Abstract
Infectious wildlife diseases have enormous global impacts, leading to human pandemics, global biodiversity declines and socio-economic hardship. Understanding how infection persists and is transmitted in wildlife is critical for managing diseases, but our understanding is limited. Our study aim was to better understand how infectious disease persists in wildlife populations by integrating genetics, ecology and epidemiology approaches. Specifically, we aimed to determine whether environmental or host factors were stronger drivers of Salmonella persistence or transmission within a remote and isolated wild pig (Sus scrofa) population. We determined the Salmonella infection status of wild pigs. Salmonella isolates were genotyped and a range of data was collected on putative risk factors for Salmonella transmission. We a priori identified several plausible biological hypotheses for Salmonella prevalence (cross sectional study design) versus transmission (molecular case series study design) and fit the data to these models. There were 543 wild pig Salmonella observations, sampled at 93 unique locations. Salmonella prevalence was 41% (95% confidence interval [CI]: 37–45%). The median Salmonella DICE coefficient (or Salmonella genetic similarity) was 52% (interquartile range [IQR]: 42–62%). Using the traditional cross sectional prevalence study design, the only supported model was based on the hypothesis that abundance of available ecological resources determines Salmonella prevalence in wild pigs. In the molecular study design, spatial proximity and herd membership as well as some individual risk factors (sex, condition score and relative density) determined transmission between pigs. Traditional cross sectional surveys and molecular epidemiological approaches are complementary and together can enhance understanding of disease ecology: abundance of ecological resources critical for wildlife influences Salmonella prevalence, whereas Salmonella transmission is driven by local spatial, social, density and individual factors, rather than resources. This enhanced understanding has implications for the control of diseases in wildlife populations. Attempts to manage wildlife disease using simplistic density approaches do not acknowledge the complexity of disease ecology.
Introduction
Infectious diseases of wildlife have caused important global pandemics in people [1], have influenced human welfare through reduced agricultural production [2], [3] and are reducing global biodiversity [4]–[6]. Management of wildlife infectious disease requires that key ecological processes and mechanisms that drive infection transmission and persistence in wildlife populations be identified, characterised and quantified [7], [8]. Despite this, little is known about disease transmission in wildlife [9]. Improved knowledge could assist management of wildlife disease thereby reducing disease emergence that threatens human and animal health and welfare, agricultural production and species conservation. In this respect, Daszak et al. [10] have proposed that emerging infectious diseases of wildlife can be classified into three major groups based on key epidemiological criteria – spillover from domestic animals to wildlife populations living in proximity; those related directly to human intervention via host or parasite translocations; and those with no overt human or domestic animal involvement.
Cross sectional surveys – where a representative sample of a population is taken, prevalence of disease measured and a contrast made between those with and without the infection to infer risk factors – have frequently been a mainstay of wildlife epidemiological study [11]. However, in recent years it has been recognised that the interface of genetics, ecology and epidemiology is poised to advance our understanding of disease ecology, such as infection transmission, in new and novel ways [12], [13].
Here, we use both the traditional and widely accepted cross sectional survey design to investigate risk factors for prevalence or persistence of Salmonella and a contemporary molecular epidemiological study design to assess risk factors for transmission of Salmonella in wild pigs. We use wild pigs (Sus scrofa) as our model as they are a species of global importance, being found on every continent except Antarctica [14]. They are a damaging invasive species or valued endemic animal depending on their location, and have frequently played an important role in the spread of infectious diseases [15]–[19], including Salmonella [20]. There are two Salmonella species, six subspecies and more than 2500 serovars [21]. In this paper Salmonella refers to serovars within Salmonella enterica subspecies enterica.
Materials and Methods
We sampled [22] and determined the Salmonella infection status of 543 wild pigs (Sus scrofa) by culturing mesenteric lymph nodes and faeces [23]. Salmonella isolates were genotyped using Pulsed field gel electrophoresis (PFGE) [24]. A range of data were collected on putative risk factors for Salmonella transmission including: pig genotype (using microsatellites) [25], spatial and remote sensing data [26] and pig demographic and morphological data [25], [27]. We took two approaches to analyse the data.
1. Cross sectional prevalence study design
A traditional cross sectional prevalence based logistic regression analysis which modelled associations between Salmonella infection and risk factors.
2. Molecular case series approach
A pair-wise molecular analysis of all Salmonella isolates which modelled Salmonella genetic relatedness for each pair of infected pigs against risk factors using linear regression. It was assumed that increasing similarity between Salmonella isolates was correlated with transmission.
We a priori identified several plausible biological hypotheses for both Salmonella prevalence in pigs (cross sectional prevalence study design) and Salmonella transmission between pigs (molecular case series study design). Two models (paired) were implemented for each hypothesis, one for the cross sectional prevalence study design and one for the molecular case series study design. We used information theoretic approaches to select the most supported models within each study design. We compared the two study design for utility for wildlife disease investigation and inferred mechanisms for Salmonella persistence and transmission between wild pigs.
