Abstract
Background
Replant disease often occurs when certain crops are “replanted” in a soil that had previously supported the same or similar plant species. This disease typically leads to reductions in plant growth, crop yields, and production duration, and its etiology remains ill-defined. The objective of this study was to identify microorganisms associated with peach replant disease symptoms at a field location in California, USA. Soil samples were subjected to treatments to create various levels of replant disease symptoms. Clonal peach seedlings were grown in the treated soils in greenhouse trials. After 6 weeks, plant growth parameters were measured, and both culture and culture-independent analyses were performed to identify root-associated bacteria, fungi and stramenopiles.
Results
A total of 295,785 bacterial operational taxonomic units (OTU) were identified by an Illumina-based, high throughput sequence analysis of rRNA genes. Among the 60 most abundant OTUs, 27 showed significant (P<0.05) negative correlation with peach shoot weights while 10 were positively correlated. Most of these OTUs belonged to the bacterial phylum Proteobacteria (96%), including the classes Gammaproteobacteria (44.4%), Betaproteobacteria (33.3%) and Alphaproteobacteria (22.2%), and the orders Pseudomonadales, Burkholderiales, Chromatiales, Rhodocyclales, and Sphingomonadales. The most abundant fungi were Trichoderma asperellum, Trichoderma virens, Fusarium oxysporum, Ceratocystis fimbriata and Fusarium solani. The most abundant stramenopiles were Pythium vexans, Pythium violae and an unidentified Aplanochytrium species. Validation experiments using sequence-selective quantitative PCR analyses identified negative and positive associations between P. vexans and Trichoderma spp. and peach shoot weights, respectively.
Conclusions
This study identified numerous microorganisms associated with peach replant symptoms, some of which have been previously identified while others represent new candidates. Subsequent Koch's postulates investigations will assess their possible roles in this replant disease.
Introduction
Replant diseases of pome, stone fruits, and closely related ornamentals have been reported in all of the major crop-growing regions of the world [1]–[3]. The problems are typically expressed as a dramatic reduction in plant growth and vigor, with a subsequent reduction in yield and a shortened production life. Annual losses in California alone are estimated to be 10–20% [4], [5]. As the causes of replant diseases are complex, may differ among regions, and are frequently ill-defined, management strategies have until recently relied on broad-spectrum fumigants such as methyl bromide. The world-wide phase-out of this fumigant, and the lack of available alternatives with similar efficacy, creates an urgent need for identifying the organism(s) that cause replant disease – a discovery that would enable the development of targeted management strategies, and therefore reduce reliance on such fumigants.
Numerous factors have been implicated in replant disease etiology. Putative causal agents have varied considerably among geographic regions and among orchards in the same region. Abiotic factors such as nutrition, soil structure, and phytotoxic metabolites from roots of previous crops have been implicated [6]–[9]. Microorganisms including a variety of bacteria, fungi, and oomycetes have also been implicated [10]–[30]. For example, replant disease in Prunus has been associated with an increase in rhizosphere bacilli and higher populations of cyanogenic microorganisms [10].
In this study, we examined a peach replant disease soil located in the center of California's San Joachim Valley near Parlier, CA, USA. We first determined that there was a biological component to the replant disease symptoms. We then used a population-based approach to identify root-associated bacteria, fungi, and stramenopiles that correlated with replant disease symptoms. Finally, sequence-selective qPCR assays were used to validate selected associations.
Results
Replant Disease Soil
A series of investigations were performed on a soil exhibiting replant disease properties located in Parlier, California, USA. When peach seedlings were planted in this field, they showed reductions in height and trunk-width 10 weeks after planting when compared to seedlings grown in soil fumigated with 1,3-dichloropropene (Telone II®) at 332 lb/acre, broadcast. In addition, the root systems had less developed feeder roots and were slightly darker in color than the ones in the fumigated soil [31].
To further examine the nature of this phenomenon, we performed greenhouse trials comparing plant growth parameters of peach seedlings grown in autoclaved and non-autoclaved portions of this soil. After 10 weeks, root weights, shoot weights, and shoot length were measured. In all cases, plant growth was better in the autoclaved portions (Table 1), indicating a biological component was contributing to the replant disease symptoms.
Table 1. Plant growth parameters of peach seedlings grown in autoclaved and non-autoclaved portions of a replant soil for 10 weeks in greenhouse experiments.
| Soil type | Plant growth parametersa | ||||
| Fresh weight (g) | Dry weight (g) | Length (cm) | |||
| Roots | Shoots | Roots | Shoots | Shoots | |
| Replant soil | 11.17±3.79 x | 12.33±5.20 x | 2.82±0.84 x | 4.36±1.82 x | 98.56±20.87 x |
| Autoclaved replant soil | 15.69±3.05 y | 22.41±3.64 y | 3.61±0.81 y | 8.28±1.52 y | 126.38±7.73 y |
Values and standard deviations are the means of 8 replicates pots. Values in columns followed by the same letter are not significantly (P<0.05) different.
Bacterial Associations
An Illumina-based, high throughput sequencing analysis of the small-subunit rRNA gene was used to examine root-associated bacteria from plants exhibiting different levels of replant disease symptoms. A total of 295,785 bacterial operational taxonomic units (OTU) were identified, 5,629 of which had significant correlation (P<0.05) with peach shoot weights. We posit that microbes are more likely to impact the host plant if their population densities are high. Thus, our subsequent analyses focused on the most abundant (>10,000 reads) OTUs, 27 of which showed significant (P<0.05) negative correlation with peach shoot weights (Table 2) while 10 were positively correlated (Table 3). Most of the 27 negatively associated OTUs belonged to the bacterial phylum Proteobacteria (96%), including the classes Gammaproteobacteria (44.4%), Betaproteobacteria (33.3%) and Alphaproteobacteria (22.2%). The predominant orders from these 37 OTUs included Pseudomonadales, Burkholderiales, Chromatiales, Rhodocyclales, and Sphingomonadales (Figure 1).
Table 2. Most abundant bacterial OTUs negatively associated with fresh peach shoot weights.
