Abstract
The Paramyxoviridae family of enveloped viruses enters cells through the concerted action of two viral glycoproteins. The receptor-binding protein, hemagglutinin-neuraminidase (HN), H, or G, binds its cellular receptor and activates the fusion protein, F, which, through an extensive refolding event, brings viral and cellular membranes together, mediating virus–cell fusion. However, the underlying mechanism of F activation on receptor engagement remains unclear. Current hypotheses propose conformational changes in HN, H, or G propagating from the receptor-binding site in the HN, H, or G globular head to the F-interacting stalk region. We provide evidence that the receptor-binding globular head domain of the paramyxovirus parainfluenza virus 5 HN protein is entirely dispensable for F activation. Considering together the crystal structures of HN from different paramyxoviruses, varying energy requirements for fusion activation, F activation involving the parainfluenza virus 5 HN stalk domain, and properties of a chimeric paramyxovirus HN protein, we propose a simple model for the activation of paramyxovirus fusion.
Keywords: fusion triggering, hemagglutinin-neuraminidase structure, protein refolding, viral membrane fusion
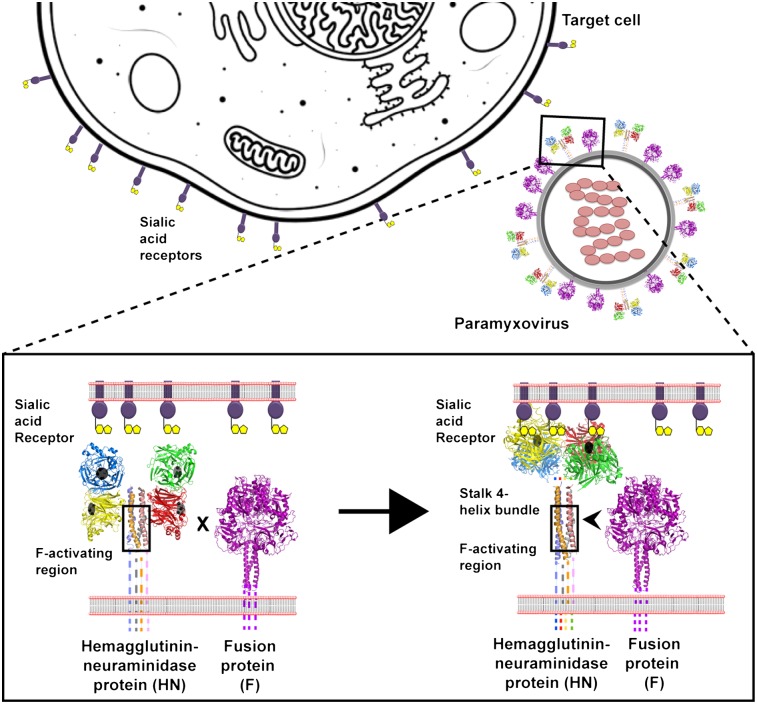
The Paramyxoviridae include some of the great and ubiquitous disease-causing viruses of humans and animals and include parainfluenza viruses (PIV) 1–5, mumps virus, Newcastle disease virus (NDV), Sendai virus, measles virus, canine distemper virus (CDV), Hendra virus, Nipah virus, respiratory syncytial virus, and metapneumoviruses. The Paramyxoviridae are enveloped, negative-stranded RNA viruses that enter cells by fusing their envelope at neutral pH with the plasma membrane of the target cell, releasing the viral genome into the cytoplasm in the form of a ribonucleoprotein complex.
Paramyxovirus-mediated fusion depends on the concerted actions of two glycoproteins, an attachment protein [hemagglutinin-neuraminidase (HN), H, or G] and its cognate fusion (F) protein, the latter initially folding into a metastable form. The attachment protein is thought to trigger the fusion protein in a receptor-dependent manner (1–5). This triggering of the metastable F (6) by the receptor-binding protein couples receptor binding of HN, H, or G to lowering the activation energy barrier of F so that F refolds into a highly stable postfusion form (7). In the process, F undergoes a series of structural rearrangements involving several intermediates and brings about membrane merger (8).
