further investigation. We speculate that a Glis3/Hnf6 protein complex together with other transcription factors, (i.e. Pdx1, Sox9, Foxa2, and Hnf1β) are a part of a larger Ngn3 regulatory network that may be directly responsible for the activation of specific genes during different steps of pancreatic development, including the generation pro-endocrine/pro-ductal progenitor cells.
MATERIALS AND METHODS
Cell culture and transient transfection
HepG2 and HEK293 cells were cultured and maintained in DMEM (10% heat inactivated FBS, penicillin-streptomycin. Rat insulinoma Ins(832/13) cells and mouse βTC6 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 10 mM HEPES, 2 mM glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM β-mercaptoethanol. Unless otherwise indicated, transfection was performed using Fugene6 (Roche, USA) in triplicate using a total of 0.5 μg DNA (0.15 μg of Ngn3 promoter reporters, the indicated amounts of the expression vectors p3 × Flag-Glis3 and pCMV-Myc-Hnf6, and 0.05 μg of pCMV-β-galactosidase or pRL-Luciferase reporters as controls).
Plasmids
Flag-tagged deletion expression constructs of Glis3 were generated by PCR amplification using Glis3 specific primer sets, deltaN-Forward; 5′-TCAGGGTCCCCCACCCCCATACCA-3′, and deltaC-Forward; 5′-CTCCTCCCAGTTACCTCCACTCAC AGC-3′ with Glis3-Reverse; 5′-GCCTTCGGTATACACAGAGG AGAG-3′, and Glis3-Forward; 5′-ATGGTTCAGCGACTGGGA CCCATT-3′ with N-reverse; 5′-GCAGTGCTTCCCCCCTGA GTCTTCCT-3′ and were subcloned into p3×Flag-CMV vector (Sigma, USA). Myc-tagged Hnf6 constructs were generated by PCR amplification. PCR products were amplified using by Takara Ex-Taq polymerase (TakaraBio, USA) with Hnf6 specific primers: Hnf6-Forward, 5′-ATGAACGCGCAGCTGACCATGG AA-3′, Hnf6-reverse 1, 5′-CTGCCCTGAATTACTTCCATTGC-3′, Hnf6-reverse 2, 5′-CGGCTCCTGCAGCCACTTCCACAT-3′, and Hnf6-reverse 3, 5′-CCACTTGTCCAGACTCCTCCTTC-3′. PCR products were subcloned into pCMV-Myc vector (Clontech, USA) with EcoRI and NotI. To generate the mNgn3 distal promoter reporter, Ngn3D-Luc, mNgn3(2.5), mNgn3(1.5), and mNgn3(0.6)-Luc, PCR amplification was performed with Ngn3 promoter specific primers and PCR products were subcloned into pTAL-luc (Clontech) or pGL4.10 luciferase reporter vector (Promega, USA) with KpnI and BglII restriction sites or KpnI and HindIII restriction sites. Ngn3D-Luc mutation constructs containing TGGAAGGGA instead of TGGGGGGGA, were generated using site-directed mutagenesis according to the manufacturer’s instructions (Stratagene, USA). Sequences were confirmed by sequencing analysis.
Generation of Glis3 expressing lentiviruses
In order to generate lentiviral Glis3 clones, pLVX-mCherry-N1 vector (Clonetech) was used to generate Glis3 lentiviral expression vectors. Glis3 was amplified with the primer sets, full length-Forward; 5′-TCGAATTCTCGCCACCATGAATGGAAG GTCATGTGG-3′, and ΔN302 Forward; 5′-TCGAATTCTCGCC ACCATGGCATTGTCCTTGTCGCCACT-3′, with Glis3 Reverse; 5′-GGTGGATCCCGGCCTTCGGTATACACAGAGG-3′ to get full length and ΔN302 clones. Amplified PCR product was digested with EcoRI and BamHI, subcloned and packaged into HEK293 cells.
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as described previously (Kim et al., 2003). In brief, double stranded-oligonucleotides corresponding to the two Glis3BSs in the promoter of mNgn3 at −3.5 kb (5′-GGGGGCAGGGCCCCTGGGGGGGAGGCCCGCTCAGT-3′) and at −2.5 kb (5′-TTGCTACCACCCCCCAGAATTCT-3′), as well as the consensus Glis3BS (5′-TCAGACGTGGGGGGT CCAACTT-3′) were 32P-labeled and incubated with the purified recombinant protein (His)6-Glis3(ZFD) in binding buffer (20 mM HEPES, pH 8.0, 5% glycerol, 2.5 mM MgCl2, 1 mM DTT, 50 mM KCl, 10 mM ZnSO4, 300 μg poly(dI-dC) for 30 min at room temperature. For cold competition analysis, mutant oligonucleotides (−3.5 kb Mut; 5′-GGGGGCAGGGCCCCTGGAAGGGAG GCCCGCTCAGT-3′, −2.5 kb Mut; 5′-TTGCTACCACCAACCA GAATTCT-3′) and WT oligonucleotides were used. The protein-DNA complexes were separated on a 10% native polyacrylamide gel and visualized by autoradiography.