| OTU designation | Nearest cultured relative (accession) (% identity)a | Nearest uncultured relative accession (% identity)a | Abundance (% of total reads) | Correlation coefficient (rb) | Probability (P) |
| 278666 | Hydrogenophaga flava (AB681848) (98%) | HQ120802 (98%) | 0.094 | −0.687 | 0.000 |
| 243054 | Aquabacterium sp. (FN692032) (98%) | HE583131 (98%) | 0.082 | −0.686 | 0.000 |
| 61 | Cupriavidus sp. (AB681843) (100%) | HQ783640 (100%) | 0.042 | −0.626 | 0.000 |
| 35800 | Pseudogulbenkiania sp. (AP012224) (98%) | AB657767 (98%) | 0.059 | −0.611 | 0.000 |
| 26781 | Pseudomonas pachastrellae (HQ425676) (94%) | FJ568592 (100%) | 0.198 | −0.567 | 0.000 |
| 129755 | Bacterium MI-37 (AB529705) (95%) | FJ568592 (97%) | 0.065 | −0.561 | 0.000 |
| 172482 | Azoarcus sp. (AP012304) (100%) | JN825463 (100%) | 0.088 | −0.554 | 0.001 |
| 234080 | Azoarcus sp. (AP012304) (96%) | JN825463 (96%) | 0.032 | −0.551 | 0.001 |
| 250441 | Thiocystis violacea (FN293059) (95%) | JF990363 (98%) | 0.111 | −0.530 | 0.002 |
| 115618 | Pseudomonas fluorescens (JN411289) (98%) | AB579016 (98%) | 0.128 | −0.520 | 0.002 |
| 273727 | Pseudomonas putida (JN411453) (96%) | AB579016 (96%) | 0.037 | −0.503 | 0.003 |
| 210082 | Dechloromonas sp. (GU202936) (100%) | GU179639 (100%) | 0.044 | −0.493 | 0.004 |
| 193280 | Pseudomonas sp. (HE586886) (100%) | JQ032435 (100%) | 0.023 | −0.484 | 0.005 |
| 236351 | Rahnella aquatilis (JQ014185) (100%) | JN998890 (100%) | 0.036 | −0.481 | 0.005 |
| 288392 | Ramlibacter sp. (HQ323427) (98%) | FQ690103 (98%) | 0.181 | −0.468 | 0.007 |
| 184527 | Rhizobacter sp. (HE616175) (100%) | FQ659876 (100%) | 0.065 | −0.458 | 0.008 |
| 244218 | Methylophaga thalassica (AB681780) (95%) | HQ697540 (100%) | 0.041 | −0.427 | 0.015 |
| 207860 | Pseudomonas taiwanensis (JQ014182) (100%) | HE650703 (100%) | 0.059 | −0.415 | 0.018 |
| 273656 | Methylobacillus sp. (EU194898) (97%) | FQ659555 (98%) | 0.155 | −0.405 | 0.022 |
| 17162 | Bradyrhizobium sp. (HQ836187) (98%) | JN540015 (98%) | 0.259 | −0.387 | 0.029 |
| 167695 | Methylophilus leisingeri (NR_041258) (100%) | AB635923 (100%) | 0.071 | −0.387 | 0.029 |
| 246943 | Pseudomonas sp. (FN995250) (94%) | FQ659619 (97%) | 0.536 | −0.374 | 0.035 |
| 166091 | Woodsholea maritima (FM886859) (97%) | HE614733 (99%) | 0.062 | −0.368 | 0.038 |
| 234039 | Terrimonas lutea (NR_041250) (100%) | FQ706675 (100%) | 0.060 | −0.367 | 0.040 |
| 11757 | Thiocystis violacea (FN293059) (97%) | FR853185 (99%) | 0.046 | −0.361 | 0.042 |
| 164910 | Pseudomonas sp. (FN995250) (92%) | FQ659619 (95%) | 0.110 | −0.361 | 0.042 |
| 32731 | Cellvibrio japonicus (CP000934) (99%) | HQ691969 (98%) | 0.119 | −0.360 | 0.043 |
% identity values are from analyses using BLAST (NCBI) where coverage was at least 96%.
r is the Pearson's correlation coefficient.
Table 3. Most abundant bacterial OTUs positively associated with fresh peach shoot weights.
| OTU designation | Nearest cultured relative (accession) (% identity)a | Nearest uncultured relative accession (% identity)a | Abundance (% of total reads) | Correlation coefficient (r b) | Probability (P) |
| 164017 | Massilia aerilata (HQ406763) (98%) | JN590660 (98%) | 0.202 | 0.546 | 0.012 |
| 101298 | Thermomonas haemolytica (GU195191) (98%) | FQ680347 (98%) | 0.339 | 0.532 | 0.017 |
| 286079 | Sphingopyxis sp. (JF297627) (99%) | HQ118566 (99%) | 0.132 | 0.447 | 0.010 |
| 66648 | Novosphingobium naphthalenivorans (AB681685) (99%) | HQ754243 (99%) | 0.154 | 0.442 | 0.011 |
| 173712 | Novosphingobium naphthalenivorans (AB681685) (98%) | HQ754243 (98%) | 0.647 | 0.431 | 0.014 |
| 259461 | Sphingopyxis sp. (JF297627) (98%) | HQ118566 (98%) | 0.427 | 0.423 | 0.016 |
| 233081 | Rhodopseudomonas palustris (AB689796) (98%) | JN863157 (98%) | 0.140 | 0.422 | 0.016 |
| 162892 | Novosphingobium subterraneum (HM032869) (98%) | FQ741870 (98%) | 0.464 | 0.398 | 0.024 |
| 275502 | Dyella sp. (GQ369135) (100%) | JF341880 (100%) | 0.259 | 0.394 | 0.025 |
| 30925 | Rhodanobacter lindaniclasticus (L76222) (100%) | JF341837 (100%) | 0.493 | 0.366 | 0.039 |
% identity values are from analyses using BLAST (NCBI) where coverage was at least 96%.
r is the Pearson's correlation coefficient.
Figure 1. Relative abundance of the most prevalent bacterial orders associated (P<0.05) with fresh peach shoot weights.
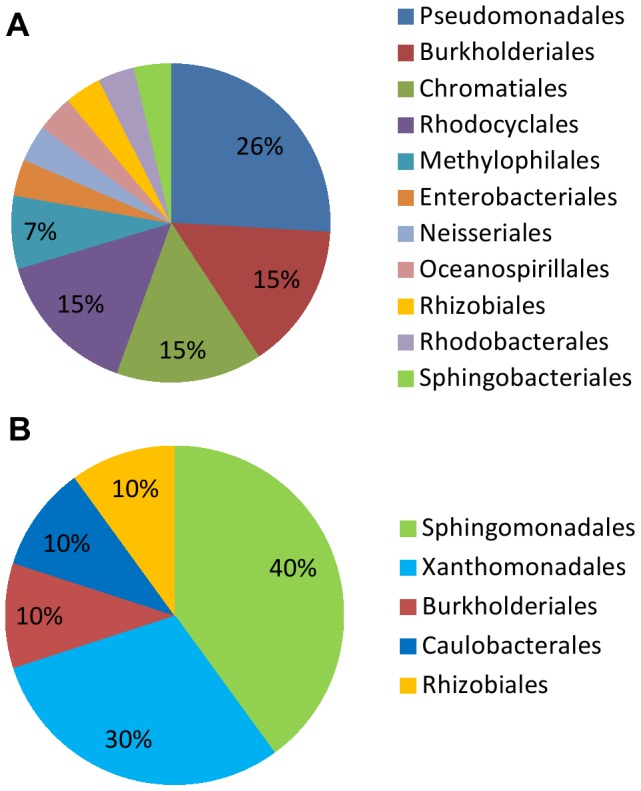
A. Negative associations. B. Positive associations.
Fungal and Stramenopile Associations
To identify fungi and stramenopiles associated with the replant disease symptoms, both culture and culture-independent analyses were performed on root-associated organisms from plants exhibiting different levels of replant disease symptoms. For the culture-based studies, 295 fungal and stramenopile isolates were obtained and identified by sequence analysis of the rRNA internal transcribed spacer (ITS). For the culture-independent analysis, 274 small-subunit rRNA gene clones were analyzed. In both the fungal and stramenopile analyses, as expected, there was a considerably greater number of phylotypes detected in the culture-independent analysis compared to the culture-based analysis.