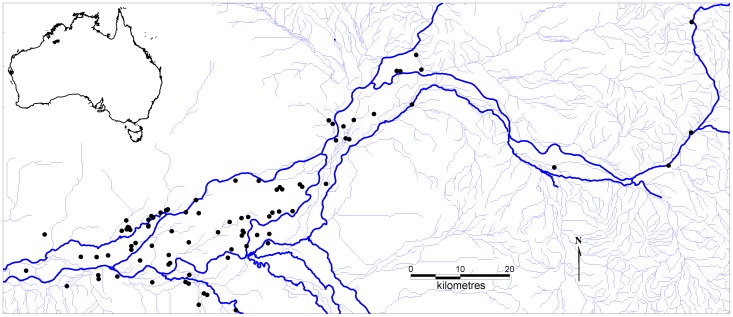
The study was undertaken on a 200 km length of the Fitzroy River floodplain in the west Kimberley region of north-western Australia (see Figure 1) in October 2010. The study area is approximately 4000 km2, lying between 18.612 to 18.037°S and 124.922 to 126.270°E. The area has a tropical monsoonal climate with mean rainfall over the last century of 484 mm per year (range 163–907) and hot temperatures (mean daily maximum temperature 30–39°C [range 18–46°C]).The sampling strategy was to systematically search water features by helicopter. All pigs observed were targeted for humane destruction according to Australian standard operating procedures [22]. Dead pigs were sampled within one hour by veterinarians. Faeces and mesenteric lymph nodes (MLN) were collected, immediately refrigerated and delivered to the laboratory chilled for culture. A sample of ear tissue was also collected from each pig and stored in salt-saturated DMSO buffer for genetic analysis.
Figure 1. The study area.
The inset shows the study area in the context of the Australian continent. The solid and dashed lines represent major and minor drainage lines respectively. The dots represent sampling locations.
Faeces and MLN were cultured according to the Australian standard for microbiology of food and animal feeding stuffs for the detection of Salmonella spp. [23] which is a national modification of the international standard (ISO 6579:2002). All Salmonella isolates confirmed by serotyping were genotyped by PFGE [24]. Gel images were imported into BioNumerics (version 6.6) for data analysis [28]. All gels were normalised using the S. Braenderup reference strain. Bands were detected automatically and verified manually. Salmonella PFGE DICE similarity coefficient was extracted for each pair-wise comparison of isolates using optimised parameters (optimisation (0.2%), tolerance (1.375%), tolerance change (0%), minimum height (9%), minimum surface (2%), uncertain bands (ignore), relaxed doublet matching (no), fuzzy logic (no), area sensitive (no) and active zones (4.3–88.6%)). DNA was extracted from pig ear tissues using the Machery Nagel NucleoSpin Tissue kit. Fourteen pig microsatellite markers [25] were amplified from DNA samples in three multiplex PCRs using the Qiagen Multiplex PCR kit. Microsatellites were genotyped on a Beckman CEQ8000 and alleles were scored using the CEQ 8000 Genetic Analysis System software Version 8.0.
Two data sets were assembled. The first data set (for the cross sectional prevalence study design) consisted of a single observation for each pig that was sampled. Presence or absence of infection was determined by culture for each pig in this dataset. Covariate data (as described in Table 1) were collected for assessment as risk factors or extraneous variables. The second data set (molecular case series study design) was a subset of the first and consisted of pair-wise comparisons between Salmonella isolates (case series with no counterfactual group). The outcome variable was the pair-wise genetic similarity (DICE) for each pair of Salmonella isolates. Covariates were the same as for the first data set except that they were the absolute difference between pair-wise covariates values, pair-wise contrasts, for example Euclidean distance between pigs, or pair-wise indicator variables (for example to indicate the same herd membership).
Table 1. Data description and hypotheses for Salmonella persistence and transmission in wild pigs (Sus scrofa) modelled using both the cross sectional prevalence study design and molecular case series study design.