Immunopulldown analysis and Western blot
Cells were transfected with p3 × Flag Glis3 and pCMV-Myc-Hnf6 expression constructs. After 48 h, cells were lysed in RIPA buffer (Upstate, USA). Lysates were incubated either with anti-Flag M2 agarose resin (Sigma) or anti-Myc agarose resin (Sigma) overnight at 4°C. Immunocomplexes were collected by centrifugation and washed with RIPA buffer 5 times. Immunocomplexes were separated on 4–12% SDS-PAGE gels, transferred to PVDF membranes and probed with anti-Flag M2 (Sigma, 1:5000), anti-Hnf6 (Santa Cruz Biotechnologies, USA, 1:100) and anti-Myc (Invitrogen, USA, 1:5000) antibodies. Secondary anti-mouse-IgG-HRP (Jackson ImmunoResearch lab, USA, 1:1000) and anti-rabbit-IgG-HRP (Jackson ImmunoResearch lab, 1:1000) conjugated antibodies were used for detection.
Reporter assay
HEK293 and HepG2 cells were plated on 96 well plates prior to performing transient transfection. After 24 h after plating, cells were transfected with Ngn3-promoter reporters (gifts from Dr. German, UCSF) with the indicated amounts of Glis3 expression vector and Hnf6 expression vectors. The reporter construct, pRL-luciferase, was used as an internal control. After 48 h, cells were lysed using 1× passive lysis buffer (Promega) and reporter activity was measured using the dual luciferase kit (Promega). INS(832/13) and βTC6 cells were plated in 12-well dishes at 1 × 105/well, incubated for 24 h at 37°C, and subsequently cotransfected with 0.15 μg of the indicated reporter, 0.05 μg pCMV-β-galactosidase as an internal control, and various amounts of expression vectors as indicated in Opti-MEM (Invitrogen) using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s instructions. Each transfection cocktail was used to transfect triplicate sets of wells. After 48 h, cells were lysed using 1× passive lysis buffer (Promega) and reporter activity was measured using a luciferase assay kit (Promega). The β-galactosidase levels were measured with a luminometric β-galactosidase detection kit (Clontech) by the manufacturer’s protocol.
RESULTS
Glis3 binds to the Ngn3 promoter
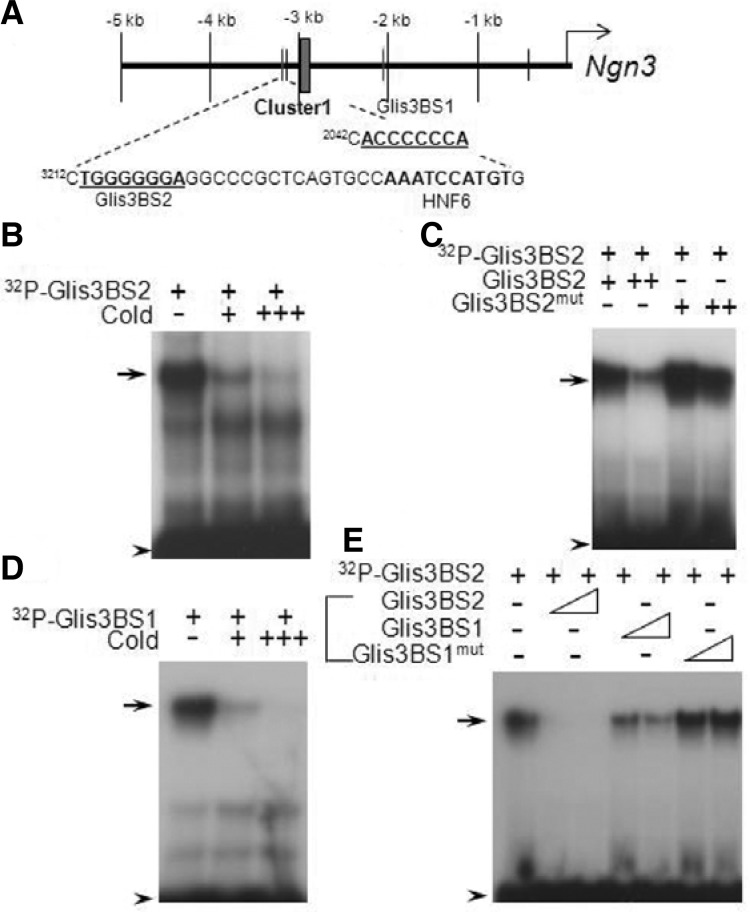
In a previous study, we found that deficiency in Glis3 causes a significant reduction in Ngn3 expression and results in defects in endocrine β-cell development. In order to determine whether Ngn3 is a direct target gene of Glis3, we examined the Ngn3 promoter region for Glis3 binding sites (Kang et al., 2009b). Ngn3 is regulated by a distal regulatory cluster that includes Hnf6 (Oc1), Onecut2 (Oc2) and Pdx1 binding elements (Jacquemin et al., 2000; Lee et al., 2001). It was previously reported that Hnf6 response elements are present in the proximal region and distal regions of Ngn3 at −453 bp and at −3187 bp, respectively (Jacquemin et al., 2000; Lee et al., 2001). Interestingly, two putative Glis3 binding sites (Glis3BS) were found at −3212 and −2042 close to the Hnf6 binding sites in both the proximal and distal regions of the Ngn3 promoter (Fig. 1A). To determine whether Glis3 was able to bind these elements, we performed EMSA (Figs. 1B-1E). Glis3 was able to bind both radiolabeled Glis3BS1 and Glis3BS2 (Figs. 1B–1E), and these interactions were greatly inhibited by competition with unlabeled Glis3BS oligos (Figs. 1B and 1D). However, mutated Glis3BS2, 5′-TGGA AGGGA-3′ (Glis3BS2mut) and Glis3BS1mut oligos were not very effective in competing for the binding of Glis3 to Glis3BS2 suggesting that Glis3 can directly bind to Ngn3 promoter via these two Glis3BS elements (Figs. 1C and 1E).