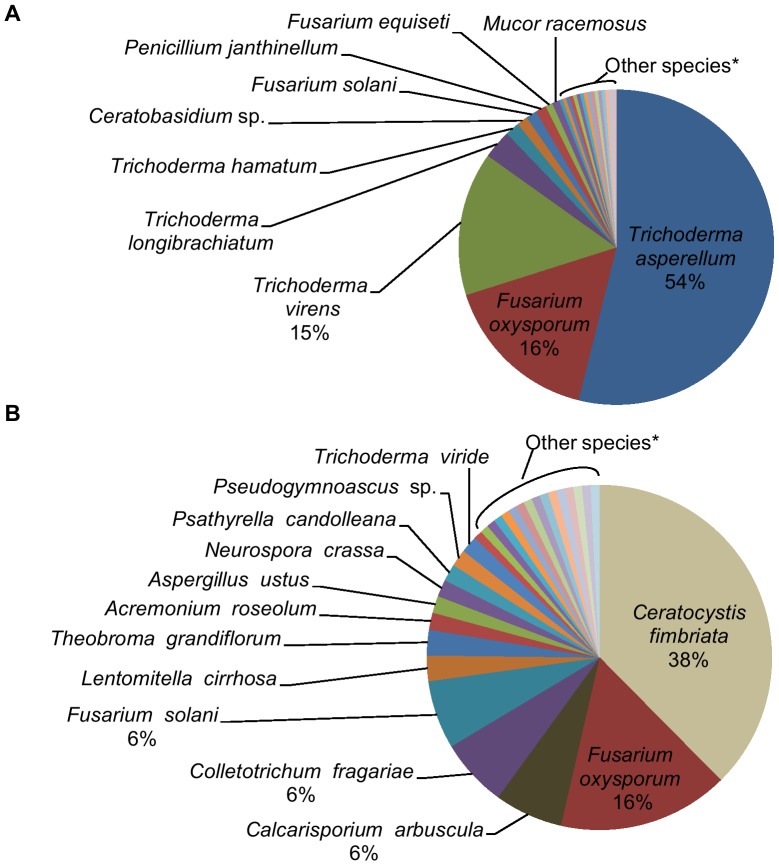
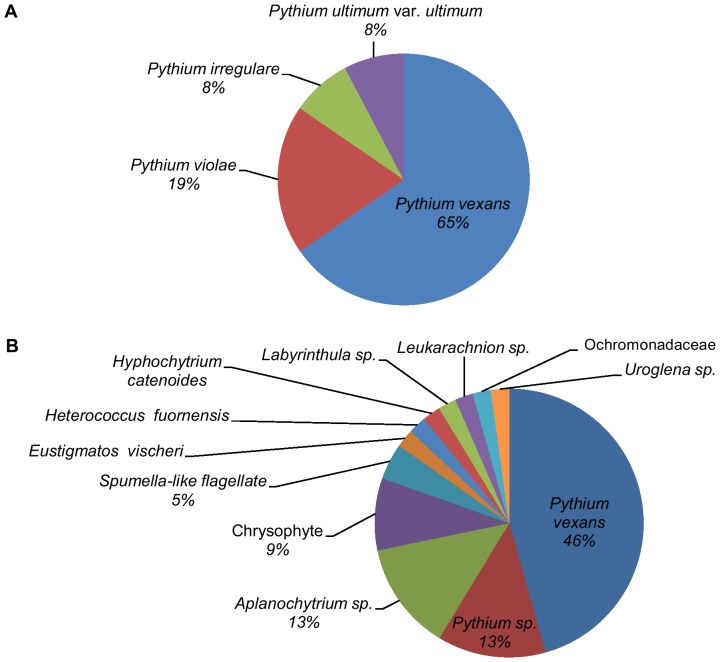
The most abundant fungi isolated from roots grown in the replant soil were Trichoderma asperellum (54%), Fusarium oxysporum (16%), and Trichoderma virens (15%) (Figure 2A). In contrast, the most abundant fungi obtained from the culture-independent analysis of roots grown in the replant soil were Ceratocystis fimbriata (38%) and F. oxysporum (16%) (Figure 2B). The most abundant stramenopiles isolated from roots grown in the replant soil were the oomycetes Pythium vexans (65%), Pythium violae (19%), and Pythium irregulare (8%) (Figure 3A). The most abundant stramenopiles obtained from the culture-independent analysis of roots grown in the replant soil were Pythium vexans (46%), an unidentified Pythium species (13%), and an unidentified Aplanochytrium species (13%) (Figure 3B).
Figure 2. Relative abundance of fungi from peach seedling roots grown in a soil exhibiting peach replant disease symptoms.
A. Cultured-based analysis; values are % of 269 isolates. *Species less than 1% included: Apodus oryzae, Ceratocystis fimbriata, Clonostachys divergens, Cunninghamella echinulata, Gelasinospora brevispora, Hypocrea koningii, Hypocrea sp., Mortierella alpina, Mortierella elongata, Mortierella minutissima, Penicillium simplicissimum, Sclerostagonospora opuntiae, Sporormia subticinensis, Trichoderma gamsii, Trichoderma pubescens, and Trichoderma viride. B. Culture-independent analysis; values are % of 130 sequences. *Species less than 1% included: Acremonium sclerotigenum, Aspergillus niger, Brachyconidiellopsis sp., Craterocolla cerasi, Elaphocordyceps ophioglossoides, Engyodontium album, Halophytophthora vesicular, Hypomyces chrysospermus, Lithothelium septemseptatum, Metarhiziopsis microspore, Neokarlingia chitinophila, Nomuraea rileyi, Ophiocordyceps konnoana, Sebacina vermifera, Verticillium dahlia, and Volutella ciliata. For A and B, taxa without numbers have relative abundance values between 1% and 5%.
Figure 3. Relative abundance of stramenopiles from peach seedling roots grown in soil exhibiting peach replant disease symptoms.
A. Cultured isolates; values are % of 26 isolates. B. Culture-independent analysis; values are % of 48 sequences. Taxa without numbers have relative abundance values of less than 5%.
The most abundant fungal and stramenopile isolates and phylotypes were subjected to further analysis using sequence-selective qPCR assays targeting the ITS region. For the phylotypes identified by the small-subunit rRNA gene analyses, chromosomal walking procedures were used to obtain the ITS sequences. Using an assay targeting both Trichoderma species, a positive association was detected with plant shoot weights [log10 rRNA copy number = 5.97+0.0648 (grams of shoots); P = 0.012; R2 = 6.5%, n = 116]. Using an assay targeting P. vexans, a negative association was detected with plant shoot weights [log10 rRNA copy number = 5.01−0.0931 (grams of shoots); P = 0.008; R2 = 8.7%, n = 116]. Using an assay targeting C. fimbriata, a negative association was detected with plant shoot weights [log10 rRNA copy number = 7.11−0.0574 (grams of shoots); P = 0.097; R2 = 2.7%, n = 104].