HN proteins bind sialic acid as their receptor and also have neuraminidase (receptor-destroying) activity. The PIV5 HN protein comprises 565 residues and has a short N-terminal cytoplasmic tail followed by a single transmembrane domain and a large ectodomain (residues 37–565). The protein consists of a stalk region (residues 1–117) that supports a large globular head domain (residues 118–565) containing the receptor-binding and neuraminidase-active site. X-ray crystal structures of the globular head domain of PIV5, NDV, Nipah virus, Hendra virus, measles virus, and human parainfluenza virus 3 (hPIV3) attachment proteins have been obtained (9–16). The PIV5 HN globular head structure (16) reveals a neuraminidase-like fold with a six-bladed β-propeller structure that is a common feature of the other paramyxovirus HN/H/G head domains, regardless of receptor specificity. The sialic acid-binding site is placed centrally within the β-propeller. PIV5 HN exists as a pair of disulfide-linked dimers with the disulfide bond at cysteine 111 (16, 17). These dimers are associated noncovalently to form a dimer-of-dimer oligomer (16, 18). The PIV5 HN dimer-of-dimer structure showed the dimers arranged at an approximately 90° angle to each other in an overall twofold symmetry creating a 657-Å2 dimer-of-dimer interface (16). This form of the tetrameric HN protein is referred henceforth to as the “four-heads-up” form. The recently obtained structure of the NDV HN globular head dimer also contains part of the stalk revealing a four-helix bundle (4HB) (15). Despite a strong structural similarity with the PIV5 HN globular head dimer (16), NDV HN showed a different arrangement of the heads in the tetramer, which we refer to hereafter as the “four-heads-down” form. In this NDV HN structure, an interaction interface was observed between residues of the globular head of NDV HN and residues 83–114 in the NDV HN 4HB stalk domain. The ability of the HN/H/G heads to adopt different interactions with respect to the stalk is supported by electron micrographs of purified PIV5 HN protein that indicate a variety of arrangements of the heads (18). Recent biochemical evidence indicates significant movement of the measles virus H protein head domains that contributes to the ability of H to trigger F (19, 20). These data suggest potential functional roles for the different conformational arrangements observed in the crystal structures of the paramyxovirus receptor-binding protein. In addition to the similarities in their HN globular heads, the stalk regions of NDV and PIV5 (15, 21) show a high degree of structural similarity and overlapping regions of putative F interaction (21). Recent studies have shown that sequences of other paramyxovirus receptor-binding protein stalks can be modeled as 4HBs based on the crystal structures of the PIV5 HN and NDV HN stalk domains (22–24).
Considerable evidence indicates that the stalk domains of paramyxovirus HN/H/G proteins have a significant role in direct interactions with the F protein (21–23, 25–34). In a number of these studies, mutations in the stalk domains of paramyxovirus receptor-binding proteins have been shown to block F activation, presumably by blocking F–HN interactions.
Assuming that interactions of HN/H/G with F probably occur via the stalk region of the receptor-binding protein and that concomitant alterations in this interaction may occur upon receptor binding to trigger F, it is possible that receptor binding by the HN/H/G head domains induces conformational changes that propagate down through the stalk on receptor binding (24, 35). Here we provide evidence for a different hypothesis. We show that residues 1–117 of the PIV5 HN stalk domain are sufficient to activate F for fusion and that the HN globular head domain is completely dispensable for PIV5 F triggering. We provide evidence suggesting that the globular head domains of paramyxovirus HN proteins play a regulatory role in fusion, possibly via the movement of the heads; and that this role is maintained even for a chimeric molecule possessing globular head and stalk domains from different paramyxoviruses. Also, we show by using previously characterized N-glycosylation mutants that the interactions of the headless PIV5 HN 1–117 protein with F are similar to the F–HN interactions of WT PIV5. To provide further evidence for the regulatory nature of the globular head, we highlight the roles of two important interfaces in the PIV5 HN globular head domains. By using mutants located in the dimer-of-dimer interface residues of the four-heads-up form (16) and in the putative head–stalk interaction residues of PIV5 HN, modeled on the four-heads-down NDV HN structure (15), we demonstrate an effect on fusion activation on the disruption of these interface interactions. We propose a simple model of paramyxovirus fusion activation in which the receptor-binding protein stalk domain provides the primary trigger in fusion and the globular head domains undergo structural changes to regulate the activation of fusion.
Results
PIV5 HN Stalk Domain 1–117 Is Sufficient to Activate Its Fusion Protein and to Mediate Cell-to-Cell Fusion.