Fig. 1.
Glis3 binds to Ngn3 promoter (A) Diagram of Glis3 binding sites (Glis3BS) in the Ngn3 promoter (Lee et al., 2001). Ngn3 Cluster 1 enhancer which contains binding sites for Hnf1β, Foxa2 and Hnf6 is indicated by the box. Putative Glis3BS1 and −2 on the Ngn3 promoter are indicated. (B-E) Electrophoretic Mobility Shift Assay (EMSA) EMSA analyses were performed with purified Glis3 protein and 32P-labeled Glis3BS oligonucleotides for the putative Glis3 binding sites 1 and 2 in the distal region of mouse Ngn3 promoter in the presence or absence of unlabeled Glis3BS2 (B), mutant Glis3BS2mut (C), Glis3BS1 (D) or mutant Glis3BS1mut (E) as indicated. The protein-DNA complexes were separated by SDS-PAGE. Arrows indicate protein-DNA binding complexes and arrow heads indicate free probes.
Glis3 induces Ngn3 reporter activity
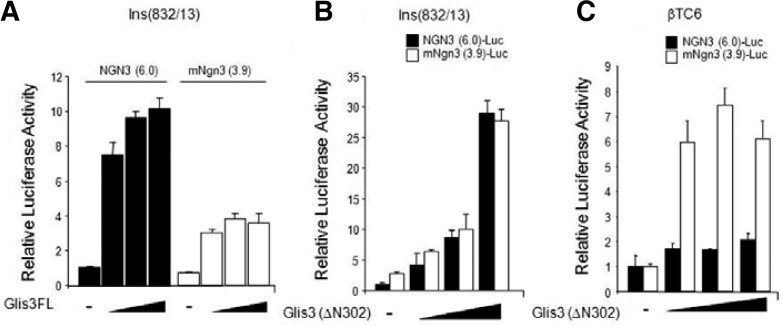
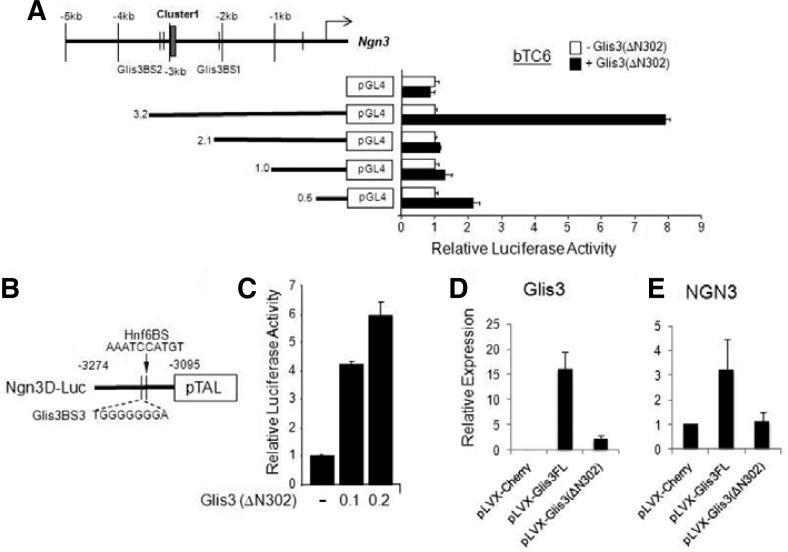
To determine whether Glis3 could activate the Ngn3 promoter, we performed reporter gene assay using rat insulinoma cell line Ins(832/13) and pancreatic endocrine βTC6 cells (Fig. 2). As shown in Fig. 2A, wild type Glis3 enhanced the Ngn3(6.0) reporter activity in a dose dependent manner in rat Ins(832/13) cells. The Ngn3(3.9) promoter reporter was also activated by Glis3 in Ins(832/13) cells. The N-terminal truncated form of Glis3, Glis3(ΔN302) can also activate Ngn3(6.0) and Ngn3(3.9) promoter in both Ins(832/13) and βTC6 cells. These data suggest that Glis3 directly regulates the Ngn3 gene in insulinoma cell lines. To further define the function of Glis3BS2 in the activation of the Ngn3 promoter, we generated several deletion promoter constructs (3.2, 2.1, 1.0 and 0.5) in the pGL4.10 luciferase reporter vector and performed reporter analysis in βTC6 cells (Fig. 3A). The Ngn3(3.2) promoter, but not the shorter promoter constructs lacking Glis3BS2, was activated by co-expression of Glis3(ΔN302). These results suggested that Glis3BS1 does not play a significant role in the activation of the Ngn3 promoter, but that Glis3BS2 is required. In order to confirm the activity by Glis3BS2 on the far distal region of Ngn3, we subcloned the Ngn3 regulatory cluster region from −3274 to −3,095 bp in the pGL4.10 reporter plasmid (Fig. 3B). Reporter analysis showed that Ngn3D was induced by Glis3(ΔN302) was able to activate the (−3274 to −3095) promoter region consistent with the conclusion that the Glis3BS2 is critical for Ngn3 gene regulation.