| Hypothesis | Rationale |
| Density of hosts | Numerous authors have presented transmission models [40], [47]. Increasingly, there is evidence that systems are more complex and that density-dependence does not hold [40], [48]. Other research has indicated that endemic marsupials near the study area have a high prevalence and diversity of Salmonella [49]. Here we hypothesised that increasing local or larger scale densities of wild pigs and native species (Macropus agilis) would increase prevalence and transmission of Salmonella. We therefore modelled Salmonella infection or Salmonella genetic similarity (DICE) against group size and density of groups of wild pigs and density of groups of agile wallabies (groups km−2) as measured during an aerial survey of pigs and agile wallabies in the study area using published aerial survey methodology [50]. |
| Environmental contamination | Salmonella survives in the environment after defecation by pigs for considerable periods of time especially in cooler conditions away from light [51]–[53]. Our environmental persistence hypothesis assumed that environmental contamination from pig defecation was responsible for Salmonella prevalence or transmission and that areas of shade such as trees would increase Salmonella prevalence or transmission. We modelled Salmonella infection or Salmonella genetic similarity (DICE) against the mean EVI [26] for the 6 months prior to sampling in each pigs home range (radius of approximately 2.5 km around each sampled pig). The distance to water resources, number of water bodies within the home range, density of pig herds, pig herd size and densities of the agile wallaby (Macropus agilis) were included as covariates to allow conditioning on these covariates for control of confounding. |
| Host immunity | There are many reasons to assume that individual animal factors such as sex, age and dominance can affect prevalence of infection [11]. We hypothesised these factors affected Salmonella prevalence or transmission and modelled age [38], condition score [36] and gender against Salmonella infection and Salmonella genetic similarity (DICE). |
| Resources | Under the hot semi arid conditions of the study area, critical resources for wild pigs are water [54], riverine habitat for thermal protection [55] and plant food resources [54], [56]. Pigs are highly social animals [23] that do not establish and defend territories [54], and as such have overlapping home ranges [22]. Distribution across a landscape therefore largely depends on resource availability. Thus, we modelled Salmonella infection or Salmonella genetic similarity (DICE) against features likely to cause aggregation of pigs (Euclidean distance to major water courses (m), the number of natural water-bodies in a pig home range and pasture availability (change in EVI over 6 months)) but included density, size of social group and condition score to control confounding. |
| Social interaction | We assumed that pigs that were more related would have greater physical contact and hence transmit Salmonella to one another more readily. We regressed pair-wise Salmonella genetic similarity [38] against pair-wise genetic distance of pigs [41] (derived from microsatellites [36]), controlling for common herd occupancy and Euclidean distance to prevent confounding. We could not assess this in the prevalence approach as little genetic structuring was detected in pigs across the study area (data not shown) and pairwise comparisons were not possible. |
Five biologically plausible hypotheses were developed a priori that sought to explain the prevalence or transmission of Salmonella between pigs within the study area (see Table 1). Two separate models were then developed for each hypothesis, one model for the cross sectional prevalence study design and one for the molecular case series study design. For the simple presence and absence data set, multivariable logistic regression models were developed, in which the covariates were related to the log odds of infection with Salmonella (i.e. generalised linear models with a logit link). For the pair-wise data set, multivariable linear models were developed that related Salmonella DICE linearly to pair-wise covariates. Permutation based methods (999 permutations [29]) were used to assess the significance of coefficients in the linear models, with coefficients ranked amongst the highest 2.5% or lowest 2.5% of permutation coefficients being considered significant at α = 0.05. Examination of the relative strength of evidence for each hypothesis within either the prevalence or molecular approach was undertaken using the approach of Burnham and Anderson [30]. The cross sectional prevalence and molecular case series study designs were then compared for consistency of outcome and relative utility. Inferences for risk factors for Salmonella prevalence (persistence) or transmission were made.
This study was approved by the University of Sydney Animal Ethics Committee (N00/6-2010/1/5319).
Results
We made 543 wild pig Salmonella observations, sampled at 93 unique locations across the study area. The mean weight of pigs was 51 kg (95% CI: 47–54) with a range of 2–150 kg. There were 264 males (49%) and 279 females (51%). Mean male weight was slightly greater than female weight 54 (95% CI: 50–58) versus 49 (95% CI: 45–52) kg, respectively. Assuming an adult wild pig is ≥30 kg [21], there were 361 adults (66%) and 182 sub adults (34%). Of the adult females, 58 were pregnant and 76 were lactating. The prevalence of Salmonella infection was 41% (95% CI: 37–45%). The median Salmonella DICE coefficient [28] (or Salmonella genetic similarity) was 52% (IQR: 42–62, range: 10.0–100). The median pig genetic dissimilarity [31] was 39% (IQR: 35–46, range: 7–71).
The only model supported using the traditional cross sectional prevalence study design, represented the hypothesis that the abundance of available ecological resources is associated with Salmonella infection in wild pigs (Table 2). All other hypotheses were highly unlikely. In the contemporary molecular study design assessing risk factors for transmission, the resource driven contact and host immunity models were equally supported models, with other models/hypotheses being unsupported by the data (Table 3).
Table 2. Akaike information criterion (AIC) values and other model selection metrics for cross sectional prevalence logistic regression models using information theoretic approaches [29].
| Model | Parameters (K) | Bias corrected AIC (AICc) | AICc differences (Δ) | Relative likelihood (evidence ratio) | Probability (Akaike weight) |
| Resource driven contact | 10 | 699.8 | 0.0 | 1.0 | 0.994 |
| Environmental contamination | 8 | 710.9 | 11.1 | 251.7 | 0.004 |
| Density dependant | 6 | 712.1 | 12.2 | 455.6 | 0.002 |
| Host immunity | 6 | 713.6 | 13.7 | 964.5 | 0.001 |
The probability of the resource transmission model is very high (>0.99) and clearly the data support this model. Models are listed in AIC ranked order for each study design.