Discussion
To our knowledge, this study presents the first use of high throughput sequencing to examine bacteria associated with replant disease. Given the large number of OTUs identified, it was not surprising that we identified considerable numbers of organisms correlating with plant growth parameters, most of which have not been previously associated with replant disease. In addition, most of these bacteria are not known to be plant pathogens or have functions that could be readily attributed to replant disease properties. Ultimately, determining the roles that these bacteria may play in replant disease will require additional experimentation, including fulfilling Koch's postulates.
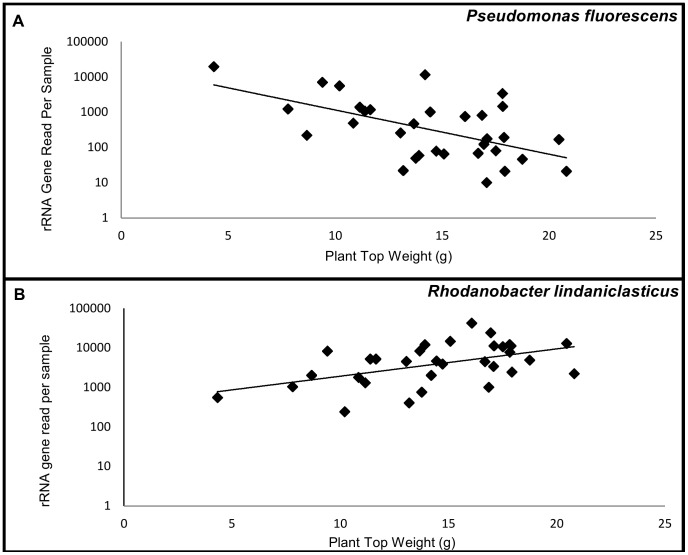
Findings from this study that are consistent with prior work include the identification of several OTUs from the genus Pseudomonas, which exhibited negative correlations with peach shoot weights. These phylotypes possess rRNA genes with high sequence identities to P. pachastrellae, P. putida, P. fluorescens, P. straminea, P. fulva, P. taiwanensis, and P. monteilii (Table 2, and example in Figure 4A). In prior investigations of root-associated bacteria and replant disease, pseudomonads were shown to be more abundant in the rhizoplane of plants grown in grapevine replant soils [32].
Figure 4. Relationships between bacteria and fresh peach shoot weights.
A. Pseudomonas fluorescens. B. Rhodanobacter lindaniclasticus. Regression equations are (A) [log10 reads per sample = 4.02−0.146 (grams of shoots); P = 0.015; R2 = 18.3%, n = 32] and (B) [log10 reads per sample = 2.59+0.069 (grams of shoots); P = 0.004; R2 = 25.0%, n = 32]. Lines are from regression analyses.
Bacteria from several taxonomic groups, including Pseudomonas, have been proposed to play a role in replant disease etiology through the production of hydrogen cyanide (HCN). When soils were amended with peach roots dried at 80°C, they caused retarded growth of peach seedlings [6]. These investigators also showed that peach roots contained considerable amounts of an HCN precursor, and roots grown in the replant soils harbored heat-resistant bacilli able to transform this precursor into HCN. While our investigation did not detect bacilli that negatively correlated with plant weights, we did identify significant negative associations with pseudomonas (Table 2), which are also known to produce HCN [33], [34]. Other investigations have reported related findings including higher levels of HCN-producing bacilli in the rhizosphere of peach plants grown in a replant soil [10]. In addition, HCN-producing Pseudomonas species were isolated from the rhizosphere in an apple replant site [35]. Variation among the results from the aforementioned studies could involve differences in the subtypes of replant diseases and/or the methods used to identify the microbial communities.
Another putatively causal taxon is the actinobacteria, formerly called actinomycetes. Prior microscopic investigations identified large numbers of actinobacteria growing in the epidermal and cortical tissue of apple seedling rootlets grown in replant disease soils, and very few actinobacteria in rootlets grown in non-replant soils [36], [37]. In addition, the amount of plant tissue damage was proportional to the number of actinobacteria [36]. In our study, we also identified several actinobacterial OTUs that were negatively associated with plant shoot weights, although they were not present in high numbers (Table 4).
Table 4. Actinobacterial OTUs negatively associated with fresh peach shoot weights.
| OTU designation | Nearest cultured relative (accession) (% identity)a | Nearest uncultured relative accession (% identity)a | Abundance (% of total reads) | Correlation coefficient (r b) | Probability (P) |
| 76253 | Catellatospora sp. (KC-EP-S6) (FJ711222) (100%) | JF982278 (100%) | <0.001 | −0.826 | 0.000 |
| 22707 | Solirubrobacter sp. L64 (FJ459990) (90%) | JF987729 (100%) | <0.001 | −0.774 | 0.004 |
| 105414 | Gaiella occulta (JF423906) (92%) | HM187195 (98%) | <0.001 | −0.744 | 0.025 |
| 66985 | Aciditerrimonas ferrireducens (AB517669) (84%) | AJ616079 (97%) | <0.001 | −0.740 | 0.027 |
| 101423 | Phycicoccus sp. P3703 (JQ419657) (98%) | CU919577 (99%) | <0.001 | −0.738 | 0.031 |
| 163353 | Virgisporangium ochraceum (AB546280) (97%) | FJ479084 (100%) | <0.001 | −0.734 | 0.037 |
| 266579 | Gaiella occulta (JF423906) (94%) | AB656050 (100%) | 0.001 | −0.732 | 0.042 |
% identity values are from analyses using BLAST (NCBI) where coverage was at least 88%.
r is the Pearson's correlation coefficient; probability values were adjusted using the Bonferroni correction method.
It was surprising to find that Xanthomonadaceae was one of the dominant taxa exhibiting a positive association with plant shoot weights (Table 3, and example in Figure 4B), because these bacteria, and specifically pathovars in the order Xanthomonas, have been shown to cause diseases on at least 124 monocotyledons and 268 dicotyledons [38]. Our results suggest a possible beneficial role of this bacterial group, where these organisms may interact with plants and/or other microbes, leading to plant growth promotion.
Numerous oomycetes also have been implicated in replant disease etiology [12], [13], [16], [18], [20], [22]–[24], [26]–[28]. In our study, P. vexans was frequently detected by both culture-based and culture-independent analyses. In addition, our sequence-selective qPCR analysis showed a negative correlation between P. vexans rRNA gene numbers and peach shoot weights. However, in another peach replant study, P. vexans was not significantly correlated with plant biomass [39]. Such site dependent variation has been observed in replant diseases of other crops. For example, P. vexans was shown to be pathogenic to apple seedlings in one study [24] yet exhibited biological control efficacy in another [20]. This phenomenon could be due to virulence differences among the P. vexans strains and/or varying interactions with abiotic or biotic features of the soils [40], [41].
Several features of P. vexans could allow it to contribute to replant disease symptoms. Zoospores of P. vexans are attracted to roots at zones of elongation and at breaks in cortical tissue associated with lateral root emersion [42]. In addition, two elicitin-like proteins (Vex1 and Vex2) secreted by P. vexans appear to induce a necrotic and hypersensitive response in a manner similar to that observed in Phytophthora species [43].