Fig. 2.
Glis3 can activate Ngn3 promoter activity (A) Ins(832/13) and βTC6 cells were transfected with pRL-luc as an internal control, human NGN3(6.0)-luc, mouse Ngn3(3.9)-luc, p3×Flag-Glis3 full length or mutant p3×Flag-Glis3(ΔN302) expression vectors as indicated. After 48 h, cells were lysed and luciferase activity was measured using the Victor3 luminometer (Perkin Elmer, USA). The experiments were performed in triplicate.
Fig. 3.
Glis3 can regulate Ngn3 via Glis3BS2 (A) βTC6 cells were plated prior to transfection with various deletion Ngn3 promoter reporters (3.2, 2.1, 1.0 and 0.5) and p3×Flag-Glis3 (ΔN302) expression vector as indicated. (B) A schematic of the Ngn3 distal promoter-reporter construct, Ngn3 (D)-Luc, which only contains Glis3BS2. (C) Reporter assay using Ngn3(D)-Luc promoter reporter. Cells were transfected with the Ngn3(D)-Luc reporter, Glis3 expression vector Glis3 (ΔN302), and with renilla luciferase reporter as internal control. Forty-eight hr post-transfection, cells were lysed and the luciferase activity was measured using dual-luciferase assay kit (Promega) and Victor3 luminometer. (D) qRT-PCR was performed using a Glis3 lentiviral expression vector in human ductal HPDE cells. Forty-eight hr after transfection, we collected RNA and evaluated the expression of Glis3 (D) and Ngn3 (E) expression by qRT-PCR.
Glis3 induces Ngn3 expression
In order to further investigate the induction of Ngn3 by Glis3, we generated Glis3 full length and Glis3(ΔN302) lentiviral over-expression vectors (pLVX-Glis3FL, pLVX-Glis3(ΔN302), respectively) and transiently transfected human pancreatic ductal HPDE cells (Supplementary Fig. 1). We confirmed the expression of Glis3 by qRT-PCR and measured endogenous Ngn3 expression by qRT-PCR (Figs. 3D and 3E). Our results showed that Glis3 was able to moderately induce the expression of Ngn3. Glis3(ΔN302) did not enhance endogenous Ngn3 expression, this might be due that it is expressed at much lower levels than Glis3. Alternatively, the data might suggest that the N-terminus of Glis3 plays a role in the activation of the endogenous Ngn3 gene.
Glis3 physically interacts with Hnf6
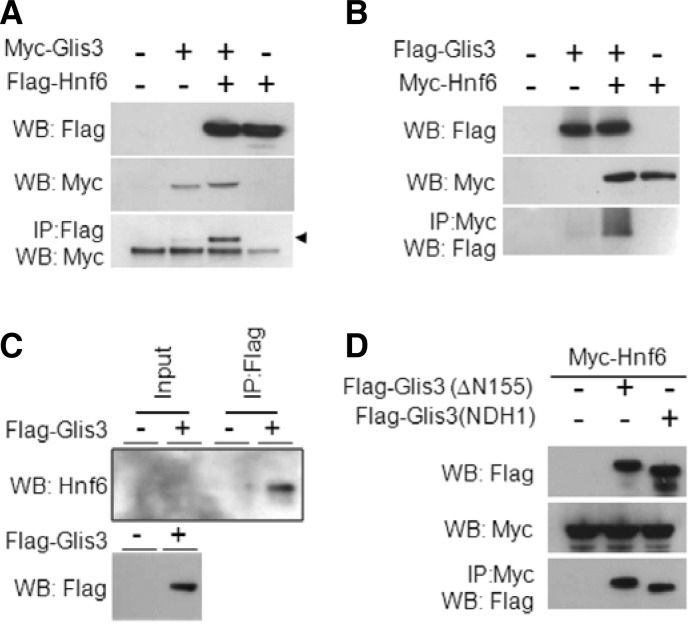
Given the similarity of the phenotypes of Hnf6 and Glis3 mutants, combined with the fact that both proteins are co-expressed within pancreatic progenitors, we sought to test whether the two factors physically interacted. We analyzed this by two-way reciprocal immunoprecipitation using Myc/FLAG-tagged full-length and truncated forms of the two proteins transiently expressed in HEK293 cells. First, we showed that full-length Glis3 interacts with full-length Hnf6 (Fig. 4A). This interaction was observed when the immunoprecipitation was performed with either the Myc or the FLAG antibody (Figs. 4A and 4B).
Fig. 4.