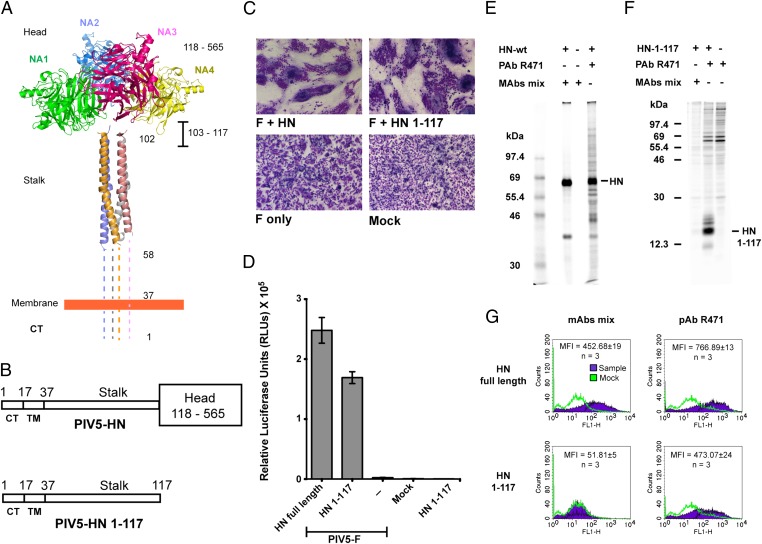
Recently we determined the structure of the PIV5 HN stalk domain from a construct expressed in insect cells containing stalk residues 56–117 (Fig. 1A) (21). This portion of the stalk is a 4HB that consists of a distinct hydrophobic core. In light of recent data indicating that paramyxovirus HN stalks have an essential role in fusion (21–23, 25, 36), we tested the function of the PIV5 HN stalk in a biological context by removing the globular head domain (Fig. 1B). We used an expression construct (PIV5 HN 1–117 stalk) lacking the entire HN head (residues 118–565). However, the HN stalk construct includes the intradimer disulfide bond at HN cysteine residue 111.
Fig. 1.
The PIV5 HN stalk domain is sufficient to activate F and mediate cell–cell fusion. (A) Structural view of the PIV5 HN full-length protein based on X-ray crystal structures of the tetrameric PIV5 HN globular head domains (16) and the 4HB of the PIV5 HN stalk domain (21). Regions for which there is no known structure (amino acids 1–58 and 103–117) are represented by dotted lines. CT, cytoplasmic tail; NA1–NA4, globular neuraminidase head domains. (B) Schematic representation of the PIV5 HN full-length WT protein (amino acids 1–565) (Upper) and the PIV5 HN stalk protein (amino acids 1–117) lacking the globular head domain (amino acids 118–565) (Lower). CT, cytoplasmic tail; TM, transmembrane domain. (C) Representative micrographs of syncytia showing cell–cell fusion in BHK-21 cells transfected with F and WT HN or HN 1–117 stalk 18 h posttransfection. (D) Luciferase reporter assay of cell–cell fusion. (E and F) Radioimmunoprecipitation of [35S]-TranS–labeled transfected 293T cell lysates, using PIV5 HN pAb R471 or a mixture of mAbs to the PIV5 HN globular head. (E) WT HN protein analyzed on a 10% (wt/vol) SDS/PAGE gel. (F) HN 1–117 stalk protein analyzed on a 15% (wt/vol) SDS/PAGE gel. Numbers to the left of each panel are molecular masses in kDa. (G) Representative graphs showing the detection of proteins at the cell surface of 293T cells transfected with WT HN or HN 1–117 stalk using flow cytometry. Surface proteins were detected using a mixture of HN mAbs or HN pAb R471. Average mean fluorescence intensities (MFI) across three independent experiments are shown.
When the PIV5 HN 1–117 stalk construct was coexpressed with PIV5 F in BHK-21 cells, significant cell–cell fusion characterized by large syncytia, similar to the syncytia observed in cells expressing WT F and HN, was observed (Fig. 1C). Quantification of this fusion using a luciferase assay indicated ∼70–75% fusion activity compared with WT F + WT HN (Fig. 1D). The ability of the PIV5 HN 1–117 stalk to activate PIV5 F is specific, because other F proteins from NDV HN, hPIV3 HN, and hPIV2 HN could not be activated by the PIV5 HN 1–117 stalk. To detect the expression of HN 1–117 stalk protein in mammalian cells, [35S] metabolically labeled proteins were immunoprecipitated from transfected cell lysates using a polyclonal HN antibody, R471 (21). Additionally, as a control, a mixture of PIV5 HN monoclonal antibodies (mAb 1b and mAb 4b) that are known to bind the globular head of PIV5 HN specifically were used (37). As expected, the PIV5 HN full-length protein could be detected by the two mAbs and by the polyclonal antibody (pAb) R471 (Fig. 1E), whereas the HN 1–117 stalk, lacking the globular head domains could be detected using only pAb R471 (Fig. 1F). A similar result was obtained when both the expressed WT PIV5 HN and PIV5 HN 1–117 stalk proteins were detected on the cell surface using pAb R471 and flow cytometry, but only the full-length PIV5 HN protein could be detected by the two mAbs (Fig. 1G). Collectively, these data indicate that fusion activation of the PIV5 F protein can be mediated by expression of the HN 1–117 stalk domain alone. A factor that could affect the level of fusion is the relative molar ratio of WT HN to HN 1–117 stalk accumulating in cells, but this effect cannot be determined readily because of the differential antibody reactivity to WT HN and the HN 1–117 stalk domain.