Table 3. Akaike information criterion (AIC) values and other model selection metrics for molecular case series linear models using information theoretic approaches [29].
| Model | Parameters (K) | Bias corrected AIC (AICc) | AICc differences (Δ) | Relative likelihood (evidence ratio) | Probability (Akaike weight) |
| Host immunity | 6 | 339132.1 | 0.0 | 0.98 | 0.580 |
| Resource driven contact | 11 | 339132.7 | 0.6 | 1.0 | 0.420 |
| Environmental contamination | 8 | 339218.0 | 85.9 | 4.4×1018 | 0.000 |
| Genetic relatedness model | 5 | 339284.7 | 152.6 | 1.4×1033 | 0.000 |
| Density dependant | 7 | 339735.4 | 603.3 | 1.0×10131 | 0.000 |
The probability of both the resources and host immunity models is high rather than the other hypothesised mechanisms of transmission. Models are listed in AIC ranked order for each study design.
Significant coefficients in the cross sectional prevalence data resource model (Table 4) indicated that resources such as the change in enhanced vegetation index (EVI) over the monsoon season (representing areas where pasture growth is marked during the wet season) and proximity to waterways (representing water to drink and riparian zone shelter) were critical features that were associated with increasing prevalence of infection. Additionally, better conditioned (fatter) pigs and increasing wild pig density (weakly: P = 0.04) were associated with increasing probability of infection (Table 4). In the contemporary molecular study design assessing risk factors for transmission, the resource driven contact (Table 5) and host immunity (Table 6) models were equally supported models, with other models/hypotheses being unsupported by the data. However, the only significant coefficients from the resources model were the extraneous variables included to control confounding of the association between Salmonella genetic similarity and the resource covariates. Thus it was apparent that resource availability had little role in transmission. Interpretation of significant coefficients for the indicator variables from the two models demonstrated that transmission of Salmonella was increased between members of the same herd and between males relative to other sexes, whilst controlling for isolates that came from the same individual (isolates from the same individual were more similar as expected). Transmission was more likely between pigs that were geographically closer with the similarity of Salmonella declining as the Euclidean distance between pigs increased. Interpretation of the remaining significant covariates from the resources and host immunity models was more complex as these covariates were the absolute differences between the value of the covariate for each pig being compared, and are thus undirected associations. There was an association between Salmonella similarity and divergent ages and densities of source population of the pigs being compared. The coefficient for condition score indicated that transmission was less likely between pigs of differing condition score.
Table 4. Resource hypothesis model formulation and coefficient estimates for cross sectional prevalence study design.
| Model | Parameter | Coefficient estimate | Standard error | Z value | P value | Odds ratio |
| log [π÷(1−π)] = β1+β2CS+β3DS+β4 |ΔEVI|+β5HS+β6NWB+β7DW+β8DP+β9X+β10Y+r.eff.(location) Calibration: le Cessie-van Houwelingen goodness of fit test (Z = 0.2, P = 0.8) Validation: AUC 0.7 Pseudo r2 = 14% | (Intercept) | 12.04 | 143.76 | 0.08 | 0.93 | … |
| Condition score (CS) | 0.76 | 0.39 | 1.93 | 0.05 | 2.13 | |
| Dist. to streams (DS) | −0.57 | 0.21 | −2.73 | 0.01 | 0.57 | |
| EVI decline (ΔEVI) | −0.38 | 0.15 | −2.59 | 0.01 | 0.68 * | |
| Herd size (HS) | 0.01 | 0.02 | 0.61 | 0.54 | 1.01 | |
| No. water bodies (NWB) | 0.06 | 0.05 | 1.26 | 0.21 | 1.06 | |
| Wallaby herd density (DW) | 0.38 | 0.38 | 1.00 | 0.32 | 1.46* | |
| Wild pig density (DP) | −0.84 | 0.40 | −2.09 | 0.04 | 0.43 * | |
| X coordinate (X) | −0.34 | 0.90 | −0.38 | 0.71 | 0.71 | |
| Y coordinate (Y) | −1.60 | 2.29 | −0.70 | 0.49 | 0.20 |
Random effects terms for herd, and fixed effect covariates for latitude and longitude were included to control clustering of data and spatial trends or autocorrelation.
These covariates were transformed (normalised z = (x−μ)÷σ) to yield more interpretable odds ratios.
Table 5. Resource hypothesis model formulation and coefficient estimates for molecular case series study design.
| Model | Parameter | Coefficient estimate | Standard error | Z value | P value |
| Salmonella DICE = β1+β2|CS|+β3|DS|+β4 ED+β5|ΔEVI|+β6|HS|+β7|DW|+β8|WB|+β9H+β10Pig+β11|DP| Adjusted r2 = 3% | (Intercept) | 55.55 | 0.25 | 224.19 | 0.001 |
| |Condition score| (|CS|) | −2.99 | 0.32 | −9.48 | <0.001 | |
| |Dist. to streams| (|DS|) | −0.22 | 0.16 | −1.33 | 0.098 | |
| Euclidean distance (ED) | −0.02 | 0.00 | −4.53 | <0.001 | |
| |EVI decline| (|ΔEVI|) | 0.00 | 0.00 | −0.46 | 0.303 | |
| |Herd Size| (|HS|) | −0.02 | 0.01 | −1.31 | 0.107 | |
| |Wallaby density| (|DW|) | −3.20 | 0.40 | −8.07 | <0.001 | |
| |water bodies| (|WB|) | 0.07 | 0.06 | 1.29 | 0.088 | |
| Same herd (Indicator: 0 = false) (H) | 1.14 | 0.24 | 4.71 | 0.001 | |
| Same pig (Indicator: 0 = False) (Pig) | 44.45 | 2.00 | 22.27 | <0.001 | |
| |Wild pig density| (|DP|) | 16.41 | 2.67 | 6.15 | <0.001 |
A fixed effect (indicator) covariate for herd and the distance between two pigs were included to control clustering and spatial autocorrelation.