Many fungi also have been implicated in peach replant diseases. For example, Fusarium equiseti, Fusarium moniliforme, Fusarium oxysporum, Fusarium solani, Alternaria tenuis, Myrothecium verrucaria, and Mycelia sterilia were frequently isolated from the rhizosphere or roots of peach trees grown in a replant soil and found to be parasitic to the roots [29]. In Kent, England, Thielaviopsis basicola was implicated in cherry and plum replant disease [15], [25], [30]. Armillaria and Verticillium were associated with peach and almond replant symptoms in California [30]. In Italy, several species of Fusarium, Penicillium, Aspergillus, and Trichoderma were common isolates from peach replant soils [17].
Several of the most abundant fungi identified in our study have been previously implicated in replant disease including the abovementioned F. oxysporum and Trichoderma spp. In addition, the most abundant fungus in our replant soil identified by the culture-independent analysis was Ceratocystis fimbriata. Although C. fimbriata has been described as the pathogen of mallet wound canker on almond, peach, apricot, and coffee [44], [45], and it is the causal agent of many other wilt and rot diseases of sweet potato, poplar, cocoa, citrus, gmelina, and Eucalyptus species [46]–[48], to our knowledge it has not been previously associated with peach replant disease.
Our investigations also identified two Trichoderma species that were frequently isolated from peach roots, and that exhibited a positive association with plant shoot weights via a sequence-selective qPCR analysis. These results suggest that these fungi may be inhibiting the causal microbes and/or acting as a plant growth promoter. Trichoderma species can inhabit a variety of soil and plant niches [49]. The genus contains members that are plant growth promoters that act through a variety of mechanisms. Trichoderma species produce several lytic enzymes and antibiotics against plant pathogens, and several products made from these fungi have been commercially marketed as biopesticides, biofertilizers, and soil amendments [50]. Trichoderma virens is one of the well-studied species, which exhibits mycoparasitic characteristics and the ability to produce several potent epithiodiketopiperazine antibiotics that inhibit oomycetes such as Pythium and Phytophthora species. It also produces a mixture of peptaibols, which are linear peptide antibiotics that might affect certain bacteria and fungi [51]. Some strains of T. asperellum and T. harzianum are capable of activating plant defense responses [52], [53]. Strains of T. asperellum also have been shown to suppress important plant pathogens including Phytophthora megakarya [54], Fusarium oxysporum f. sp. lycopersici [55], Rhizoctonia solani [56], and Meloidogyne javanica [57].
In sum, this study identified large numbers of microorganisms associated with peach replant disease symptoms in a Californian soil. Such associations point toward organisms that could be causing or inhibiting the replant disease, or that are simply responding to changes in the environment caused by the disease. Future investigations that assess cause and effect, such as Koch's postulates experimentation, will be needed to further define the roles of these organisms.
Materials and Methods
Greenhouse Trials
Greenhouse trials were performed to (i) determine if there was a biological component causing the replant disease symptoms and to (ii) create soils with various levels of the replant disease symptoms for microbial community analyses. Soil (upper 30-cm) was collected from a field at the University of California Kearney Agricultural Research and Extension Center in Parlier, California, USA, where replant disease symptoms were observed on Nemaguard rootstocks 10 weeks after planting [31]. Soil texture parameters were 62% sand, 30% silt, and 8% clay. Soil was passed through a metal sieve with 12-mm openings prior to use in greenhouse experiments.
To determine if there was a biological component causing the replant disease symptoms, peach seedling clones were planted in the untreated replant soil and replant soil that had been autoclaved for 2 hours at 121°C. Plastic pots with drain holes were double-cupped and filled with 800-cm3 of the two soils. Each pot was planted with one two-month-old Nemaguard peach seedling clone, donated by Duarte Nursery, Hughson, CA. Each pot was fertilized with 7-g of slow-release fertilizer (Sierra 17-6-10 plus Minors, Scotts-Sierra Horticultural Products Company, Marysville, OH) and watered as needed. Trials were performed in a greenhouse, arranged in a randomized complete design with 8 replicates for each soil treatment. After 10 weeks, root weights, shoot weights and shoot length were measured.
To create soils with various levels of the replant disease symptoms for the microbial community analyses, soils were (i) temperature-treated or (ii) diluted with various amounts of autoclaved soil. For the temperature treatments, soils were exposed to room temperature, 40°C, 50°C, 60°C and 70°C. Soil samples (∼1 kg) were double-bagged and submerged in a water bath, and held for 30 minutes once the center of the sample reached the target temperature. The bags were then cooled to room temperature under running tap water. All samples of the same treatment were pooled and mixed. For the dilution treatments, soils were mixed with different percentages of autoclaved soil (121°C for 2 hours) at ratios of non-treated to autoclaved soil of 100∶0, 10∶90, 1∶99, 0.1∶99.9 and 0∶100. Treated soils were aerated at room temperature for 2 days prior to use. As described above, pots were filled with soil, planted with peach seedling clones, fertilized and watered. Trials were performed in a greenhouse, arranged in a randomized complete design with six replicates for each soil treatment, and performed twice. After 6 weeks, plant shoots were cut off 10-cm above the soil level and weighed. Shoot lengths of each branch were measured from the main stem. Plant dry weights were measured after 3-days in a drying oven at 125°C. Root tip samples (200 mg) from each plant were collected and stored at −20°C for DNA extraction. Fine root tips were collected and stored in sterile tubes at room temperature for culturing of fungi and oomycetes. Plant growth parameters obtained from the greenhouse trials were subjected to ANOVA and two-tailed student t-tests using Microsoft Excel 2007 (Microsoft, Redmond, WA).
Isolation of Fungi and Stramenopiles
Pieces of fine root tips from each of the non-treated replant soils were collected at the end of the trials, stored at room temperature and processed for culturing within 24 hours after sampling. From each replicate pot, 12 pieces of 3-cm-long root tips were rinsed with ultrapure water for 15 seconds, dried by pressing between paper towels, placed on 1% water agar, and incubated at room temperature. Fungi and oomycetes that emerged from the root surfaces during the first 36 hours were sub-cultured on new 1% water agar plates. The hyphal-tip method was used to obtain pure cultures. Isolates were identified by rRNA gene sequence analysis (described below).
DNA Extraction
DNA was extracted from root tip samples collected at the end of the greenhouse trials and fungi and oomycetes cultured from the roots. Two hundred milligrams of root tips or fungal hyphae were used for each DNA extraction. Genomic DNA was extracted using the FastDNA Spin Kit for Soil (Qbiogene, Carlsbad, CA) as described by the manufacturer using a 90 second bead-beating step in a FastPrep Instrument (Qbiogene) and a 5.5 setting. The extraction product was further purified by electrophoresis in 1% agarose gels. DNA larger than 3 Kb was isolated by using a MinElute Gel Extraction Kit (Qiagen, Valencia, CA), without use of UV lighter or ethidium bromide.
