Abstract
Genome packaging is an essential step in virus replication and a potential drug target. Single-stranded RNA viruses have been thought to encapsidate their genomes by gradual co-assembly with capsid subunits. In contrast, using a single molecule fluorescence assay to monitor RNA conformation and virus assembly in real time, with two viruses from differing structural families, we have discovered that packaging is a two-stage process. Initially, the genomic RNAs undergo rapid and dramatic (approximately 20–30%) collapse of their solution conformations upon addition of cognate coat proteins. The collapse occurs with a substoichiometric ratio of coat protein subunits and is followed by a gradual increase in particle size, consistent with the recruitment of additional subunits to complete a growing capsid. Equivalently sized nonviral RNAs, including high copy potential in vivo competitor mRNAs, do not collapse. They do support particle assembly, however, but yield many aberrant structures in contrast to viral RNAs that make only capsids of the correct size. The collapse is specific to viral RNA fragments, implying that it depends on a series of specific RNA–protein interactions. For bacteriophage MS2, we have shown that collapse is driven by subsequent protein–protein interactions, consistent with the RNA–protein contacts occurring in defined spatial locations. Conformational collapse appears to be a distinct feature of viral RNA that has evolved to facilitate assembly. Aspects of this process mimic those seen in ribosome assembly.
Keywords: fluorescence correlation spectroscopy, RNA folding, RNA condensation, kinetics, hydrodynamic radius
Positive-sense, single-stranded (ss)RNA viruses are ubiquitous pathogens in all kingdoms of life (1), causing significant human disease and major financial losses (2). Therapeutic strategies are currently limited and the ideal of vaccination will only ever be practical in a minority of the human and animal viruses. In addition, recent work has highlighted the potential problems that might arise by misincorporation of nonviral RNAs in virus-like particles (VLPs) (3) that are being considered as synthetic vaccines against both pathogens and oncogenic viruses (4). A more thorough understanding of the molecular events central to viral lifecycles is therefore needed. Such studies may reveal novel strategies for therapeutic intervention.
Genomic RNAs play essential roles during the viral lifecycle, adopting different metastable conformations in order to be replicated, translated and packaged into the virion (5, 6). The latter process, the final step in the production of infectious viral progeny, is the topic of the experiments described here. For isometric, nonenveloped virions, protein capsids self-assemble around their genomes, resulting in their confinement at relatively high packing densities (7). It has been proposed that this is a spontaneous process because RNA molecules are branched polymers and there is no barrier to compaction due to large persistence lengths, as seen in dsDNA phages. Electrostatic neutralisation of the nucleic acid charge has been seen as the principal driving force for this confinement step (8–12), consistent with the packaging of noncognate RNAs or protein-alone assembly in vitro (13).
Here we have investigated the molecular mechanism controlling this vital step in the viral lifecycle by taking advantage of single molecule fluorescence correlation spectroscopy (smFCS) to selectively monitor coat protein (CP) or viral RNA components in in vitro reassembly reactions. Such assays can be carried out at low concentrations (≤ 1 μM), allowing observation of mechanistic features that are not dominated by high CP concentrations. We have applied the smFCS assays to two viral model systems (Fig. 1), the RNA bacteriophage MS2 of the Leviviridae family and Satellite Tobacco Necrosis virus (STNV), which is representative of a large number of plant viruses. These were chosen because our previous studies (14–22) have revealed potential roles played by the RNA in coat protein quasi-conformer switching, required for correct assembly of the MS2 T = 3 capsid and the presence of multiple putative preferred coat protein binding sites (packaging signals) within the STNV genome, which gets packaged into a T = 1 capsid in which all protein conformers are identical. The two viral coat protein architectures are distinct, the STNV CP having a positively charged N-terminal extension to its globular body which is essential for assembly (17) and partially disordered in X-ray structures, whilst the MS2 CP lacks these features. These differences allow us to identify conserved and distinct mechanistic processes.
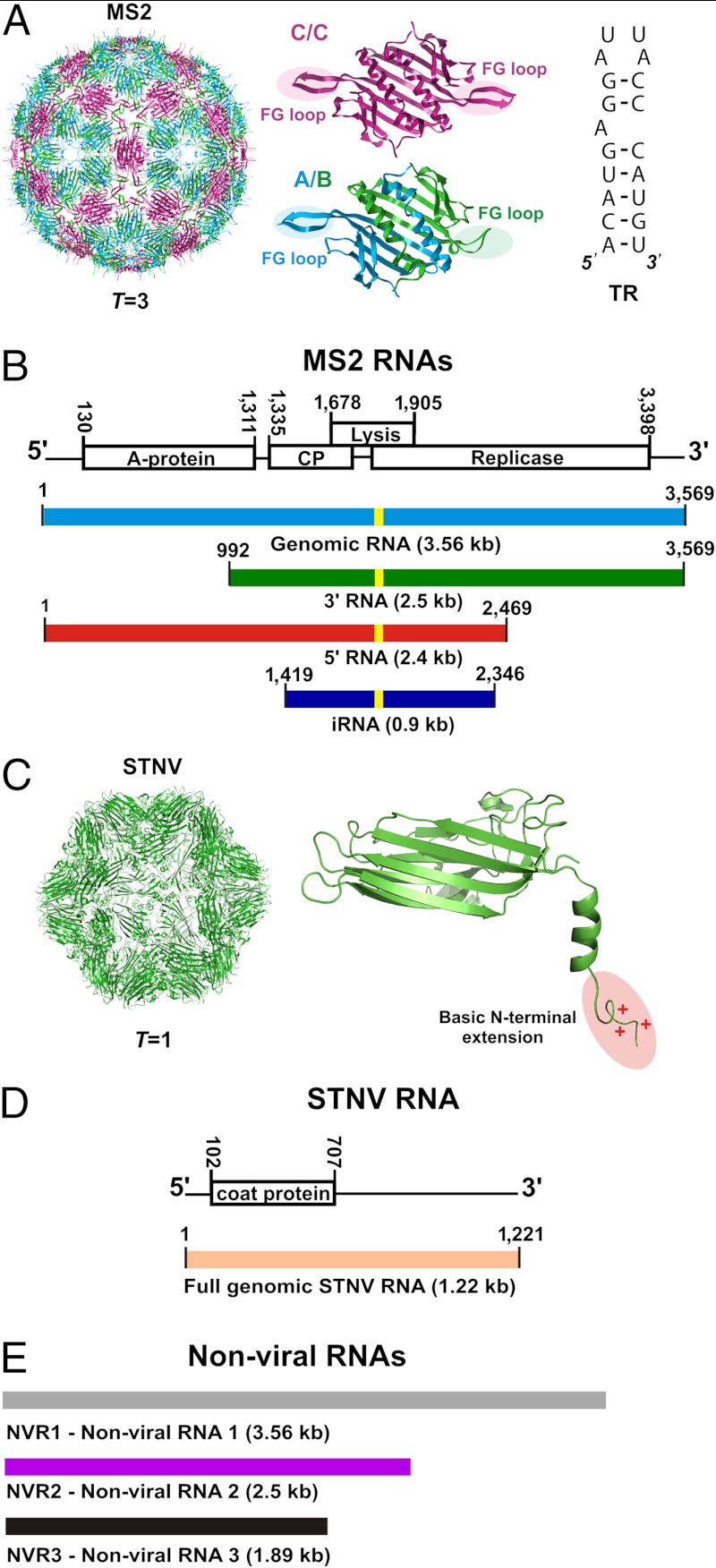
Fig. 1.