Absence of the PIV5 HN Globular Head Domain Energetically Favors Fusion Activation.
To explore further the mechanism by which the HN 1–117 stalk domain activates fusion in the absence of the head domain, the level of fusion activation by the PIV5 HN 1–117 stalk protein and the WT HN protein at a suboptimal temperature was compared. In a quantitative luciferase fusion assay performed at 33 °C, fusion activation by WT HN was reduced significantly compared with fusion at 37 °C. However, there was very little reduction in the amount of fusion mediated by the PIV5 HN 1–117 stalk at this suboptimal temperature (Fig. 2A). These data suggest an energy requirement of WT PIV5 HN for F activation that is observed only at a suboptimal temperature and that this energy is required only when the head domains are present.
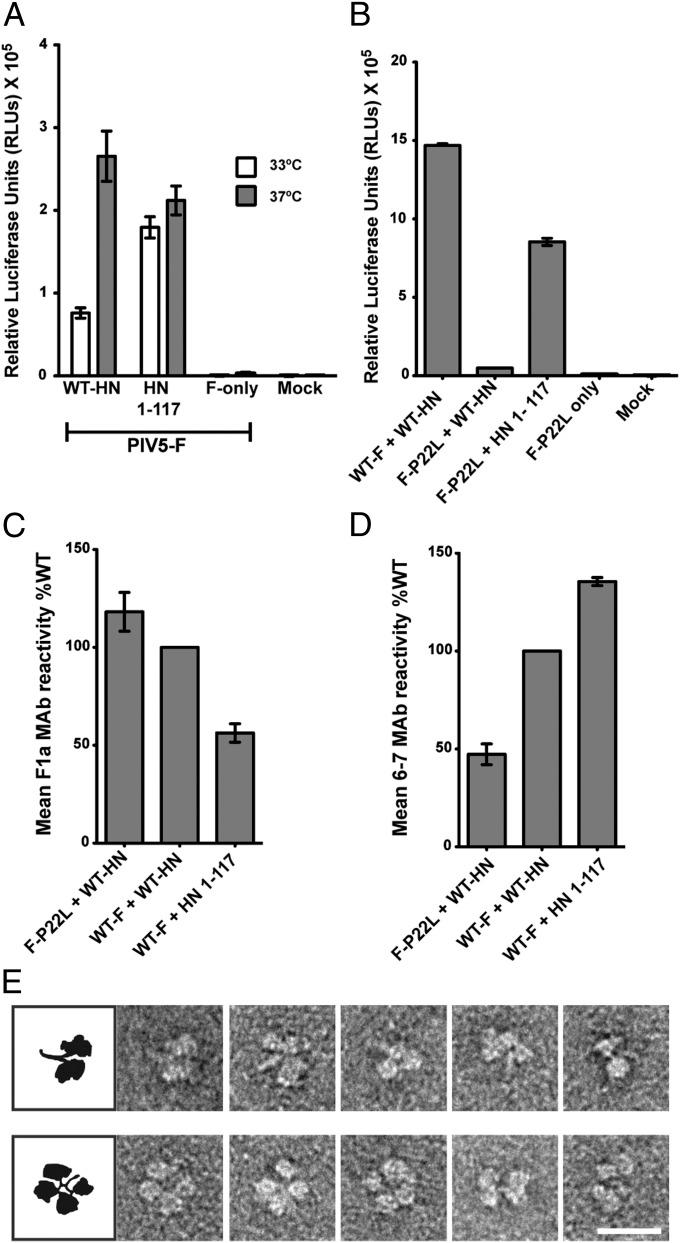
Fig. 2.
Absence of the PIV5 HN globular head domain energetically favors fusion activation. (A) Fusion activation at 33 °C (white bars) and 37 °C (gray bars) by the PIV5 HN WT and HN 1–117 stalk protein. Fusion was measured by a luciferase assay. (B) Luciferase reporter assay showing relative activation of a hypofusogenic mutant of PIV5 F (F-P22L) by WT PIV5 HN protein in comparison with activation by the PIV5 HN 1–117 stalk protein. (C and D) Detection of pre- and postfusion PIV5 F and PIV5 F P22L proteins on the surface of cells cotransfected with WT PIV5 HN or the PIV5 HN 1–117 stalk, using flow cytometry. The amounts of surface antigen detected by an F-prefusion conformation–specific mAb, F1a, (C) and an F-postfusion conformation–specific mAb, 6-7, (D) are shown as a percentage MFI of the WT F + WT HN sample (n = 3). (E) Representative electron micrographs of purified PIV5 HN ectodomain showing different conformations of the dimer-of-dimer globular heads with respect to the stalk domain. (Upper) Conformations of HN in which the stalk is exposed (four-heads-up–like arrangements). (Lower) Examples of HN with the heads juxtaposed with the stalk domain (four-heads-down–like arrangements). (Scale bar, 20 nm.)