Table 6. Host immunity hypothesis model formulation and coefficient estimates for molecular case series study designs.
| Model | Parameter | Coefficient estimate | Standard error | Z value | P value |
| Salmonella DICE = β1+β2|Age|+β3|CS|+β4ED+β5H+β6Pig+β7Sex Adjusted r2 = 2% | (Intercept) | 56.81 | 0.26 | 216.33 | <0.001 |
| |Age| | −0.58 | 0.00 | −3.01 | 0.001 | |
| |Condition score| (|CS|) | −3.01 | 0.31 | −9.57 | <0.001 | |
| Euclidean distance (km) (ED) | −0.03 | 0.00 | −11.39 | <0.001 | |
| Same herd (Indicator: 0 = false) (H) | 0.71 | 0.23 | 3.04 | 0.001 | |
| Same pig (Indicator: 0 = False) (Pig) | 44.45 | 1.99 | 22.32 | 0.001 | |
| Sex Indicator variable (reference Male: Male) (Sex) | |||||
| Female/Female | −1.89 | 0.24 | −7.88 | <0.001 | |
| Female/Male) | −0.92 | 0.22 | −4.19 | <0.001 |
A fixed effect (indicator) covariate for herd and the distance between two pigs were included to control clustering and spatial autocorrelation.
Discussion
It is interesting to posit the reasons that Salmonella prevalence was higher in well conditioned (fatter) pigs and in resource rich areas across the landscape. Prevalence of an infection in a population depends on several factors, especially transmission rates, but also disease induced mortality, duration of infection and the length of time an infection has been present in a population [11]. Given that resources were not observed to be important to transmission in the molecular approach, it is likely that wild pigs in resource rich areas may have had higher prevalence for reasons other than transmission. The last of these alternate explanations above, namely that prevalence is increased in populations where infection has been present in populations for longer is intriguing. It suggests that resource rich areas across the landscape may act as areas for persistence of Salmonella in pig populations. With regards to condition score as a risk factor, it is possible that those pigs that were fatter exhibited some specific behaviour that increased exposure to infection. Given their better body condition, these pigs may have been travelling further and foraging more effectively and widely for food. This may have exposed them to more infection, resulting in higher prevalence in these pigs through more effective contacts. This is consistent with prior research in human infections [32], and recently demonstrated in wildlife [33] that super-spreaders (or individuals responsible for the majority of transmission) are disproportionately important for disease transmission. However, reverse causality concepts associated with cross sectional surveys also suggests alternate explanations, such as that better conditioned pigs simply survived better with Salmonella infection.
The molecular study design demonstrated that transmission was more likely within social groups and to other pigs within close proximity. This was expected as wild pigs have been shown to be generally very sedentary [34], highly social [35] and with overlapping home ranges [36]. This suggests the mechanism for transmission is largely social and foraging behaviour between local pigs that increases effective contact. Transmission was also more common between males. Adult male pigs have larger home ranges than females [34] and are often found singly or associating in small male groups in the study area [37]. These characteristics may account for the observed greater male to male transmission. It also suggests that older males may be relatively more important in the transmission of Salmonella in our study area.
An important result from the molecular study design were the low R2 values associated with both the risk and resources model, indicating the majority of the variability in the Salmonella genetic relatedness was not due to direct transmission between wild pigs. Instead complex phylodynamic processes [38] or the presence of other species involved in Salmonella ecology likely introduced considerable Salmonella genetic diversity that we could not model under the assumption of transmission between pigs. Phylodynamic processes include mechanisms associated with the persistence of Salmonella infections within individual pigs, leading to increased selection pressure and interplay between host immune responses and mutations [38]. Were we to rely only on the traditional cross sectional study design and assume that infection status was correlated with infection transmission we would have overestimated the direct transmission between pigs that occurred in our study population, with the R2 value of the cross sectional resources model an order of magnitude greater than the resources or risk model in the molecular study. Additionally, an assumption that transmission was associated with resources would have been made, instead of the likely reason that resources affected prevalence through other reasons such as persistence of Salmonella in pig populations in resource rich areas.