The bacteriophage MS2 and Satellite Tobacco Necrosis Virus (STNV) systems. (A) Structure of the T = 3 MS2 bacteriophage (PDB ID 2ms2). Structures of the A/B and C/C quasiequivalent coat protein dimers (PDB ID 1zdh) that differ in the conformations of their FG-loops (highlighted). Sequence and secondary structure of the high-affinity 19 nt TR stem-loop, which is known to cause a conformational switch in coat protein from the C/C to the A/B dimer. Capsid assembly is believed to be initiated at this site on genomic RNA. (B) Genetic map of the MS2 genome (GenBank Accession NC001417) and the RNAs used in assembly studies (color-coded here and throughout). The locations of TR are indicated by the yellow stripes. (C) Structure of the T = 1 STNV capsid. Structure of the STNV coat protein monomer (PDB ID 3RQV) shown with its positively charged N-terminal extension (highlighted). (D) Genetic map of the STNV genome (strain C, GenBank Accession AJ000898) and the RNA used in assembly studies. (E) Nonviral RNA controls used in the study (color-coded here and throughout).
SmFCS reveals that there is no simple correlation between RNA length and hydrodynamic radius (Rh) and this property cannot be used to discriminate between viral and nonviral RNAs for these systems. The viral RNAs are larger in the absence of their CPs than the capsids into which they must eventually fit. Remarkably instead of a steady condensation of the RNA, which would be expected by a charge neutralisation mechanism, addition of CPs to their cognate RNAs results in a rapid collapse (< 1 min) in the solution conformation. Collapse depends on protein–protein interactions, and does not occur on nonviral RNA controls or with the noncognate viral RNA, showing that it depends on specific RNA-CP interactions mediated by the sequence and structure of each genome. The collapsed state is smaller than the capsid and appears to consist of complexes with substoichiometric amounts of coat proteins with respect to capsids, but with roughly the correct shell curvature. The full complement of CPs is recruited in a second slower stage of assembly. Nonviral RNAs support assembly inefficiently and with much lower fidelity.
Results and Discussion
During assembly the sizes and hydrodynamic properties of viral RNAs and CP aggregates change as a result of their co-polymerisation. SmFCS is particularly suitable for determining the hydrodynamic sizes of species in solution (see SI Methods for a brief introduction and further references). By fluorescent labeling of the CP component one can selectively follow capsid shell polymerisation, whilst labeling of the viral RNA monitors any conformational changes, such as compaction, that occur (23). As described in Materials and Methods and Fig. S1, we labeled MS2 coat proteins and transcript RNAs with Alexa-fluor-488. These molecules retained the ability to self-assemble into T = 3 capsids, as described in SI Materials and Methods and in Fig. S2. In experiments using TR oligonucleotides we were able to show that the smFCS assays reproduced the features of the MS2 assembly reaction seen previously by mass spectrometry (20). The derived hydrodynamic radii of the species involved matched very closely values for the same species obtained by other techniques (Table S1).
Investigating the Solution Conformations of RNAs Via smFCS.
Unlike ensemble methods smFCS data can be obtained in very dilute solutions, effectively eliminating artefacts due to aggregation, including nonspecific interactions between large RNA molecules. Fig. 2A shows the average Rh values for the dye-labeled genomic and subgenomic RNAs from MS2 and STNV, together with three nonviral RNA controls (Fig. 1 and Figs. S1 and S2) as a function of their nucleotide lengths. The nonviral control RNAs (NVR1-3) were chosen because they had similar lengths to some of the viral RNAs being used. NVR1 is a fragment of the E.coli RNA polymerase B subunit (rpoB), a high copy number transcript present in the same cells in which MS2 replicates. NVR2 is a transcript from pGEM-3Zf(+) encompassing most of the plasmid. NVR3 is a transcript encompassing a eukaryotic ORF.