Interaction between Glis3 and Hnf6 (A) HEK293 cells were transiently transfected with Myc-tagged Glis3 and Flag-tagged Hnf6. After 48 h, cells were lysed with RIPA buffer and immunocomplexes precipitated by anti-Flag M2 agarose resin (Sigma). Proteins were separated by SDS-PAGE (4–12%) and probed with anti-Flag M2 (Sigma) and anti-Myc (Invitrogen) antibodies. Arrow head indicates Myc-tagged Glis3 that was pulled down by anti-Flag antibody. (B) HEK293 cells were transiently transfected with Flag-tagged Glis3 and Myc-tagged Hnf6. Immunocomplexes were purified by anti-Myc agarose resin (Sigma) and proteins were detected by anti-Flag M2 (1:5000) and anti-Myc (Invitrogen, 1:5000) antibodies. (C) HepG2 cells were transiently transfected with Flag-tagged Glis3. After 48 h, cells were lysed by RIPA buffer (Upstate) and lysates incubated with anti-Flag M2 agarose resin for pulldown of the complexes. Proteins were separated on 4–12% SDS-PAGE gel and transferred to PVDF membrane. Proteins were detected using anti-Hnf6 (Santa Cruz, 1:100) and anti-Flag M2 (Sigma, 1:5000) antibodies. (D) The Glis3 mutant Flag-Glis3(NDHI) containing a frameshift mimicking the human NDH1 mutant, can still interact with Hnf6.
To assess if this interaction was biologically relevant, and not an artifact based on excessively high amounts of both proteins achieved through transient transfection, we next performed in vivo pull-down of endogenous Hnf6 using transiently transfected Flag-Glis3 in HepG2 cell line (Fig. 4C). These results showed that FLAG-Glis3 was able to pulldown endogenous Hnf6 supporting the conclusion that the proteins interact (Fig. 4C).
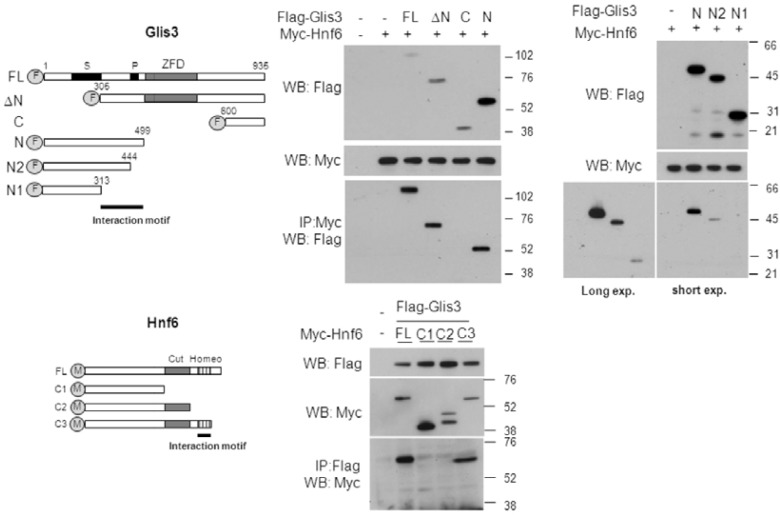
To map the regions required for the interaction of Glis3 and Hnf6, we first focused on the domain requirements of Glis3. To do this, we performed immuno-pulldown using various deletion constructs of FLAG-tagged Glis3 (Fig. 5A). We observed that the Glis3 deletion mutants (ΔN) and (N) were still able to interact with Hnf6, but that carboxyl-terminus deletion mutant of Glis3 (C) did not. This was supported by the observation that the mutant (Glis3(NDHI)), containing a frameshift similar to the Glis3 mutation responsible for human NDH1, also interacted well with Hnf6 (Fig. 4D). These data suggested that the region at the N-terminus of Glis3 between aa 307–499 is important for the interaction with Hnf6 (Figs. 5B and 5C). Interestingly, the amino terminus of Glis3 contains a proline-rich and serine-rich domain between amino acids 444–461 and 196–310, respectively. In order to investigate the role of these domains in this interaction, we examined the ability of the Glis3 mutants (N2) and (N3), which contains shorter region of the N-terminus on the interaction with Hnf6 (Fig. 5C). This analysis indicated that the proline-rich region between 444–499 is important for the interaction and the region between 313–444 may also play a role to a lesser degree.
Fig. 5.
The mapping of interaction domain in Glis3 (A) Schematic of the various Glis3 constructs: FL spans aa 1–935, DeltaN (ΔN) aa 305–935, C aa 600–935, N aa 1–499. N2 spans aa 1–444 lacking the putative proline-rich region, and N1 spans aa 1–313. The amino-terminus truncated construct were shown. S and P indicate putative serine-rich and proline-rich domains, respectively. (B, C) HEK293 cells were transfected with various Flag-tagged Glis3 and Myc-Hnf6 expression vectors as indicated. Forty-eight h post-transfection, cells were harvested and immunocomplexes purified by anti-Myc resin (Sigma). Immunocomplexes were separated on the 4–12% SDS-PAGE gels and transferred to a PVDF membrane. Proteins were detected with Flag M2 and Myc antibodies. (D) A schematic showing the Hnf6 domains and deletion constructs. The putative interacting motif is marked by a black bar. (E) Cells were transiently transfected with various deletions constructs of Hnf6 with Flag-tagged Glis3 expression vector. Immunocomplexes were detected by anti-Myc (Invitrogen, 1:5000) and anti-Flag M2 (Sigma, 1:5000) antibodies.