PIV5 F-P22L is a hypofusogenic mutant of PIV5 F that needs to overcome an increased activation barrier to transition into the more stable postfusion form (38). PIV5 F-P22L is activated very poorly by WT PIV5 HN at 37 °C (38). However, when F-P22L was cotransfected with the PIV5 HN 1–117 stalk, a significant amount of fusion (∼50% of WT HN), compared with minimal F-P22L activation by the PIV5 HN full-length protein, was observed in a quantitative fusion assay (Fig. 2B).
To examine the comparative populations of the prefusion and the postfusion conformations of F or F-P22L at the cell surface on activation by the PIV5 WT HN or the HN 1–117 stalk protein, flow cytometry was performed using a F prefusion conformation-specific mAb (F1a) (Fig. 2C) or a F postfusion conformation-specific mAb 6-7 (Fig. 2D). The comparable amounts of prefusion F detected on the surface of transfected cells were in the order F-P22L + WT HN > WT F + WT HN > WT F + HN 1–117 stalk (Fig. 2C). On the other hand, the amounts of postfusion F detected on the cell surface were in the reverse order compared with the amounts of prefusion F: WT F + HN 1–117 stalk > WT F + WT HN > F-22L + WT HN (Fig. 2D). Taken together the data indicate that loss of the HN head domain to generate the HN stalk 1–117 leads to an increased ability to convert prefusion F to the postfusion form. Furthermore, the data shown in Fig. 2 A and B indicate an energy requirement for HN for fusion activation, possibly to move the head domains with respect to the stalk. Multiple arrangements of the head domains with respect to the stalk domains are possible, as seen in electron micrographs of purified PIV5 HN ectodomain protein (Fig. 2E).
N-Linked Carbohydrate Chains in the Putative F-Interacting Region of the PIV5 HN 1–117 Stalk Block Fusion Activation.
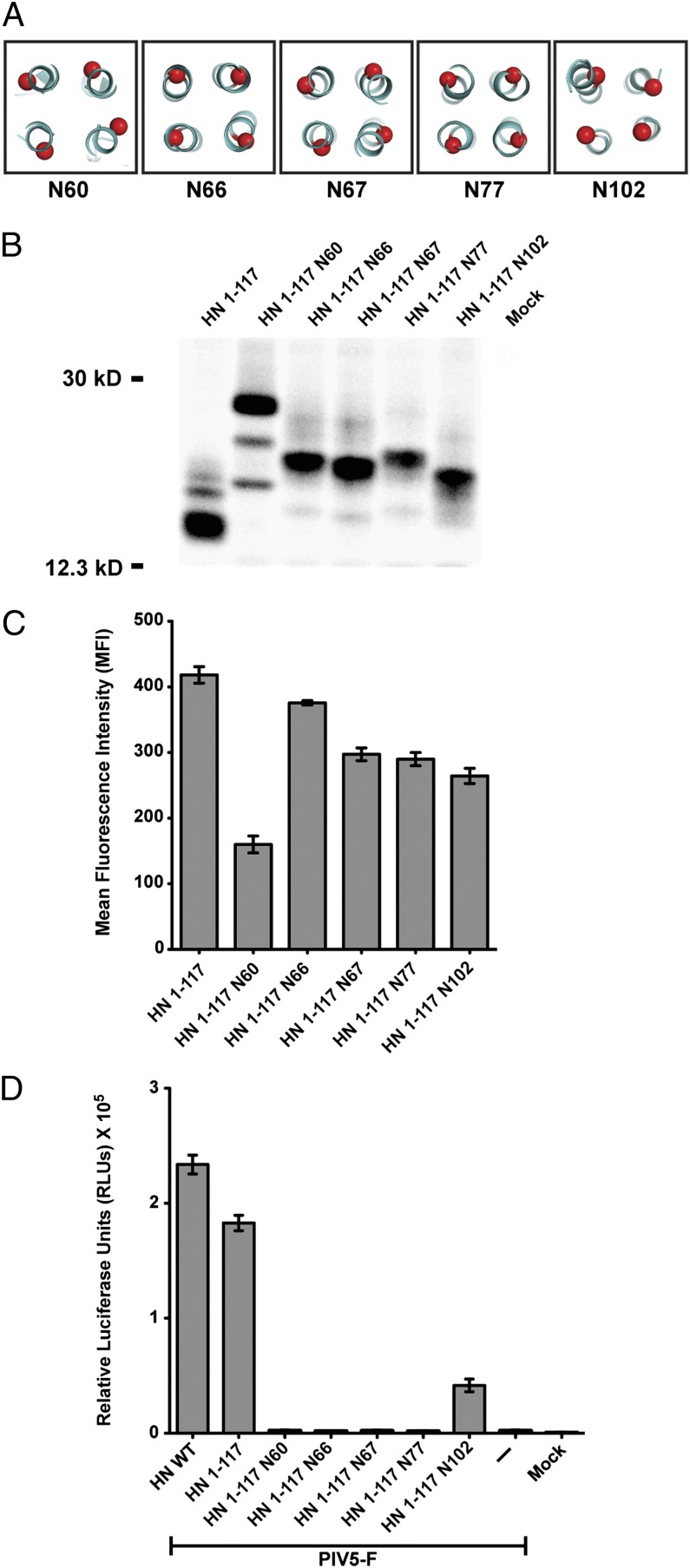
Previously we described PIV5 HN mutants that introduced sites for the addition of N-linked carbohydrate chains into the HN stalk; these mutants were designed to block F and HN interactions physically and were based on a set of functional assays in which we defined a putative region of F interaction on the PIV5 HN stalk (21). All the PIV5 HN mutants showed a defect in fusion except one (N102) in which an N-linked glycan chain was added to residue 102. Using the information gained from the HN mutants (21), we introduced mutations in the HN 1–117 stalk to add glycosylation sites at residues 60, 66, 67, 77, and 102 that mapped to the outer surface of the 4HB (Fig. 3A). The HN 1–117 stalk N-glycosylation mutants were expressed in cells and were metabolically labeled, and the proteins were immunoprecipitated using HN pAb R471 (Fig. 3B). The decrease in electrophoretic mobility of these mutants compared with HN 1–117 stalk on a SDS/PAGE gel