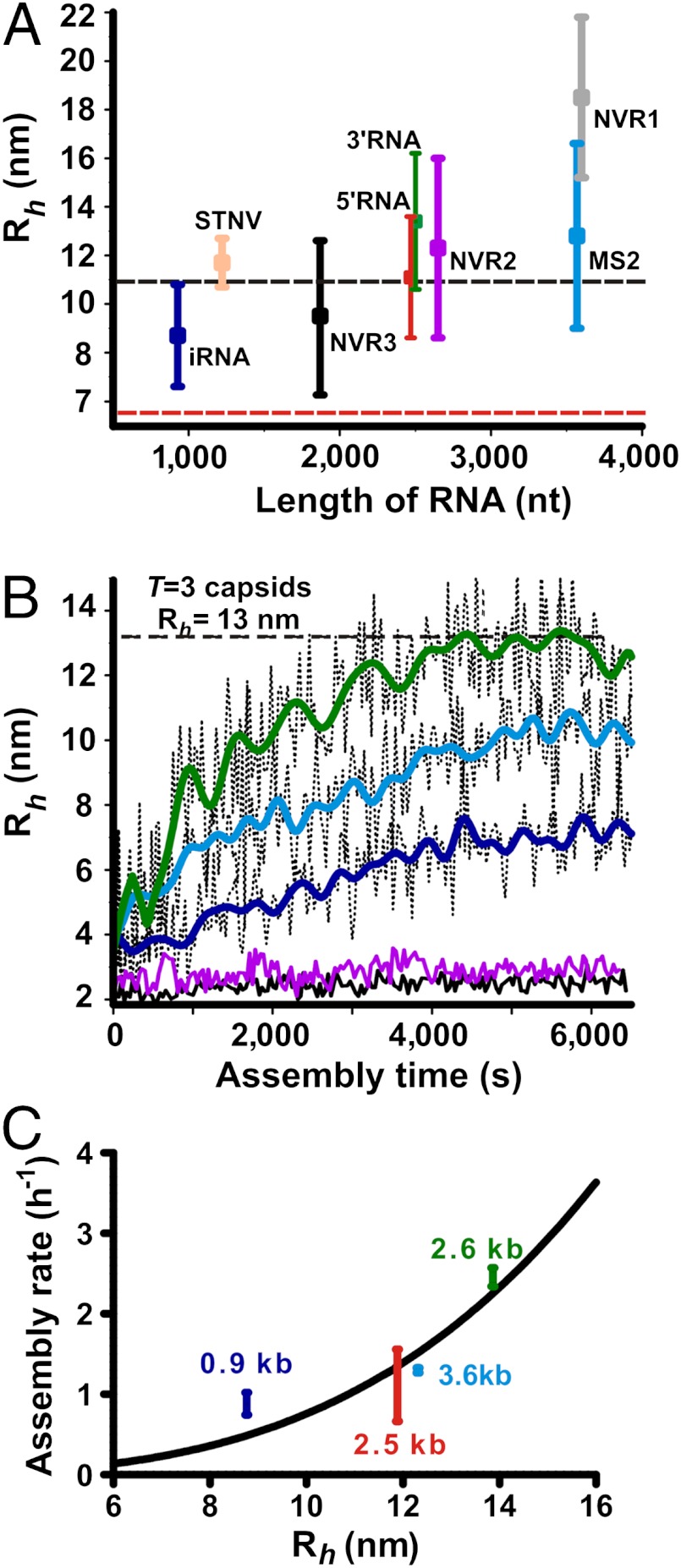
Fig. 2.
(A) Size distribution of the test RNAs (color-coded as on Fig. 1) measured by FCS as a function of their lengths in nucleotides. Vertical error bars correspond to the full width at half maximum of Rh distributions for each of the RNAs. The black dashed line represents the inner capsid radius of MS2, whilst the red one corresponds to the inner radius of STNV capsid. (B) Assembly assays with fluorescently labeled MS2 CP2. Capsid assembly kinetics with genomic (blue), and subgenomic 3’RNA (green) and iRNA (dark blue). The kinetics with nonviral RNA controls are shown in magenta (NVR2) and black (NVR3). Apparent hydrodynamic radii, Rh, are plotted as a function of time from assembly initiation (assembly time). FFT smoothed data are shown as solid lines overlaid on the original traces (thin dotted lines). The gray dashed line represents the hydrodynamic radius of the MS2 virion. (C) Apparent assembly rates obtained from a single exponential approximation of Rh(t) for different MS2 RNAs as a function of their hydrodynamic size. Vertical error bars (rate) represent the standard error of repeated experiments.
There is a general increase of Rh with the length of the RNAs, but they are not directly proportional. For example gRNA and the 5’RNA have similar average Rh values (approximately 12 nm) despite the fact that the latter is approximately 30% shorter. Similarly, the 3’RNA fragment has an even larger Rh (approximately 14 nm), implying that there are long range RNA-RNA contacts that are preserved in both the gRNA and 5’RNAs that stabilize their more compact conformations (24, 25). Contrary to previous predictions (8) the viral RNAs are not significantly more compact than their nonviral counterparts (c.f. 3’RNA and NVR2, STNV and NVR3).
SmFCS is sensitive to variations in Rh due to different conformations and these can be seen in time dependent Rh distributions (Fig. S3 and Table S2). The distribution width is a measure of the conformational heterogeneity for each RNA and is shown as error bars in Fig. 2A. It is clear that even the relatively compact and folded MS2 genomic RNA is an ensemble of molecules with a wide range of conformations.(Rh approximately 10–16 nm). Even for conformers that are relatively compact the sizes of the majority of viral RNAs are larger than the volumes of their cognate capsids. Hence, the genomic RNAs need to be condensed during packaging.
Assembly Kinetics are Dependent on the Sizes and Sequences of the RNA Being Encapsidated.