Our empirical findings, that Salmonella persistence in pig populations is associated with resource abundance, and conversely that density has little role in persistence, have implications for control of infection in pigs in Northern Australia and in wildlife more generally. It indicates that control of wildlife infections may not always be achieved through simplistic application of threshold density concepts, as indicated by prior theoretical models [39], [40]. Based on this principal, many authors have proposed that simply reducing abundance or the susceptible proportion of wildlife (e.g. by culling or vaccination) will lead to disease fadeout because transmission cannot be maintained [41]. Other authors argue that empirical evidence of such an effect is lacking [42], [43], or that percolation thresholds better explain empirical data of threshold densities [43]. Our results do not disprove a threshold effect but do indicate that targeting areas for control simplistically based on density may not be a useful strategy. Instead careful consideration of resource distribution across the landscape and spatial targeting of control to those areas of greatest risk would be more efficient at reducing prevalence than control targeted at wildlife density alone. Additionally, the local transmission observed in our study suggests that in the event of a spreading epidemic in a naive population where vaccination or culling zones are implemented [44], these zones can be structured on the probable movements of local pigs (especially males). Transmission was also more likely between pigs in areas where the density of their local population differed markedly. The role of density in control may thus be to allow targeting of areas where transmission may occur from population to population (i.e. at areas of divergent density).
Salmonella is an important human pathogen. It is also important in livestock production, both due to its effect on health and productivity and due to its role as a foodborne zoonosis. Salmonella has been isolated from the carcasses of wild pigs harvested for human consumption in Australia [45], [46]. Salmonella infection of wild pig populations might represent a reservoir of infection for grazing livestock (sheep and cattle) or pose a direct (wild pigs are hunted as a recreational pursuit) and a foodborne (wild pigs are commercially harvested) zoonotic hazard [35]. The role that wild pig populations might play as reservoirs of Salmonella for domestic livestock apparently has not been investigated. Whether this is an ecosystem with no overt domestic animal (or human) involvement i.e. wild pigs as a reservoir, or spillover from domestic animals to wildlife populations living in proximity [10], is an open question. In the current study evidence was found to support ecological resources as a driver of Salmonella transmission; in addition, spatial proximity and other host factors influenced transmission between pigs. These results suggest that a Salmonella pig-pig ecosystem exists but does not answer the open question posed above. However, we have commenced research to determine whether the wild pig population described in this study is a reservoir of Salmonella for co-grazing cattle by using molecular and spatial epidemiological methods.
We conclude that molecular epidemiological approaches and traditional cross sectional surveys are complementary and can enhance the understanding that can be achieved using either approach alone. Even in a complex hyper-endemic Salmonella ecological system, strong signals were evident and greater inferences were possible than using either approach alone. Our analyses indicated that the abundance of ecological resources critical for wildlife influences Salmonella prevalence, likely through greater persistence of Salmonella in wild pig populations. Importantly the use of a molecular approach allowed differentiation between persistence and transmission of Salmonella, revealing that transmission is influenced by local spatial, social and individual factors, rather than just resources. Additionally, reliance on only cross sectional data for evaluating Salmonella transmission would have overestimated the proportion of variability of Salmonella data that could be explained, with R2 values orders of magnitude greater than with the molecular approaches. The integration of molecular and cross sectional approaches also allows nuanced inferences for control. Implementation of control zones for wildlife disease management should be structured on complex spatial, social, density and resource distribution principals that aim to reduce prevalence as well as transmission, rather than on simple host density principals outlined in previous theoretical models.
Acknowledgments
We thank Mick Everett for his skill in shooting wild pigs, and the Northern Territory branch of the Sporting Shooters Association of Australia for collecting samples during phase 2 of the study. We also thank the University of Sydney Animal Ethics Committee for advice on animal welfare and approval (N00/6-2010/1/5319). Our sincere thanks go to Lyn O'Reilly for her assistance with PFGE.
Funding Statement
The authors gratefully acknowledge funding from the Cattle Council of Australia (www.cattlecouncil.com.au), Meat and Livestock Australia (www.mla.com.au; grant B.AHE.0053), Australian Pork Ltd. (www.australianpork.com.au; grant 1012.361), the Australian Government Department of Agriculture, Fisheries and Forestry (www.daff.gov.au) and the Western Australian Department of Agriculture and Food (www.agric.wa.gov.au). This research was also supported under Australian Research Council's Linkage Program (www.arc.gov.au) funding scheme (grant LP100200110). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, et al. (2008) Global trends in emerging infectious diseases. Nature 451: 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thomson GR (2009) Currently important animal disease management issues in sub-Saharan Africa. Onderstepoort J Vet Res 76: 129–134. [DOI] [PubMed] [Google Scholar]
- 3. Donnelly CA, Woodroffe R, Cox DR, Bourne FJ, Cheeseman CL (2006) Positive and negative effects of widespread badger culling on tuberculosis in cattle. Nature 439: 843–846. [DOI] [PubMed] [Google Scholar]
- 4. Miller W, Hayes VM, Ratan A, Petersen DC, Wittekindt NE, et al. (2011) Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil). Proc Natl Academy Sci USA 108: 12348–12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Skerratt LF, Berger L, Speare R, Cashins S, MacDonald KR, et al. (2007) Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. Ecohealth 4: 125–134. [Google Scholar]
- 6. McCallum H (2008) Tasmanian devil facial tumour disease: lessons for conservation biology. Ecol Evol 23: 631–637. [DOI] [PubMed] [Google Scholar]
- 7.Delahay RJ, Smith GC, Hutchings MR (2009) The Science of Wildlife Disease Management. New York: Springer, 1–8 pp.