Bacterial rRNA Gene Sequencing
A high throughput sequencing analysis of the bacterial small-subunit rRNA genes was performed using genomic DNA extracted from the root samples collected at the end of the greenhouse trials as templates. Root samples used were from seedlings grown in the non-treated to autoclaved soil ratios of 100∶0, 10∶90, 1∶99, and the temperature treatments of 40°C and 50°C. One hundred microliter amplification reactions were performed in an MJ Research PTC-200 thermal cycler (Bio-Rad Inc., Hercules, CA) and contained: 50 mM Tris (pH 8.3), 500 µg/ml bovine serum albumin (BSA), 2.5 mM MgCl2, 250 µM of each deoxynucleotide triphosphate (dNTP), 400 nM of each primer, 4 µl of DNA template, and 2.5 units JumpStart Taq DNA polymerase (Sigma-Aldrich, St. Louis, MO). The PCR primers (F515/R806) targeted a portion of the 16S rRNA gene containing the hypervariable V4 region, with the reverse primers including a 12-bp barcode (Tables S1 and S2) [58]. Thermal cycling parameters were 94°C for 5 minutes; 35 cycles of 94°C for 20 seconds, 50°C for 20 seconds, and 72°C for 30 seconds, and followed by 72°C for 10 minutes. PCR products were purified using a MinElute Gel Extraction Kit (Qiagen) except PB buffer was substituted with QG buffer. PCR products were diluted to 20 ng/µl. DNA sequencing was performed using an Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA). One hundred base sequencing reads of the 5′ end of the amplicons and seven base barcode reads were obtained using the sequencing primers listed in Table S3. De-multiplexing, quality control and OTU binning were performed using QIIME [59]. The 5′ base of the barcodes was not used for de-multiplexing because of low quality reads at this position; however, all 32 codes (one for each of the 32 samples) were distinguishable using the last six bases. Low quality sequences were removed using the following parameters: Q20, minimum number of consecutive high quality base calls = 100 bp, maximum number of N characters allowed = 4, maximum number of consecutive low quality base calls allowed before truncating a read = 4. The total number of sequencing reads was 71,320,513. Numbers of sequences removed using the aforementioned quality control parameters were: barcode errors (3,893,840), reads too short in length (17,427,455), and too many Ns (4,072,554). OTUs were binned at 97% identity.
PCR Amplification of Stramenopile and Fungal rRNA Genes
For the culture-independent analysis, root DNA extracted from plants exhibiting a wide range of replant disease symptoms from greenhouse trials were used as templates. Ten microliter amplification reactions were performed in 10-µl glass capillary tubes using a RapidCycler (Idaho Technologies, Salt Lake City, UT) containing the following reagents: 50 mM Tris (pH 8.3), 500 mg/ml bovine serum albumin (BSA), 2.5 mM MgCl2, 250 mM of each dNTP, 400 nM of each forward and reverse primer, 1-µl (∼66 ng) of peach root DNA and 0.5 units Taq DNA polymerase. Fungi-selective primers were nu-SSU-0817-5 (TTAGCATGGAATAATRRAATAGGA) and nu-SSU-1536-3 (ATTGCAATGCYCTATCCCCA) [60] while stramenopiles were StramenoSSUF1 (GATGATTAGATACCATCGTA) and StramenoSSUR2 (AAAGGGCAGGGACGT) [39], with PCR products being ∼762 bp and ∼638 bp, respectively. Thermal cycling parameters were 94°C for 5 minutes; 35 cycles of 94°C for 20 seconds, X°C for 30 seconds and 72°C for 40 seconds; followed by 72°C for 5 minutes, where X = 55 for fungi and 59 for stramenopiles.
For the culture-based analyses, fungal and stramenopile (which were all oomycetes) isolates were identified by rRNA gene analyses. rRNA gene primers ITS1FUSER (GGGAAAGUCTTGGTCATTTAGAGGAAGTAA) [61] and ITS4USER (TCCTCCGCTTATTGATATGC) [62] were used with the following thermal cycling conditions: 94°C for 5 minutes, followed by 40 cycles of 94°C for 20 seconds, 52°C for 20 seconds, 72°C for 40 seconds, and a final incubation at 72°C for 5 minutes. PCR master mixes were prepared as described above in this subsection.
To obtain the sequences of the internal transcribed spacer (ITS) region for P. vexans and C. fimbriata for the quantitative PCR assays, chromosome walking was conducted as follows. PCR was performed on peach root DNA using the following forward primers combined with ITS4 (TCCTCCGCTTATTGATATGC) [62]: PvexansSSUF3 (GGGACTTTTGGGTAATC) or CfimbSSUF1 (AGGTCCAGACACAG). The forward primers were designed using PRISE [63]. The thermal cycling conditions were 94°C for 5 minutes; 40 cycles of 94°C for 20 seconds, 52°C for 30 seconds and 72°C for 90 seconds; followed by 72°C for 10 minutes. Amplification products were gel isolated and cloned as described previously [39], and the nucleotide sequences were obtained as described below.
Nucleotide sequences of fungi and stramenopile rRNA gene fragments were determined using the ABI BigDye Terminator v3.1 Cycle Sequencing Kit and an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA). Sequence identities were determined by an analyses using BLAST (NCBI) [64].
Quantitative PCR
Real-time qPCR assays targeting P. vexans, C. fimbriata, and Trichoderma (targeting species T. asperellum and T. virens) were performed using a Bio-Rad iCycler MyiQTM Real-Time Detection System (Bio-Rad Laboratories, Inc). The templates were genomic DNA extracted from the root samples collected at the end of the greenhouse trials. Sequence-selective primers developed in this study were designed using PRISE software [63]. The selective primers for P. vexans were VexansITSF31 (GCTGCTGGCGCTTGAT) and VexansITSR31 (TTCGTCCCCACAGTATACTT). The primers for C. fimbriata were CfimbITSF2 (TCTTCCTTGACAGAGATG) and CfimbITSR9 (TCACTGAGCCATCCAA). The primers for Trichoderma species were TricoITSF9 (TCCGAGCGTCATTTCAA) and TricoITSR3 (GTGCAAACTACTGCGC). The targets were fragments of the ITS rRNA gene with sizes of 131-bp, 181-bp, and 126-bp, respectively. The thermal cycling conditions were 94°C for 5 minutes; X cycles of 94°C for 20 seconds, Y°C for 30 seconds and 72°C for Z seconds; followed by 72°C for 10 minutes; where (X, Y, Z) = (44, 69.5, 40) for P. vexans, (38, 66.5, 30) for C. fimbriata, and (42, 65, 30) for Trichoderma. The amplification reactions were performed in iCycler iQ PCR Plates with Optical Flat 8-Cap Strips (Bio-Rad Laboratories, Inc.). PCR amplifications were performed in 25-µl reactions contained the following reagents: 50 mM Tris (pH 8.3), 500 ug/ml bovine serum albumin (BSA), 2.5 mM MgCl2, 250 mM of each dNTP, 400 nM of each primer, 1-µl of template DNA (∼176 ng), 2-µl of 10X SYBR Green I (Invitrogen, Carlsbad, CA) and 1.25 units Taq DNA polymerase. rRNA gene levels in root DNA were quantified by interpolation using a standard curve comprised of a dilution series of cloned rRNA genes.
Nucleotide Sequence Data
The nucleotide sequences of the following rRNA genes reported in this paper have been deposited in GenBank (JQ973613-JQ973615) and Sequence Read Archives (SRA049976.1).
Supporting Information
Reverse PCR primers used in the Illumina-based high throughput sequence analysis of bacterial 16S rRNA genes. Each reverse PCR primer is comprised of the 4 adjoining segments in each row shown above.