We then examined the assembly of labelled MS2 CPs in the presence of genomic and various su-genomic RNAs (Fig. 1 and 2B), all of which encompass the TR site and presumably initiate assembly from that point. These fragments have been shown previously to support assembly of T = 3 capsids (23) and EMs of the smFCS reaction end-points confirmed this (Fig. S2). Assembly time courses were measured by acquiring time-dependent correlation functions, which were then reduced to plots of apparent Rh vs. time (Fig. 2B). As expected, there is time-dependent variation in the apparent Rh values (shown by the black dotted lines) but the trends of each data set are obvious after data smoothing (solid lines, see Materials and Methods). The apparent rates of assembly vary between RNA fragments but are not directly related to RNA fragment length. Instead, the rates increase roughly with the (Rh)3 values of each RNA; i.e., their volumes (Fig. 2C), suggesting that the rate is determined by complex factors such as compactness and shape. In contrast, coat protein assembles very slowly if at all with two of the nonviral RNAs, confirming that RNA sequence/structure determines assembly efficiency.
RNA Conformational Changes During Encapsidation.
We then examined the process of RNA compaction using labelled RNAs. Fig. 3 shows time-resolved Rh(t) values for RNAs in assembly buffer. Fluctuations in the initial Rh(t) values correspond to the conformational variations discussed above for protein-free RNA. At the times indicated by arrows in Fig. 3A–C, capsid assembly was initiated on each RNA by the addition of a 200-fold molar excess of MS2 CP2; i.e., enough CP to complete capsid formation around each RNA. Fig. 3A compares the data for gRNA (approximately 3.5 kb) and the NVR1 control (approximately 3.5 kb). Both protein-free RNAs have dramatically different Rh values although they are roughly the same length. Although the viral RNA is the smaller, it is larger than the inner capsid radius (approximately 10 nm). Strikingly, these RNAs behave very differently upon addition of coat protein. There is an immediate collapse by approximately 30% of the Rh of the viral RNA, whereas very little happens to NVR1. The smoothing of the Rh(t) traces disguises the kinetics of the collapse with the viral RNA, which happens within the experimental dead time (< 1 min) and is presumably a consequence of the rapid formation of RNA-CP interactions.
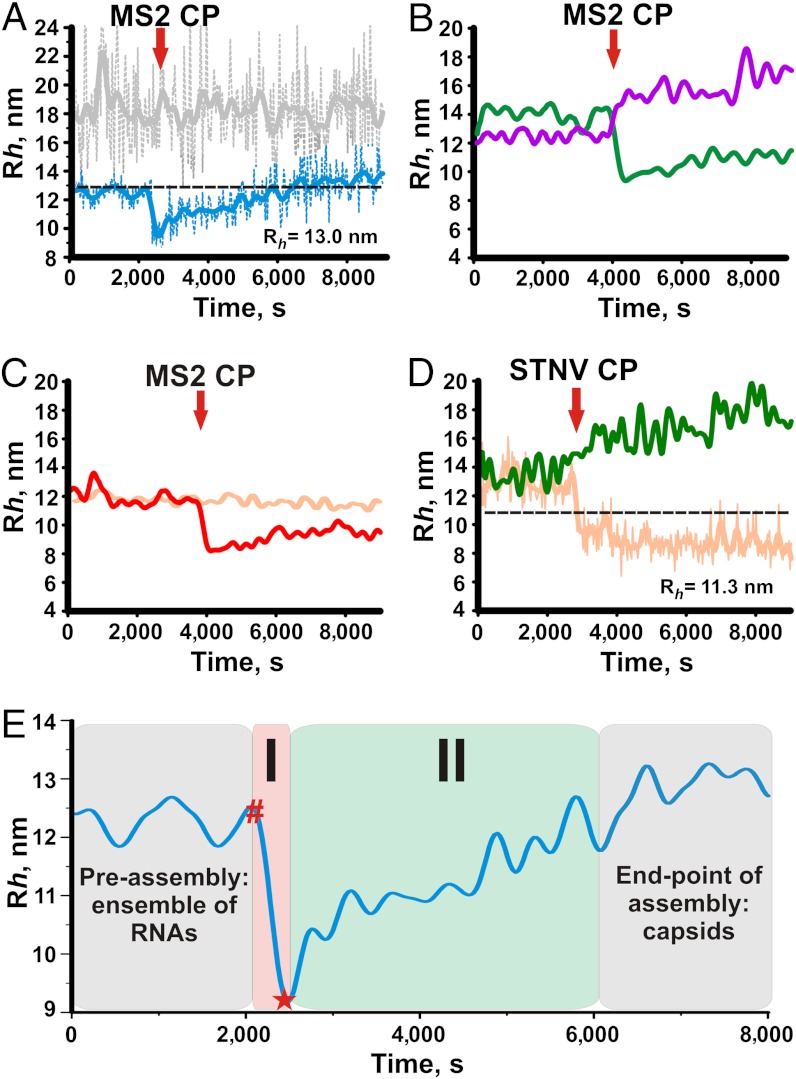
Fig. 3.
Assembly with labelled long RNA was followed by Rh(t) and performed at 1∶200 molar ratio RNA∶CP2 for MS2 or at 1∶60 molar ratio for STNV under the conditions described in Materials and Methods . Data collection was started for RNA alone and the coat protein (CP) was mixed with RNA at the time points indicated by the red arrows. (A) gRNA (light blue) and NVR1 (gray). The black dashed line represents the Rh of the MS2 virion. (B) 3’RNA (green) and NVR2 (magenta). (C) 5’RNA (red) and STNV RNA (orange). (D) 3’RNA (green) & STNV RNA (orange) upon addition of STNV CP. The black dashed line represents the Rh of the STNV virion. (E) Changes in Rh for viral RNAs upon addition of cognate coat protein. Red hash (#) indicates the precollapse state of the RNA-protein complex, which is not detectable by FCS due to the fast kinetics of the CP binding reaction. The initial (ensemble of RNA molecules) and end-points of the assembly reaction (capsids) are highlighted in gray. The collapse stage (I) is highlighted in pink and the collapsed RNA intermediate is marked with a red star (*). The capsid assembly stage (II) is highlighted in green.