indicates that these mutants are glycosylated. The minor HN-specific bands possibly are the result of partial glycosylation at a natural glycosylation site at N110. Glycosylation of these mutants was confirmed by analyzing mutant HN proteins that had been metabolically labeled in the presence of the N-linked glycosylation inhibitor tunicamycin (5 μg/mL). All mutant proteins showed faster electrophoretic mobility, as expected for unglycosylated proteins. All HN 1–117 stalk N-glycosylation mutants except N60 had relatively robust levels of protein expression on the surface of transfected cells, ranging between 75–90% of that of the HN 1–117 stalk (Fig. 3C). The aberrant migration of HN 1–117 N60 protein may be caused by altered glycosylation.
Fig. 3.
N-linked carbohydrate chains in the putative F-interacting region of PIV5 HN 1–117 block fusion activation. (A) Schematic representation of positions of residues on the PIV5 HN 1–117 stalk to which sites for the addition of N-linked carbohydrate chains were added by mutagenesis. The mutants are named according to the residue carrying the N-glycosylation. (B) Migration pattern of N-glycosylation mutants in the PIV5 HN 1–117 stalk on a 15% (wt/vol) SDS PAGE gel. Polypeptides were immunoprecipitated from radiolabeled lysates of transfected 293T cells using PIV5 HN pAb R471. Numbers on the left are molecular masses in kDa. (C) Cell-surface expression of PIV5 HN 1–117 N-glycosylation mutants determined using HN pAb R471 as measured by flow cytometry (n = 3). (D) Fusion activation of PIV5 F by the HN 1–117 stalk and its N-glycosylation mutants. Fusion was quantified using a luciferase reporter assay as described in Experimental Procedures.
To demonstrate the specificity of the HN 1–117 stalk domain in activating fusion, the HN 1–117 stalk N-glycosylation mutants described above were coexpressed with F, and a quantitative fusion assay was performed. As shown in Fig. 3D, mutants N60, N66, N67, and N77 abolished fusion completely, whereas mutant N102 had a low but significant level of fusion reflecting the results obtained using full-length mutant HN proteins (21). These data suggest that the stalk interacts with the F protein in a similar manner, regardless of the presence or absence of the HN globular head.
F Activation Mediated by the PIV5 HN 1–117 Stalk Does Not Depend on Engagement of the Sialic Acid Receptor.
For PIV5 WT HN the globular head domains bind sialic acid, and this binding generally is believed to act as the signal for F activation. To understand fusion activation by the headless HN 1–117 stalk, we first eliminated the possibility that HN 1–117 stalk binds sialic acid. PIV5 HN 1–117 stalk showed no specific retention of chicken RBCs, indicating that it had no specific sialic acid-binding capability (Fig. 4A).