To identify the domain requirements for the interaction of Hnf6 with Glis3, we created three different Myc-tagged Hnf6 mutants truncated at the C-terminus (Fig. 5D) and performed immunoprecipitation analysis. As shown in Fig. 5B, only the full-length and the C3 mutant of Hnf6 interacted with Glis3, whereas the C1 and C2 deletion mutants did not (Fig. 5E). The data indicate that the homeodomain of Hnf6 is critical for its interaction with Glis3.
DISCUSSION
Recent human studies linked genetic alterations in the GLIS3 gene to neonatal diabetes, type 1 and type 2 diabetes (Barrett et al., 2009; Dupuis et al., 2010; Senee et al., 2006). Mice deficient in Glis3 also develop neonatal diabetes in addition to polycystic kidney disease (Kang et al., 2009a; 2009b; Watanabe et al., 2009; Yang et al., 2009). The expression of a key pro-endocrine marker, Ngn3, was shown to be dramatically decreased with its downstream target genes, but several early progenitor markers, such as Pdx1, Sox9, and Nkx6.1, were not altered in Glis3 null mice. Although Ngn3 is known as a key transcription factor for endocrine lineage development, the regulation of Ngn3 is not well-elucidated. Previous studies demonstrated that a distal region in the Ngn3 promoter (cluster 1) containing binding sites for Hnf1β, Foxa2, and Hnf6, and Pdx1 plays a critical role in Ngn3 transcriptional regulation (Oliver-Krasinski et al., 2009). Pdx1 and Hnf6 have been shown to physically interact and synergistically activate Ngn3 gene transcription (Oliver-Krasinski et al., 2009). These observations suggest that the regulation of Ngn3 is complex and that many factors are involved in controlling Ngn3 expression and endocrine progenitor development. Based on the pancreatic phenotype of Glis3 deficient mice and the down-regulation of Ngn3 expression, we speculated that Glis3 might be directly involved in the regulation of Ngn3.
Glis3 functions as a transcription activator by binding to the classical GLI response elements (GRE, 5-ACCACCCAC-3′) and closely-related Glis3 binding sites (Glis3BS, 5′-(G/T)ACCC CCCA(G/C)-3′) (Beak et al., 2008). To investigate the regulation of Ngn3 by Glis3, we checked the 5′-flanking sequence region of mouse Ngn3 for Glis3BS sequences (Jacquemin et al., 2000; Lee et al., 2001). Interestingly, we found two putative Glis3BSs: Glis3BS1 (5′-ACACCCCCCAG-3′) positioned at - 2041bp and Glis3BS2 (5′-CTGGGGGGAG-3′) positioned at - 3211bp within the distal promoter region of the Ngn3 gene. EMSA revealed that Glis3 can bind to both response elements, but appeared to exhibit a higher affinity for Glis3BS2 than Glis3BS1. The human NGN3(6.0) and mouse Ngn3 promoter were activated by full length Glis3 in two insulinoma cell lines. The mNgn3(3.2) promoter region, which contains both Glis3BS1 and GlisBS2, was activated by Glis3, but promoters, including mNgn3(2.0), which did not contain Glis3BS2, were not. Furthermore, Glis3 was able to induce activation of the reporter under the control of mNgn3D, which only contains Glis3BS2. Although we cannot rule out that both Glis3 BS play a role, these observations indicated that the activation of Ngn3 transcription by Glis3 is mediated through Glis2BS2 binding rather than Glis3BS1.
Hnf6 is known as a critical regulator in early endoderm development through regulation of vHNF1/HNF1β, and subsequently, Hnf6 functions as a key determinant of pancreas specification via the regulation of Pdx1 (Jacquemin et al., 2000; Lee et al., 2001; Oliver-Krasinski et al., 2009; Poll et al., 2006). Hnf6 regulates Ngn3 by binding to Hnf6 response elements in the Ngn3 promoter (Jacquemin et al., 2000; Lee et al., 2001). Hnf6 null mutant mice exhibit a severe defect in endoderm differentiation, and are born with a hypoplastic pancreas due to insufficient Pdx1 expression (Jacquemin et al., 2000; 2003). Although Hnf6 can regulate Ngn3, it is not the sole driving force in controlling endocrine lineage development. Stoffers et al. demonstrated that Pdx1 is also a critical player in the regulation of Ngn3 expression through the distal region in the Ngn3 promoter (Oliver-Krasinski et al., 2009). Interestingly, although Hnf6 mRNA expression was slightly elevated in Glis3 null mice, Ngn3 expression was dramatically reduced (Kang et al., 2009b). However, Glis3 expression is not changed in Hnf6 null mice (Kang et al., 2009b). Data suggest that in pancreatic progenitor cells Glis3 acts in parallel with Hnf6 or upstream of Hnf6.
Hnf6 binding sites in the distal region of the Ngn3 promoter, which includes the Ngn3 enhancer cluster 1, exhibit a higher affinity for Hnf6 than the binding site in the proximal region (Jacquemin et al., 2000; Lee et al., 2001). Interestingly, we found that two putative Glis3 binding sites are localized close to the Hnf6 binding sites in the distal region of the Ngn3 promoter. Both EMSA and reporter gene analysis demonstrated that Glis3 regulates Ngn3 transactivation by binding to these Glis3BS.