Following the collapse, gRNA increase its apparent hydrodynamic radius reaching a plateau close to the size expected for T = 3 capsids (13 nm, Fig. 3A). In contrast, the nonviral control, NVR1, appears largely unaffected by the presence of coat protein. Both RNAs lead to the formation of capsids or capsid-like aggregates, presumably with different yields, as judged by TEM, with the nonviral RNA showing many more aggregates than gRNA. The Rh distributions of the assembly reactions show that gRNA results in a tight distribution similar to bona fide samples of capsid (Fig. S3). In contrast, the NVR1 distribution is slightly larger than that for the free RNA with a peak much larger than the diameter of the T = 3 MS2 capsid. The data support the ideas that faithful assembly requires compaction of the protein-free RNA and that this is facilitated by the viral RNA sequence compared to a similar length control.
In order to assess the roles of viral RNA sequences in this collapse and the faithful assembly process we then examined two subgenomic RNA fragments in similar assays. The MS2 3’RNA and the control NVR2 are similar lengths but the MS2 RNA has a larger initial Rh. Strikingly it also shows a CP-induced collapse whilst the control does not collapse (Fig. 3B). In this case both RNAs show a gradual Rh(t) increase after addition of the coat protein consistent with further assembly of RNA-protein complexes, which was confirmed by TEMs (Fig. S3). Once again the viral RNA sequence leads to faithful production of T = 3 shells whilst the nonviral RNA control yields large fused-shell structures or aggregates (Fig. S3). This result confirms that it is the viral RNA sequence rather than its size that governs the initial collapse and subsequent assembly. Similar behavior, for example, lack of initial collapse and aberrant assembly, occurs for the other nonviral RNA NVR3 (Fig. S3).
As a final control we compared two viral RNA fragments, the MS2 5’RNA and the STNV genomic RNA, that both must undergo compaction during assembly of their cognate capsids (see Fig. 2). The data (Fig. 3C) show that the cognate interaction between MS2 coat protein and its RNA causes the collapse to occur whilst the STNV RNA behaves similarly to the nonviral RNA controls (Fig. S3). This confirms that the initial collapse requires sequence-specific RNA recognition by the coat protein. Above a certain threshold CP2 concentration (approximately 50 nM) the magnitude of this collapse does not depend on protein concentration (Fig. S4). This result is not compatible with a simple charge neutralization in which increasing protein concentration would produce a gradual compaction of the RNA. Instead it suggests a cooperative binding of a subset of CP2 to the RNA. Using an MS2 coat protein mutant (W82R) that retains the ability to bind TR sequences but not to assemble beyond that point (26), we were able to show that the collapse depends on protein–protein as well as protein–RNA interactions (Fig. S5).
Thus, it appears that specific recognition of multiple sites on MS2 viral RNAs by a subset of the coat protein subunits required to form the capsid triggers a collapse, which facilitates assembly. In order to assess whether this assembly mechanism is specific to MS2 and related bacteriophages, or also occurs more widely, we carried out similar assays using STNV CP and STNV genomic RNA (Fig. 3D). The comparison in this case was between STNV RNA and MS2 3’RNA which is of a similar Rh, although it is much longer. In this case, a rapid collapse occurs when STNV coat is mixed with its cognate RNA whilst the MS2 RNA behaves similarly to the previous nonviral controls. TEMs and Rh distributions (Fig. S3) show that the cognate protein–RNA interaction yields mostly T = 1 capsids, whereas reassembly of MS2 RNA by STNV protein yields mostly fused shells and aggregates. These very similar outcomes for two different viruses, especially since they represent very different coat protein folds, MS2 lacking the positively charged N-terminal region of STNV CP, suggests that viral RNA collapse is likely to be a widespread phenomenon and is a prerequisite for correct assembly.
A Two-Stage Assembly Mechanism.
A typical path of a viral RNA through packaging is epitomised by MS2 gRNA (Fig. 3E). In the preassembly state, the protein-free RNA exists in an ensemble of conformations. These are relatively compact and likely possess folded structures that favor assembly, since heat denaturation followed by rapid quenching of this RNA on ice produces an RNA with decreased assembly rate and yield (Fig. S6). Rapid coat protein binding to RNA initiates the first stage of assembly (region I in Fig. 3E), namely a cooperative RNA collapse, which results in a compact initiation complex. This stage is dependent both on specific recognition of viral RNA sequences and protein–protein interactions. In the second stage of assembly (II in Fig. 3E), additional CP subunits are recruited to the growing shell in a process that is CP concentration dependent (Fig. S4). The similarity in behavior of the MS2 and STNV genomic RNAs in the presence of their cognate coat proteins suggests that this two-stage mechanism is likely adopted by many ssRNA viruses. Nonspecific RNAs, on the other hand, may be encapsidated using a different mechanism which bypasses stage I via formation of heterologous nucleation events from multiple sites, often leading to fused or partial capsids.
Biological Implications.
The data described above, together with our extensive previous studies on the assembly of MS2 (20–23, 27), allow us to propose a molecular model for capsid assembly (Fig. 4) that amounts to a paradigm shift with respect to ssRNA viruses. Current views of assembly are dominated by protein-centric models based on the observations that many viral coat proteins can assemble in the absence of RNA, around nonviral RNA or polyanions (28, 29) and even nanoparticles (30). This has led to the conclusion that viral proteins are the key component in the assembly process (31). In vivo, however, most viruses package their own genomes highly efficiently into defined capsids even in the presence of competitor cellular RNA. There must therefore be some mechanism promoting both specificity and assembly fidelity. We have argued that coat proteins and RNA act as mutual chaperones altering their conformations due to complex formation in a context specific way as the capsid shell grows from an initiation complex (23).