(DOCX)
Forward PCR primer used in the Illumina-based high throughput sequence analysis of bacterial 16S rRNA genes. The forward PCR primer is comprised of the 3 adjoining segments shown above.
(DOCX)
Sequencing primers used in the Illumina-based high throughput sequence analysis of bacterial 16S rRNA genes.
(DOCX)
Acknowledgments
The authors thank John Darsow for his technical assistance, and the Duarte Nursery (Hughson, CA) for their generous donation of the peach seedlings.
Funding Statement
This project was supported by Cooperative State Research, Education, and Extension Service, United States of America Department of Agriculture, under Award No. 2005-51102-02340, and by the University of California Agricultural Experiment Station. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Koch LW (1955) The peach replant problem in Ontario: I. Symptomatology and distribution. Can J Botany 33: 450–460. [Google Scholar]
- 2. Mai WF, Abawi GS (1981) Controlling replant diseases of pome and stone fruits in Northeastern United States by preplant fumigation. Plant Dis 65: 859–864. [Google Scholar]
- 3. Utkhede RS, Smith EM (2000) Impact of chemical, biological and cultural treatments on the growth and yield of apple in replant-disease soil. Australas Plant Path 29: 129–136. [Google Scholar]
- 4. Koenning SR, Overstreet C, Noling JW, Donald PA, Becker JO, et al. (1999) Survey of crop losses in response to phytoparasitic nematodes in the United States for 1994. J Nematol 31: 587–618. [PMC free article] [PubMed] [Google Scholar]
- 5.McKenry MV (1999) The replant problem and its management. Catalina Publishing, Fresno, CA. [Google Scholar]
- 6. Gur A, Cohen Y (1989) The peach replant problem - some causal agents. Soil Biol Biochem 21: 829–834. [Google Scholar]
- 7. Mizutani F, Hirota R, Kadoya K (1988) Growth inhibiting substances from peach roots and their possible involvement in peach replant problems. Acta Horticulturae 233: 37–43. [Google Scholar]
- 8. Patrick ZA (1955) The peach replant problem in Ontario. II. Toxic substances from microbial decomposition products of peach root residues. Can J Botany 33: 461–486. [Google Scholar]
- 9. Proebsting E, Gilmore A (1941) The relation of peach root toxicity to the re-establishing of peach orchards. Proc Am Soc Hort Sci 38: 21–26. [Google Scholar]
- 10. Benizri E, Piutti S, Verger S, Page L, Vercambre G, et al. (2005) Replant diseases: bacterial community structure and diversity in peach rhizosphere as determined by metabolic and genetic fingerprinting. Soil Biol Biochem 37: 1738–1746. [Google Scholar]
- 11. Gu YH, Mazzola M (2003) Modification of fluorescent pseudomonad community and control of apple replant disease induced in a wheat cultivar-specific manner. Appl Soil Ecol 24: 57–72. [Google Scholar]
- 12. Hendrix FF, Powell WM (1970) Control of root pathogens in peach decline sites. Phytopathology 60: 16–19. [Google Scholar]
- 13. Hendrix FF, Powell WH, Owen JH (1966) Relation of root necrosis caused by Pythium species to peach decline. Phytopathology 56: 1229–1232. [Google Scholar]
- 14. Hine RB (1961) Role of fungi in peach replant problem. Plant Dis Rep 45: 462–465. [Google Scholar]
- 15. Hoestra H (1965) Thielaviopsis basicola, a factor in the cherry replant problem in the Netherlands. Eur J Plant Pathol 71: 180–182. [Google Scholar]
- 16. Kouyeas H (1971) On the apoplexy of fruit trees caused by Phytophthora spp. Ann Inst Phytopath Benaki 10: 163. [Google Scholar]
- 17. Manici LM, Caputo F (2010) Soil fungal communities as indicators for replanting new peach orchards in intensively cultivated areas. Eur J Agron 33: 188–196. [Google Scholar]
- 18. Mazzola M (1998) Elucidation of the microbial complex having a causal role in the development of apple replant disease in Washington. Phytopathology 88: 930–938. [DOI] [PubMed] [Google Scholar]
- 19. Mazzola M (2007) Manipulation of rhizosphere bacterial communities to induce suppressive soils. J Nematol 39: 213–220. [PMC free article] [PubMed] [Google Scholar]
- 20. Mazzola M, Andrews PK, Reganold JP, Levesque CA (2002) Frequency, virulence, and metalaxyl sensitivity of Pythium spp. isolated from apple roots under conventional and organic production systems. Plant Dis 86: 669–675. [DOI] [PubMed] [Google Scholar]
- 21. Mazzola M, Gu YH (2000) Impact of wheat cultivation on microbial communities from replant soils and apple growth in greenhouse trials. Phytopathology 90: 114–119. [DOI] [PubMed] [Google Scholar]
- 22. Mircetich SM (1971) The role of Pythium in feeder roots of diseased and symptomless peach trees and in orchard soils in peach tree decline. Phytopathology 61: 357–360. [Google Scholar]
- 23. Mircetich SM, Matheron ME (1976) Phytophthora root and crown rot of cherry trees. Phytopathology 66: 549–558. [Google Scholar]
- 24. Mulder D (1969) The pathogenicity of several Pythium species to rootlets of apple seedlings. Neth J Plant Pathol 75: 178–181. [Google Scholar]
- 25. Sewell GWF, Wilson JF (1975) The role of Thielaviopsis basicola in the specific replant disorders of cherry and plum. Ann Appl Biol 79: 149–169. [Google Scholar]
- 26. Sitepu D, Wallace HR (1974) Diagnosis of retarded growth in an apple orchard. Austral J Expt Agric and Animal Husb 14: 577–584. [Google Scholar]
- 27. Sutton T, Wayne D, Sullivan W, Nardacci J, Klimstra D (1981) Causes of apple tree death in Henderson county, North Carolina. Plant Dis 65: 330–331. [Google Scholar]
- 28. Traquair J (1984) Etiology and control of orchard replant problems: a review. Can J Plant Pathol 6: 54–62. [Google Scholar]
- 29. Wensley RN (1956) The peach replant problem in Ontario: IV. Fungi associated with replant failure and their importance in fumigated and nonfumigated soils. Can J Botany 34: 967–981. [Google Scholar]
- 30.Yadava UL, Doud SL (1980) The short life and replant problems of deciduous fruit trees, p. 1–116, Horticultural Reviews. John Wiley & Sons, Inc. [Google Scholar]
- 31.McKenry MV, Buzo T (2009) Evaluation of “starve and switch” approach to replanting trees. Annu Int Res Conf Methyl Bromide Altern Emiss Reduct, San Diego, CA, Abstract 48. Website for 2012 Annual International Research Conference on Methyl Bromide Alternatives and Emissions Reductions. Available: http://mbao.org. Accessed 2012, September 12.