Previously, we reported that Glis3 contains several domains; the C-terminus was found to contain the activation domain. Loss of this domain in humans or Glis3 null mice leads to the development of polycystic kidney disease and neonatal diabetes (Kang et al., 2009b). However, the function of the amino-terminus of Glis3 has not yet been established (Kang et al., 2010; Zeruth et al., 2011). Interestingly, in this study we provide evidence for a physical interaction between Glis3 and Hnf6. The amino-terminus of Glis3 was found to be important in the interaction with Hnf6, while the homeodomain of Hnf6 is crucial for its interaction with Glis3. Interestingly, the amino terminus of Glis3 contains a serine-rich and a proline-rich domain between amino acids 196–310 and 444–461, respectively. Deletion of the proline-rich region greatly diminished the ability of Glis3 to interact with Hnf6 suggesting that this region is important in the interaction of Glis3 with Hnf6 (Fig. 5C, long exposure).
The expression of several pancreatic transcription factors, including Pdx1, Ngn3, and Hnf6, is highly restricted spatially and temporally during pancreas development. Specifically, Hnf6 and Ngn3 expression are dramatically reduced at later stages of endocrine development. Sustained expression of Hnf6 has been shown to cause a loss of pancreatic beta-cells by apoptosis (Hara et al., 2007). In contrast to Hnf6 expression, expression of Pdx1 and Glis3 persists postnatally and in adults. Previous studies demonstrated that Glis3 regulates Insulin gene expression directly by binding Glis3BS elements in the proximal region of the Ins gene or indirectly through its interaction with other transcription factors, such as Pdx1, MafA, and NeuroD (Kang et al., 2009b; Yang et al., 2009). In addition, Glis3 is expressed in pancreatic ducts and deficiency in Glis3 leads to pancreatic duct dilation (Kang et al., 2009b). The observations indicate that Glis3 exhibits several unique functions in the pancreas. Glis3 appears not only to be involved in the regulation of endocrine progenitor determination but also in the maintenance of adult pancreatic β-cells and the control of pancreatic ductal cell functions. It is likely that the regulation of these functions involve multiple, interacting regulatory networks including the crosstalk between Pdx1, Hnf6, and Glis3. Similarly the regulation of Ngn3 and endocrine cell fate by Glis3 also may involve the formation of Glis3 complexes containing HNF6. In addition, the crosstalk between Pdx1, NeuroD, MafA, and Glis3 complexes may play a role in mature endocrine cells, including the regulation of Insulin transcription. However, many questions about the function of Glis3 in pancreas development and gene regulation still remain unanswered.
In summary, in this study we provide evidence for Glis3-mediated transcriptional regulation of Ngn3, which is critical for the generation of pancreatic endocrine progenitors. This activation appears mediated through Glis3BS elements within the distal region of the Ngn3 promoter. In addition, we provide evidence that this may involve crosstalk between Glis3 and another endocrine factor, Hnf6. As such, this control of Ngn3 expression might be part of the regulation of endocrine progenitor fate determination.
Supplementary Material
Acknowledgments
Authors specially thank Dr. M. German (UCSF) and Dr. F. Lemaigre (Université Catholique de Louvain) for the human Ngn3 promoter and Hnf6 constructs, respectively. Authors also thank to Dr. M. Schmerr (Cleveland Clinic), Kristin Lichti-Kaiser, and Gary Zeruth (NIEHS) for their critical review and comments. This research was supported by the Intramural Research Program of the NIEHS, NIH (Z01-ES-100485).
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Apelqvist A., Li H., Sommer L., Beatus P., Anderson D.J., Honjo T., Hrabe de Angelis M., Lendahl U., Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A., Julier C., Morahan G., Nerup J., Nierras C., et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beak J.Y., Kang H.S., Kim Y.S., Jetten A.M. Functional analysis of the zinc finger and activation domains of Glis3 and mutant Glis3(NDH1) Nucleic Acids Res. 2008;36:1690–1702. doi: 10.1093/nar/gkn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes G.K. Developmental biology of the pancreas: a comprehensive review. Dev. Biol. 2009;326:4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Dierich A., LeMeur M., Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. USA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J., Melton D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Hara M., Shen J., Pugh W., Polonsky K.S., Le Beau M.M., Bell G.I. Sustained expression of hepatocyte nuclear factor-6 leads to loss of pancreatic beta-cells by apoptosis. Exp. Clin. Endocrinol. Diabetes. 2007;115:654–661. doi: 10.1055/s-2007-982514. [DOI] [PubMed] [Google Scholar]
- Jacquemin P., Lannoy V.J., Rousseau G.G., Lemaigre F.P. OC-2, a novel mammalian member of the ONECUT class of homeodomain transcription factors whose function in liver partially overlaps with that of hepatocyte nuclear factor-6. J. Biol. Chem. 1999;274:2665–2671. doi: 10.1074/jbc.274.5.2665. [DOI] [PubMed] [Google Scholar]