Fig. 4.
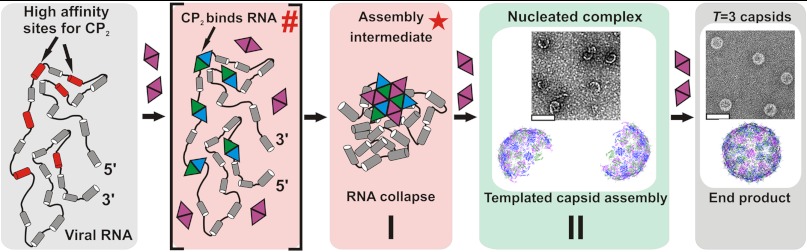
Proposed two-stage assembly model for MS2. A pre-assembly ensemble of protein-free genomic RNAs (gray box) presents a number of dispersed high affinity sites for CP2 (red, not to scale), having an Rh approximately 20–30% larger than the inner space of the T = 3 capsid. CP2 binds to these putative packaging signals creating an initiation complex denoted as a (hash) as in Fig. 3E. Protein–protein interactions within this complex trigger RNA collapse into the assembly intermediate with an Rh small enough to fit within the confines of the capsid shell, denoted by the (red star). This completes stage I of the assembly process (pink box). During stage II the stable collapsed complex recruits additional CP2 to populate shell-like species of the correct curvature (green box, see inset for EM) and allows efficient completion of capsids (end-point of assembly, gray box).
The smFCS data are entirely consistent with this view and reveal several novel features to the assembly mechanism. In the first stage of assembly, these RNAs undergo a rapid collapse in their solution conformations. The threshold concentration at which this occurs for MS2 suggests that this is driven by the formation of multiple CP-RNA contacts, presumably at TR and TR-like degenerate sites (shown in red in Fig. 4). The Kd value of the highest affinity MS2 CP binding site, TR, is low nanomolar and secondary sites would be expected to be lower affinity (32), consistent with the effect being mediated by RNA-protein affinities in this concentration range (i.e., 50 nM, see Fig. S4). This idea is further supported by the lack of collapse for the smallest iRNA (Fig. S6B and C), which encompasses the high affinity TR site but may lack some secondary binding sites. Indeed, a set of dispersed, TR-like binding sites located throughout the genome has recently been identified for MS2 and a related phage GA [Dykeman, Stockley and Twarock, unpublished]. That bioinformatics analysis also demonstrated that such a disposition of binding sites is unlikely to occur in cellular RNAs. Similarly, degenerate CP binding sites have been identified in STNV (17) and other ssRNA viruses (33). We propose that such coat protein binding sites distinguish viral RNAs from their cellular competitors by facilitating the initial collapse, which also promotes packaging at coat protein concentrations significantly lower than those necessary for formation of empty shells in vitro. Assembly at low protein concentrations would also minimize nonspecific packaging of cellular RNAs in infected cells. Assembly has an intrinsic entropic cost as the ensemble of solution RNA conformations and multiple CP subunits become ordered in the virion. Clearly for the two test viral RNAs used here, most of this cost is compensated by the enthalpic gains from the initial CP-RNA contacts and further augmented by protein–protein interactions in the initiation complex. Hence, most of the energy cost may be recouped during the formation of the specific initiation complex and consequently stage II may be more favorable for packaging of specific rather than for nonspecific RNAs. This is clearly not the case for noncognate RNAs where coat proteins would bind at more or less random places along each RNA, resulting in heterologous nucleation and complexes of low stability. The constant break-down and reformation of weak nucleation complexes would slow the rate of capsid formation (34) for such noncognate RNAs, which is indeed the case (Fig. 2B). This mechanism also explains why viral RNA gets efficiently packaged even in the presence of a large number of cellular competitors. The viral RNA sequence and structure, fine tuned by evolution, decreases the entropic penalty for the formation of the collapsed/compact initiation complex, which is stable and competes efficiently for additional coat proteins from the less stable heterologously initiated cellular competitors.
The collapsed RNAs have sizes (Rh ∼ 9–10 or 8 nm, for MS2 and STNV, respectively) similar to that of their cognate capsid interiors [Rinner ∼ 11 and 6 nm (15, 18, 35)] and may act as a scaffold during stage II, facilitating formation of complete capsids with the correct curvature. Negative stain EMs of the early products of assembly with MS2, equivalent to the start of stage II, show incomplete shells of approximately the correct curvature, consistent with growth from an initiation complex (Fig. 4). At later times all the viral RNAs result in high yields of T = 3 capsids with few misassembled species. Because this is also true of subgenomic RNA fragments, it implies that for the collapsed complex CP-CP interactions are sufficient to complete the capsid shell.
This behavior is remarkably similar to results emerging from studies of RNA folding and particularly ribosome assembly (36). Folding of the Tetrahymena ribozyme shows that it can undergo very rapid collapse into its native conformation (in approximately 1 s). However, the bulk of the RNA collapses into nonnative folds that then rearrange over a much longer time period (min). In contrast, ribosomal 16S rRNA folding is both rapid and mediated by interactions with the 30S proteins. The result is a highly ordered and accurate assembly reaction brought about by induced conformational changes within the ribonucleoprotein complex, although in that case there are few direct protein–protein contacts. It appears that with significantly larger RNAs to fold into the restricted volumes of their capsids, RNA viruses exploit similar basic mechanisms but with the additional co-operativity of the CP lattice that forms around the RNA. The smFCS assays described here will be an important tool in the armoury of techniques needed to investigate the behaviour of both viral and other large RNAs (37).
Materials and Methods
MS2 virus-like particles (VLPs) were produced and purified as described previously (38). Purified VLPs were covalently labeled with Alexa Fluor 488-SDP ester (Invitrogen) via surface lysines (39). RNA-free CP, having approximately 0.3 mole dye/mole CP2, was isolated as previously described (40) and used in assembly assays.