- 8. Tompkins DM, Dunn AM, Smith MJ, Telfer S (2011) Wildlife diseases: from individuals to ecosystems. J Animal Ecol 80: 19–38. [DOI] [PubMed] [Google Scholar]
- 9.Wobeser GA (2007) Disease in wild animals: investigation and management. 2nd Ed. Berlin: Springer-Verlag.
- 10. Daszak P, Cunningham AA, Hyatt AD (2000) Emerging infectious diseases of wildlife – threats to biodiversity and human health. Science 287: 443–449. [DOI] [PubMed] [Google Scholar]
- 11.Cross PC, Drewe J, Patrek V, Pearce G, Samuel MD, et al.. (2009) Wildlife Population Structure and Parasite Transmission: Implications for Disease Management. New York: Springer. 9–29 pp.
- 12. Biek R, Real LA (2010) The landscape genetics of infectious disease emergence and spread. Mol Ecol 19: 3515–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Archie EA, Luikart G, Ezenwa VO (2009) Infecting epidemiology with genetics: a new frontier in disease ecology. Ecol Evol 24: 21–30. [DOI] [PubMed] [Google Scholar]
- 14.Oliver W, Leus K (2008) Sus scrofa. In IUCN 2010. IUCN Red List of Threatened Species. Version 2010.4.
- 15. Costard S, Wieland B, de Glanville W, Jori F, Rowlands R, et al. (2009) African swine fever: how can global spread be prevented? Philos Trans R Soc Lond B Biol Sci 364: 2683–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hone J, Pech R, Yip P (1992) Estimation of the dynamics and rate of transmission of classical swine fever (hog cholera) in wild pigs. Epidemiol Infect 108: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Corn JL, Stallknecht DE, Mechlin NM, Luttrell MP, Fischer JR (2004) Persistence of pseudorabies virus in feral swine populations. J Wildl Dis 40: 307–310. [DOI] [PubMed] [Google Scholar]
- 18.Khomenko S (2011) Silence of the wild boar. In: Rome, 39th General Session of the European Commission for the control of Foot-And-Mouth Disease (EuFMD). Food and Agriculture Organisation, 559–619 pp.
- 19. Van Der Leek ML, Becker HN, Humphrey P, Adams CL, Belden RC, et al. (1993) Prevalence of Brucella sp. antibodies in feral swine in Florida. J Wildlife Dis 29: 410–415. [DOI] [PubMed] [Google Scholar]
- 20. Methner U, Heller M, Bocklisch H (2010) Salmonella enterica subspecies enterica serovar Choleraesuis in a wild boar population in Germany. European J Wildlife Res 56: 493–502. [Google Scholar]
- 21.Malorny B, Hauser E, Dieckmann R (2011) New Approaches in Subspecies-level Salmonella Classification. Wymondham UK: Caister Academic Press, 1–23 pp.
- 22.Sharp T, Saunders G (2005) Standard Operating Procedure: PIG002 aerial shooting of feral pigs. Orange, NSWL Department of Primary Industries (Unit VPR).
- 23. Standards Australia (2009) AS 5013.10-2009. Microbiology of food and animal feeding stuffs - Horizontal method for the detection of Salmonella spp. (ISO 6579:2002, MOD). Standards Australia [Google Scholar]
- 24. Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, et al. (2006) Standardization of Pulsed-Field Gel Electrophoresis Protocols for the Subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Path Dis 3: 59–67. [DOI] [PubMed] [Google Scholar]
- 25. Cowled B, Lapidge SJ, Hampton JO, Spencer PBS (2006) Measuring the demographic and genetic effects of pest control in a highly persecuted feral pig population. J Wildl Manage 70: 1690–1697. [Google Scholar]
- 26.NASA (2012) Measuring vegetation (NDVI and EVI): Enhanced vegetation index (EVI). Washington: EOS Project Science Office.
- 27. Choquenot D, Saunders G (1993) A comparison of three ageing techniques for feral pigs from subalpine and semi-arid habitats. Wildl Res 20: 163–171. [Google Scholar]
- 28.Applied_Maths (2012) Bionumerics (Applied Maths, Ghent), 6.6.
- 29.Manly BFJ (2007) Randomization, Bootstrap And Monte Carlo Methods in Biology (Chapman & Hall/CRC).
- 30.Burnham KP, Anderson DR (2002) Model selection and multimodel inference: A practical information-theoretic approach. 2nd Ed. New York: Springer-Verlag, 488 p.
- 31. Kosman E, Leonard KJ (2005) Similarity coefficients for molecular markers in studies of genetic relationships between individuals for haploid, diploid, and polyploid species. Mol Ecol 14: 415–424. [DOI] [PubMed] [Google Scholar]
- 32. Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM (2005) Superspreading and the effect of individual variation on disease emergence. Nature 438: 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoye BJ, Fouchier RAM, Klaassen M (2012) Host behaviour and physiology underpin individual variation in avian influenza virus infection in migratory Bewick's swans. Proc R Soc B-Biol Sci 279: 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caley P (1997) Movements, activity patterns and habitat use of feral pigs (Sus scrofa) in a tropical habitat. Wildl Res 24: 77–87. [Google Scholar]
- 35.Choquenot D, McIlroy J, Korn T (1996) Managing Vertebrate Pests: Feral Pigs. Canberra: Bureau of Resource Sciences, Australian Government Publishing Service.