- Jacquemin P., Durviaux S.M., Jensen J., Godfraind C., Gradwohl G., Guillemot F., Madsen O.D., Carmeliet P., Dewerchin M., Collen D., et al. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol. Cell. Biol. 2000;20:4445–4454. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin P., Lemaigre F.P., Rousseau G.G. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev. Biol. 2003;258:105–116. doi: 10.1016/s0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- Jensen J. Gene regulatory factors in pancreatic development. Dev. Dyn. 2004;229:176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- Jensen J., Pedersen E.E., Galante P., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Kang H.S., Beak J.Y., Kim Y.S., Herbert R., Jetten A.M. Glis3 is associated with primary cilia and Wwtr1/TAZ and implicated in polycystic kidney disease. Mol. Cell. Biol. 2009a;29:2556–2569. doi: 10.1128/MCB.01620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.S., Kim Y.S., ZeRuth G., Beak J.Y., Gerrish K., Kilic G., Sosa-Pineda B., Jensen J., Foley J., Jetten A.M. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol. Cell. Biol. 2009b;29:6366–6379. doi: 10.1128/MCB.01259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.S., ZeRuth G., Lichti-Kaiser K., Vasanth S., Yin Z., Kim Y.S., Jetten A.M. Gli-similar (Glis) Kruppel-like zinc finger proteins: insights into their physiological functions and critical roles in neonatal diabetes and cystic renal disease. Histol. Histopathol. 2010;25:1481–1496. doi: 10.14670/hh-25.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Lewandoski M., Perantoni A.O., Kurebayashi S., Nakanishi G., Jetten A.M. Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J. Biol. Chem. 2002;277:30901–30913. doi: 10.1074/jbc.M203563200. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Nakanishi G., Lewandoski M., Jetten A.M. GLIS3, a novel member of the GLIS subfamily of Kruppel-like zinc finger proteins with repressor and activation functions. Nucleic Acids Res. 2003;31:5513–5525. doi: 10.1093/nar/gkg776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.C., Smith S.B., Watada H., Lin J., Scheel D., Wang J., Mirmira R.G., German M.S. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes. 2001;50:928–936. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- Lemaigre F.P., Durviaux S.M., Truong O., Lannoy V.J., Hsuan J.J., Rousseau G.G. Hepatocyte nuclear factor 6, a transcription factor that contains a novel type of homeodomain and a single cut domain. Proc. Natl. Acad. Sci. USA. 1996;93:9460–9464. doi: 10.1073/pnas.93.18.9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Krasinski J.M., Kasner M.T., Yang J., Crutchlow M.F., Rustgi A.K., Kaestner K.H., Stoffers D.A. The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J. Clin. Invest. 2009;119:1888–1898. doi: 10.1172/JCI37028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierreux C.E., Poll A.V., Kemp C.R., Clotman F., Maestro M.A., Cordi S., Ferrer J., Leyns L., Rousseau G.G., Lemaigre F.P. The transcription factor hepatocyte nuclear factor-6 controls the development of pancreatic ducts in the mouse. Gastroenterology. 2006;130:532–541. doi: 10.1053/j.gastro.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Poll A.V., Pierreux C.E., Lokmane L., Haumaitre C., Achouri Y., Jacquemin P., Rousseau G.G., Cereghini S., Lemaigre F.P. A vHNF1/TCF2-HNF6 cascade regulates the transcription factor network that controls generation of pancreatic precursor cells. Diabetes. 2006;55:61–69. [PubMed] [Google Scholar]
- Rukstalis J.M., Habener J.F. Neurogenin3: a master regulator of pancreatic islet differentiation and regeneration. Islets. 2009;1:177–184. doi: 10.4161/isl.1.3.9877. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel V.M., Scheel D.W., Conners J.R., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- Senee V., Chelala C., Duchatelet S., Feng D., Blanc H., Cossec J.C., Charon C., Nicolino M., Boileau P., Cavener D.R., et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat. Genet. 2006;38:682–687. doi: 10.1038/ng1802. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Hiramatsu K., Miyamoto R., Yasuda K., Suzuki N., Oshima N., Kiyonari H., Shiba D., Nishio S., Mochizuki T., et al. A murine model of neonatal diabetes mellitus in Glis3-deficient mice. FEBS Lett. 2009;583:2108–2113. doi: 10.1016/j.febslet.2009.05.039. [DOI] [PubMed] [Google Scholar]
- Xu X., D’Hoker J., Stange G., Bonne S., De Leu N., Xiao X., Van de Casteele M., Mellitzer G., Ling Z., Pipeleers D., et al. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Yang Y., Chang B.H., Samson S.L., Li M.V., Chan L. The Kruppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res. 2009;37:2529–2538. doi: 10.1093/nar/gkp122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeruth G.T., Yang X.P., Jetten A.M. Modulation of the transactivation function and stability of Kruppel-like zinc finger protein gli-similar 3 (Glis3) by suppressor of fused. J. Biol. Chem. 2011;286:22077–22089. doi: 10.1074/jbc.M111.224964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Nakanishi G., Kurebayashi S., Yoshino K., Perantoni A., Kim Y.S., Jetten A.M. Characterization of Glis2, a novel gene encoding a Gli-related, Kruppel-like transcription factor with transactivation and repressor functions. Roles in kidney development and neurogenesis. J. Biol. Chem. 2002;277:10139–10149. doi: 10.1074/jbc.M108062200. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.