RNA 19-mer, TR, encompassing the high affinity packaging signal (Fig. 1C and ref. 20, 27) and carrying a 3’-amino group, was produced by solid-phase synthesis and labeled using Alexa Fluor 488-SDP ester. The genomic and sub-genomic RNAs (Fig. 1D) were produced by transcription of cDNA clones (23). Genomic RNA extracted from the phage using RNeasy mini kit (QIAGEN) and produced similar results in assembly assays to the corresponding transcript; hence transcripts were used throughout unless otherwise indicated. Nonviral control RNAs were transcribed from linearized templates as described in Supporting Information. All RNAs were purified using RNeasy mini kit (QIAGEN). 3’-end fluorescent labelling of RNA transcripts was achieved via ligation of a 5’-phosphorylated-oligo-A(25-mer)-3’-AF488 oligonucleotide (subgenomic and nonviral RNAs) or incorporation of a reactive amine group at the 3’-end using poly-A polymerase, followed by chemical coupling of the amine to AF488 SDP-ester (MS2 full genomic RNA, STNV). STNV genome was also trace labelled by transcription in the presence of amino-allyl UTP followed by chemical coupling of the amine to AF488 SDP-ester. Denaturing formaldehyde agarose gels were used to assess RNA integrity before and after experiments.
Assembly conditions (23) were optimised for the single molecule assays. RNA (0.5 nM) was mixed with AF488-labeled CP2 (final concentrations 25 nM–100 nM) to achieve the desired CP2∶RNA ratio. Similarly, labeled RNAs (long RNAs at 1 nM, TR at 10 nM) were mixed with unlabeled CP2 (50 nM-5 μM). All assembly reactions were performed in assembly buffer [50 mM ammonium acetate, 1 mM EDTA, 1 mM DTT, 0.05% (v/v) Tween20, pH 7.5] at 21 °C. Manual mixing caused approximately 1 min delay at the start of time-resolved FCS data collection. FCS measurements were made using a custom-built FCS setup (41) with 10 s data accumulation per each autocorrelation function (CF). Individual CFs were decomposed into triplet state relaxation and quenching (if necessary) and diffusion (characterized by diffusion time, tD) components and the latter was converted into an apparent hydrodynamic radius Rh (42). Electron microscopy grids were prepared at the end of FCS data collection to monitor formation of assembly products by imaging in negative stain. Further details are given in SI Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Professor Dave Rowlands and Professor Reidun Twarock and her group (University of York) for many helpful comments on these results, Dr. Robert Coutts for providing the STNV-C genome and Amy Barker for synthesis of RNA oligonucleotides. We thank Dr. Tomas Fessl (University of Dundee) for help with processing of FCS data. AB thanks the Wellcome Trust for support of his postgraduate studentship [089310/Z/09/Z] and support of the facilities used [062164][090932/Z/09/Z]. Single molecule work within the Astbury Centre has been supported by The University of Leeds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204357109/-/DCSupplemental.
References
- 1.Schneemann A. The structural and functional role of RNA in icosahedral virus assembly. Annu Rev Microbiol. 2006;60:51–67. doi: 10.1146/annurev.micro.60.080805.142304. [DOI] [PubMed] [Google Scholar]
- 2.Lomonossoff GP. Pathogen-derived resistance to plant-viruses. Annu Rev Phytopathol. 1995;33:323–343. doi: 10.1146/annurev.py.33.090195.001543. [DOI] [PubMed] [Google Scholar]
- 3.Routh A, Domitrovic T, Johnson JE. Host RNAs, including transposons, are encapsidated by a eukaryotic single-stranded RNA virus. Proc Natl Acad Sci USA. 2012;109:1907–12. doi: 10.1073/pnas.1116168109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tumban E, Peabody J, Peabody DS, Chackerian B. A pan-HPV vaccine based on bacteriophage PP7 VLPs displaying broadly cross-neutralizing epitopes from the HPV minor capsid protein, L2. Plos One. 2011;6:e23310. doi: 10.1371/journal.pone.0023310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon AE, Gehrke L. RNA conformational changes in the life cycles of RNA viruses, viroids, and virus-associated RNAs. Biochim Biophys Acta. 2009;1789:571–583. doi: 10.1016/j.bbagrm.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venter PA, Krishna NK, Schneemann A. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J Virol. 2005;79:6239–6248. doi: 10.1128/JVI.79.10.6239-6248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Speir JA, Johnson JE. Nucleic acid packaging in viruses. Curr Opin Struc Biol. 2012;22:65–71. doi: 10.1016/j.sbi.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoffe AM, et al. Predicting the sizes of large RNA molecules. Proc Natl Acad Sci USA. 2008;105:16153–16158. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruinsma RF. Physics of RNA and viral assembly. Eur Phys J E. 2006;19:303–310. doi: 10.1140/epje/i2005-10071-1. [DOI] [PubMed] [Google Scholar]
- 10.Rudnick J, Bruinsma R. Icosahedral packing of RNA viral genomes. Phys Rev Lett. 2005;94:038101–038104. doi: 10.1103/PhysRevLett.94.038101. [DOI] [PubMed] [Google Scholar]
- 11.van der Schoot P, Bruinsma R. Electrostatics and the assembly of an RNA virus. Phys Rev E. 2005;71:061928–061939. doi: 10.1103/PhysRevE.71.061928. [DOI] [PubMed] [Google Scholar]
- 12.Belyi VA, Muthukumar M. Electrostatic origin of the genome packing in viruses. Proc Natl Acad Sci USA. 2006;103:17174–17178. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balint R, Cohen SS. The incorporation of radiolabeled polyamines and methionine into turnip yellow mosaic-virus in protoplasts from infected plants. Virology. 1985;144:181–193. doi: 10.1016/0042-6822(85)90316-2. [DOI] [PubMed] [Google Scholar]