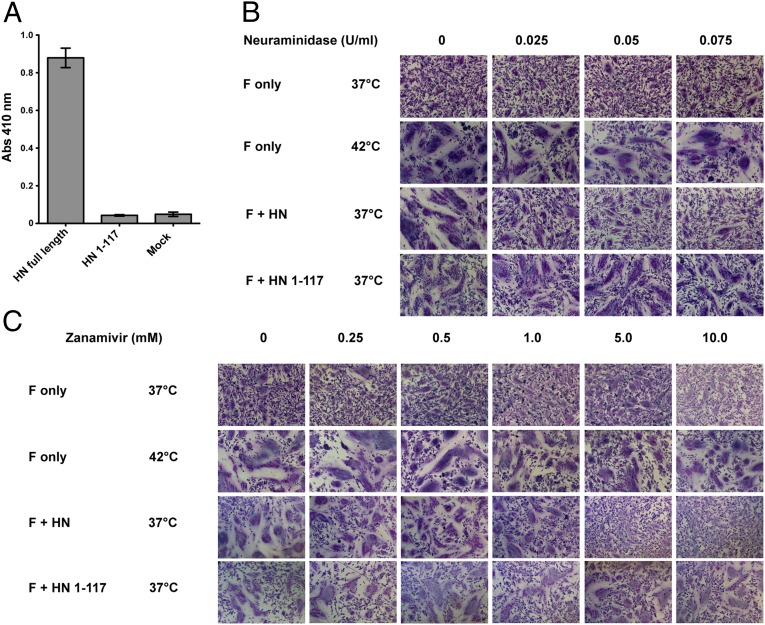
Fig. 4.
F activation mediated by the PIV5 HN 1–117 stalk does not depend on engagement of the sialic acid receptor. (A) Receptor-binding activity of the PIV5 HN 1–117 stalk. Chicken RBCs bound onto surfaces of transfected 293T cells were quantified by lysing the bound RBCs after extensive PBS washes and measuring absorbance of chicken hemoglobin at 410 nm. (B and C) BHK-21 cells transfected with PIV5 F only or cotransfected with PIV5 F and PIV5 HN or PIV5 HN 1–117 stalk were incubated with C. perfringens neuraminidase or zanamivir at 37 °C or 42 °C. Cells were fixed, stained, and imaged 18 h posttransfection. (B and C) Fusion activation in the presence of 0 U/mL, 0.025 U/mL, 0.05 U/mL, and 0.075 U/mL C. perfringens neuraminidase to remove most sialic acid linkages (B) and with the addition of 0 mM, 0.25 mM, 0.5 mM, 1 mM, 5 mM, and 10 mM zanamivir to inhibit PIV5 HN catalytic sites specifically (C).
Unlike most paramyxoviruses, for PIV5 (isolate W3A) expression of F alone can mediate some fusion of transfected cells in the absence of HN; however, coexpression of F and HN greatly increases the extent of fusion (2). Additionally, we have shown previously that heat can be used as a surrogate for HN in triggering fusion (39). As shown in Fig. 4B, cells expressing F alone that were incubated at 37 °C produced small syncytia, but cells expressing F alone that were incubated at 42 °C produced syncytia that were comparable in size to those produced by cells coexpressing F and HN that were incubated at 37 °C. Treatment of cells with increasing concentrations of exogenous Clostridium perfringens neuraminidase (0.025–0.075 U/mL), as expected, blocked extensive syncytia formation in cells coexpressing F and HN that were incubated at 37 °C (Fig. 4B). However, neuraminidase treatment of cells expressing F alone that were incubated at 37 °C or 42 °C had no effect on syncytia formation. At 37 °C the formation of syncytia in cells coexpressing PIV5 HN 1–117 stalk and F was unaffected by neuraminidase treatment, contrary to the results obtained for WT F + WT HN expression.
To confirm that fusion activation did not require sialic acid binding to the specific receptor-binding sites on the HN head domains, cells expressing F alone, F and HN, or F and HN 1–117 stalk were treated with 0–10 mM 4-guanadino-2-deoxy-2,3-didehydro-N-acetylneuraminic acid (zanamivir), the specific inhibitor of the HN catalytic site. In cells coexpressing F and HN incubated at 37 °C, 5 mM zanamivir inhibited syncytia formation (Fig. 4C). In contrast, in cells expressing either F alone incubated at 42 °C or F and HN 1–117 stalk incubated at 37 °C, 10 mM zanamivir did not inhibit syncytia formation (Fig. 4C). Taken together, these data show that the promotion of fusion by the HN 1–117 stalk (or by F alone when incubated at 42 °C) does not depend on the engagement of the sialic acid receptor. These data do not address whether PIV5 F has a cellular receptor molecule or whether cell–cell contact with F expression is sufficient to allow the formation of syncytia on the expression of HN 1–117 stalk or when F is heated to 42 °C.
Domain-Specific Functions of the HN Stalk and Energy Requirement by the Globular Head During F Activation by a Chimeric Molecule.