- 32. Waschkies C, Schropp A, Marschner H (1994) Relations between grapevine replant disease and root colonization of grapevine (Vitis sp.) by fluorescent pseudomonads and endomycorrhizal fungi. Plant Soil 162: 219–227. [Google Scholar]
- 33. Blumer C, Haas D (2000) Mechanism, regulation and ecological role of bacterial cyanide biosynthesis. Achievements Microbiol 173: 170–177. [DOI] [PubMed] [Google Scholar]
- 34. Pal KK, Tilak KV, Saxena AK, Dey R, Singh CS (2000) Antifungal characteristics of a fluorescent Pseudomonas strain involved in the biological control of Rhizoctonia solani . Microbiol Res 155: 233–243. [DOI] [PubMed] [Google Scholar]
- 35. Rumberger A, Merwin IA, Thies JE (2007) Microbial community development in the rhizosphere of apple trees at a replant disease site. Soil Biol Biochem 39: 1645–1654. [Google Scholar]
- 36. Otto G, Winkler H (1977) Investigations about cause of specific replant disease of fruit trees. VI. Proof of actinomycetes in feeder roots of apple seedlings in soils with different degrees of soil sickness. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg 132: 593–606. [PubMed] [Google Scholar]
- 37. Szabo K, Winkler H, Petzold H, Marwitz R (1998) Evidence for the pathogenicity of actinomycetes in rootlets of apple seedlings from soils conducive to specific apple replant disease. Acta hortic 477: 53–65. [Google Scholar]
- 38. Leyns F, De Cleene M, Swings JG, De Ley J (1984) The host range of the genus Xanthomonas . Bot Rev 50: 308–356. [Google Scholar]
- 39. Bent E, Loffredo A, Yang J, McKenry MV, Becker JO, et al. (2009) Investigations into peach seedling stunting caused by a replant soil. FEMS Microbiol Ecol 68: 192–200. [DOI] [PubMed] [Google Scholar]
- 40. Cantrell HF, Dowler WM (1971) Effects of temperature and pH on growth and composition of Pythium irregulare and Pythium vexans . Mycologia 63: 31–37. [PubMed] [Google Scholar]
- 41. Gardner DE, Hendrix FF (1973) Carbon dioxide and oxygen concentrations in relation to survival and saprophytic growth of Pythium irregulare and Pythium vexans in soil. Can J Bot 51: 1593–1598. [Google Scholar]
- 42. Biesbroc JA, Hendrix FF (1970) Influence of soil water and temperature on root necrosis of peach caused by Pythium spp. Phytopathology 60: 880–882. [Google Scholar]
- 43. Huet JC, Le Caer JP, Nespoulous C, Pernollet JC (1995) The relationships between the toxicity and the primary and secondary structures of elicitin-like protein elicitors secreted by the phytopathogenic fungus Pythium vexans . Mol Plant Microbe Interact 8: 302–310. [DOI] [PubMed] [Google Scholar]
- 44. DeVay JE, Lukezic FL, English WH, Moller WJ, Parkinson BW (1965) Controlling: Ceratocystis canker of stone fruit trees. California Agriculture 19: 2–4. [Google Scholar]
- 45. Marin M, Castro B, Gaitan A, Preisig O, Wingfield BD, et al. (2003) Relationships of Ceratocystis fimbriata isolates from Colombian coffee-growing regions based on molecular data and pathogenicity. J Phytopathol 151: 395–405. [Google Scholar]
- 46. Roux J, Wingfield MJ, Bouillet JP, Wingfield BD, Alfenas AC (2000) A serious new wilt disease of Eucalyptus caused by Ceratocystis fimbriata in Central Africa. Forest Pathology 30: 175–184. [Google Scholar]
- 47. Van Wyk M, Wingfield BD, Marin M, Wingfield MJ (2010) New Ceratocystis species infecting coffee, cacao, citrus and native trees in Colombia. Fungal Diversity 40: 103–117. [Google Scholar]
- 48. Lewthwaite SL, Wright PJ, Triggs CM (2011) Sweetpotato cultivar susceptibility to infection by Ceratocystis imbriata . New Zealand Plant Protection 64: 1–6. [Google Scholar]
- 49. Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004) Trichoderma species - opportunistic, avirulent plant symbionts. Nat Rev Micro 2: 43–56. [DOI] [PubMed] [Google Scholar]
- 50. Vinale F, Sivasithamparam K, Ghisalberti EL, Marra R, Woo SL, et al. (2008) Trichoderma–plant–pathogen interactions. Soil Biol Biochem 40: 1–10. [Google Scholar]
- 51. Howell CR (2006) Understanding the mechanisms employed by Trichoderma virens to effect biological control of cotton diseases. Phytopathology 96: 178–180. [DOI] [PubMed] [Google Scholar]
- 52. Yedida I, Shorest M, Karem Z, Benhamou N, Kapulnik Y, et al. (2003) Concomitant induction of systemic resistance to Pseudomonas syringae pv. lacrymans in cucumber by Trichoderma asperellum (T-203) and accumulation of phytoalexins. Appl Environ Microbiol 69: 7343–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yedidia I, Benhamou N, Chet I (1999) Induction of defense response in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum . Appl Environ Microbiol 653: 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tondje PR, Roberts DP, Bon MC, Widmer T, Samuels GJ, et al. (2007) Isolation and identification of mycoparasitic isolates of Trichoderma asperellum with potential for suppression of black pod disease of cacao in Cameroon. Biol Control 43: 202–212. [Google Scholar]
- 55. Cotxarrera L, Trillas-Gay MI, Steinberg C, Alabouvette C (2002) Use of sewage sludge compost and Trichoderma asperellum isolates to suppress Fusarium wilt of tomato. Soil Biol Biochem 34: 467–476. [Google Scholar]
- 56. Trillas MI, Casanova E, Cotxarrera L, Ordovás J, Borrero C, et al. (2006) Composts from agricultural waste and the Trichoderma asperellum strain T-34 suppress Rhizoctonia solani in cucumber seedlings. Biol Control 39: 32–38. [Google Scholar]
- 57. Sharon E, Chet I, Viterbo A, Bar-Eyal M, Nagan H, et al. (2007) Parasitism of Trichoderma on Meloidogyne javanica and role of the gelatinous matrix. Eur J Plant Pathol 118: 247–258. [Google Scholar]
- 58. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, et al. (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Nat Acad Sci 108: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Meth 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Borneman J, Hartin RJ (2000) PCR primers that amplify fungal rRNA genes from environmental samples. Appl Environ Microbiol 66: 4356–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol 2: 113–118. [DOI] [PubMed] [Google Scholar]
- 62.White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Sninsky JJ, White TJ, editors. PCR Protocols - A Guide to Methods and Applications. San Diego CA: Academic Press. pp. 315–32. [Google Scholar]
- 63. Fu Q, Ruegger P, Bent E, Chrobak M, Borneman J (2008) PRISE (PRImer SElector): Software for designing sequence-selective PCR primers. J Microbiol Methods 72: 263–267. [DOI] [PubMed] [Google Scholar]
- 64. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reverse PCR primers used in the Illumina-based high throughput sequence analysis of bacterial 16S rRNA genes. Each reverse PCR primer is comprised of the 4 adjoining segments in each row shown above.
(DOCX)
Forward PCR primer used in the Illumina-based high throughput sequence analysis of bacterial 16S rRNA genes. The forward PCR primer is comprised of the 3 adjoining segments shown above.
(DOCX)
Sequencing primers used in the Illumina-based high throughput sequence analysis of bacterial 16S rRNA genes.
(DOCX)