- 36. Saunders G, Kay B (1991) Movements of feral pigs Sus-scrofa at Sunny Corner New South Wales Australia. Wildl Res 18: 9–62. [Google Scholar]
- 37. Twigg LE, Lowe T, Martin G, Everett M (2005) Feral pigs in north-western Australia: basic biology, bait consumption, and the efficacy of 1080 baits. Wildl Res 32: 281–296. [Google Scholar]
- 38. Grenfell BT, Pybus OG, Gog JR, Wood JLN, et al. (2004) Unifying the Epidemiological and Evolutionary Dynamics of Pathogens. Science 303: 327–332. [DOI] [PubMed] [Google Scholar]
- 39. Anderson RM, May RM (1979) Population biology of infectious diseases: Part I. Nature 280: 361–367. [DOI] [PubMed] [Google Scholar]
- 40. McCallum H, Barlow N, Hone J (2001) How should pathogen transmission be modelled? Ecol Evol 16: 295–300. [DOI] [PubMed] [Google Scholar]
- 41. Pech RP, Hone J (1988) A model of the dynamics and control of an outbreak of foot and mouth disease in feral pigs in Australia. J Appl Ecol 25: 63–78. [Google Scholar]
- 42. Lloyd-Smith JO, Cross PC, Briggs CJ, Daugherty M, Getz WM, et al. (2005) Should we expect population thresholds for wildlife disease? Ecol Evol 20: 511–519. [DOI] [PubMed] [Google Scholar]
- 43. Davis S, Trapman P, Leirs H, Begon M, Heesterbeek JAP (2008) The abundance threshold for plague as a critical percolation phenomenon. Nature 454: 634–637. [DOI] [PubMed] [Google Scholar]
- 44. Cowled BD, Garner MG, Negus K, Ward MP (2012) Controlling disease outbreaks in wildlife using limited culling: modelling classical swine fever incursions in wild pigs in Australia. Vet Res 43: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eglezos S, Stuttard E, Huang BiXing, et al. (2008) A survey of the microbiological quality of feral pig carcasses processed for human consumption in Queensland, Australia. Foodborne Path Dis 5: 105–109. [DOI] [PubMed] [Google Scholar]
- 46. Bensink JC, Ekaputra I, Taliotis C (1991) The isolation of Salmonella from kangaroos and feral pigs processed for human consumption. Aust Vet J 68: 106–107. [DOI] [PubMed] [Google Scholar]
- 47. Anderson RM, Jackson HC, May RM, Smith AM (1981) Population dynamics of fox rabies in Europe. Nature 289: 765–771. [DOI] [PubMed] [Google Scholar]
- 48. Smith MJ, Telfer S, Kallio ER, Burthe S, Cook AR, et al. (2009) Host-pathogen time series data in wildlife support a transmission function between density and frequency dependence. Proc Natl Academy Sci USA 106: 7905–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. How RA, Bradley AJ, Iveson JB, Kemper CM, Kitchener DJ, et al. (1983) The natural history of salmonellae in mammals of the tropical Kimberley region, Western Australia. Ecol Dis 2: 9–32. [PubMed] [Google Scholar]
- 50. Fleming PJS, Tracey JP (2008) Aerial surveys of wildlife: Theory and applications – Preface. Wildl Res 35: III–IV. [Google Scholar]
- 51. Jensen AN, Dalsgaard A, Stockmarr A, Nielsen EM, Baggesen DL (2006) Survival and transmission of Salmonella enterica serovar typhimurium in an outdoor organic pig farming environment. Appl Environ Microbiol 72: 1833–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arrus KM, Holley RA, Ominski KH, Tenuta M, Blank G (2006) Influence of temperature on Salmonella survival in hog manure slurry and seasonal temperature profiles in farm manure storage reservoirs. Livestock Sci 102: 226–236. [Google Scholar]
- 53.Choquenot D, McIlroy J, Korn T (1996) Managing Vertebrate Pests: Feral Pigs. Canberra: Bureau of Resource Sciences, Australian Government Publishing Service, 163 p.
- 54. Hutchison ML, Walters LD, Moore A, Avery SM (2005) Declines of zoonotic agents in liquid livestock wastes stored in batches on-farm. J Appl Microbiol 99: 58–65. [DOI] [PubMed] [Google Scholar]
- 55. Choquenot D, Ruscoe WA (2003) Landscape complementation and food limitation of large herbivores: Habitat-related constraints on the foraging efficiency of wild pigs. J Animal Ecol 72: 14–26. [Google Scholar]
- 56.Giles JR (1980) The ecology of the feral pig in western New South Wales. Doctor of Philosophy thesis. Sydney: University of Sydney.