- 14.Toropova K, Stockley PG, Ranson NA. Visualising a viral RNA genome poised for release from its receptor complex. J Mol Biol. 2011;408:408–419. doi: 10.1016/j.jmb.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 15.Lane SW, et al. Construction and crystal structure of recombinant STNV capsids. J Mol Biol. 2011;413:41–50. doi: 10.1016/j.jmb.2011.07.062. [DOI] [PubMed] [Google Scholar]
- 16.Dykeman EC, et al. Simple rules for efficient assembly predict the layout of a packaged viral RNA. J Mol Biol. 2011;408:399–407. doi: 10.1016/j.jmb.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 17.Bunka DHJ, et al. Degenerate RNA packaging signals in the genome of satellite tobacco necrosis virus: Implications for the assembly of a T = 1 capsid. J Mol Biol. 2011;413:51–65. doi: 10.1016/j.jmb.2011.07.063. [DOI] [PubMed] [Google Scholar]
- 18.Toropova K, et al. The three-dimensional structure of genomic RNA in bacteriophage MS2: Implications for assembly. J Mol Biol. 2008;375:824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 19.Rolfsson O, et al. RNA packing specificity and folding during assembly of the bacteriophage MS2. Comput Math Methods Med. 2008;9:339–349. doi: 10.1080/17486700802168445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stockley PG, et al. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T = 3 viral capsid. J Mol Biol. 2007;369:541–552. doi: 10.1016/j.jmb.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basnak G, et al. Viral genomic single-stranded RNA directs the pathway toward a T = 3 capsid. J Mol Biol. 2010;395:924–936. doi: 10.1016/j.jmb.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dykeman EC, Stockley PG, Twarock R. Dynamic allostery controls coat protein conformer switching during MS2 phage assembly. J Mol Biol. 2010;395:916–923. doi: 10.1016/j.jmb.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Rolfsson O, Toropova K, Ranson NA, Stockley PG. Mutually-induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid. J Mol Biol. 2010;401:309–322. doi: 10.1016/j.jmb.2010.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodish HF, Zinder ND. Mutants of the bacteriophage f2. 8. Control mechanisms for phage-specific syntheses. J Mol Biol. 1966;19:333–348. doi: 10.1016/s0022-2836(66)80008-6. [DOI] [PubMed] [Google Scholar]
- 25.Klovins J, Berzins V, Van Duin J. A long-range interaction in Q(beta) RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA. 1998;4:948–957. doi: 10.1017/s1355838298980177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peabody DS, Ely KR. Control of translational repression by protein–protein interactions. Nucleic Acids Res. 1992;20:1649–1655. doi: 10.1093/nar/20.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morton VL, et al. The impact of viral RNA on assembly pathway selection. J Mol Biol. 2010;401:298–308. doi: 10.1016/j.jmb.2010.05.059. [DOI] [PubMed] [Google Scholar]
- 28.Cadena-Nava RD, et al. Exploiting fluorescent polymers to probe the self-assembly of virus-like particles. J Phys Chem B. 2011;115:2386–2391. doi: 10.1021/jp1094118. [DOI] [PubMed] [Google Scholar]
- 29.Hu YF, et al. Packaging of a polymer by a viral capsid: The interplay between polymer length and capsid size. Biophys J. 2008;94:1428–1436. doi: 10.1529/biophysj.107.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C, et al. Nanoparticle-templated assembly of viral protein cages. Nano Lett. 2006;6:611–615. doi: 10.1021/nl0600878. [DOI] [PubMed] [Google Scholar]
- 31.Zlotnick A, Mukhopadhyay S. Virus assembly, allostery and antivirals. Trends Microbiol. 2011;19:14–23. doi: 10.1016/j.tim.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lago H, et al. Dissecting the key recognition features of the MS2 bacteriophage translational repression complex. Nucleic Acids Res. 1998;26:1337–1344. doi: 10.1093/nar/26.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim DY, et al. Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution. J Virol. 2011;85:8022–8036. doi: 10.1128/JVI.00644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prevelige PE, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chauvin C, Jacrot B, Witz J. Structure and molecular-weight of satellite tobacco necrosis virus—neutron small-angle scattering study. Virology. 1977;83:479–481. doi: 10.1016/0042-6822(77)90199-4. [DOI] [PubMed] [Google Scholar]
- 36.Woodson SA. RNA folding pathways and the self-assembly of ribosomes. Acc Chem Res. 2011;44:1312–1319. doi: 10.1021/ar2000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gopal A, Zhou ZH, Knobler CM, Gelbart WM. Visualizing large RNA molecules in solution. RNA. 2012;18:284–299. doi: 10.1261/rna.027557.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stonehouse NJ, Stockley PG. Effects of amino-acid substitution on the thermal-stability of Ms2 capsids lacking genomic RNA. Febs Lett. 1993;334:355–389. doi: 10.1016/0014-5793(93)80711-3. [DOI] [PubMed] [Google Scholar]
- 39.Hermanson GT, Hermanson GT. Bioconjugate techniques. San Diego: Academic Press; 1996. p. 785. [Google Scholar]
- 40.Sugiyama T, Nakada D. Control of translation of Ms2 Rna cistrons by Ms2 coat protein. Proc Natl Acad Sci USA. 1967;57:1744–1750. doi: 10.1073/pnas.57.6.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gell C, et al. Single-molecule fluorescence resonance energy transfer assays reveal heterogeneous folding ensembles in a simple RNA stem-loop. J Mol Biol. 2008;384:264–278. doi: 10.1016/j.jmb.2008.08.088. [DOI] [PubMed] [Google Scholar]
- 42.Gell C, Brockwell D, Smith A. Handbook of Single Molecule Fluorescence Spectroscopy. Oxford: Oxford University Press; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.