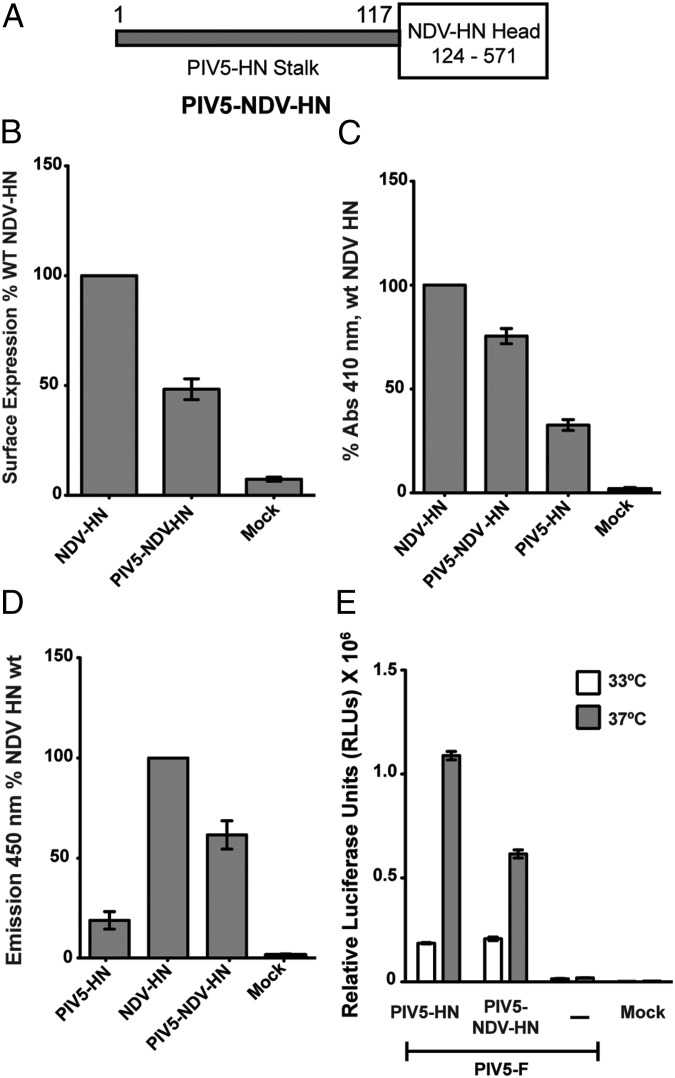
The facts that the PIV5 HN 1–117 stalk alone can activate F and that the PIV5 HN globular heads require energy for F activation at 33 °C led us to investigate if the head and the stalk of paramyxovirus HN proteins could function as independent domains linked through a flexible linker. To test this notion, we created a chimeric HN protein by fusing the PIV5 HN 1–117 stalk to the NDV HN protein head (residues 124–571) (Fig. 5A). The chimeric protein was expressed in cells and was detected at the cell surface by flow cytometry using a mixture of NDV HN monoclonal antibodies (40). The PIV5-NDV HN chimera was expressed at the cell surface at ∼50% of WT NDV HN levels (Fig. 5B). Receptor binding by the NDV HN head in the chimeric protein, as measured by a hemadsorption assay using chicken RBCs, was 80% of WT NDV HN (Fig. 5C). The neuraminidase activity of the PIV5-NDV HN chimera was similar to that of the NDV HN protein when the differences in cell-surface expression are taken into account (Fig. 5D).
Fig. 5.
Chimeras and NDV HN stalk activation. (A) Schematic representation of the chimeric protein PIV5-NDV-HN, consisting of the PIV5 HN stalk (amino acids 1–117) followed by the NDV HN head (amino acids 124–571). (B) Surface expression of PIV5-NDV-HN as measured by flow cytometry using a mixture of NDV HN 4a and 2b mAbs and expressed as a percentage of WT NDV HN expression. (C) Receptor-binding activity of PIV5-NDV-HN compared with PIV5 HN and NDV HN and expressed as a percentage of WT NDV HN bound to chicken RBCs at 4 °C. (D) Neuraminidase activity of the PIV5-NDV-HN chimera compared with PIV5 HN and NDV HN proteins, as a measure of fluorescent product formation, expressed as a percentage of WT NDV HN. (E) Fusion promotion of the PIV5-NDV-HN chimera as measured by luciferase reporter assays at 33 °C and 37 °C.
When tested for its ability to activate F protein as measured by a luciferase reporter assay, PIV5-NDV HN showed significant amounts of fusion with PIV5 F. NDV F was not activated by PIV5-NDV HN, showing that the specificity of the F protein resides in the stalk (Fig. 5E and Fig. S1A). Because the PIV5 HN 1–117 stalk domain attached to a noncognate globular head could activate F successfully, we investigated whether the PIV5-NDV HN chimera would require energy to activate F at 33 °C, as was observed in the full-length PIV5 HN protein (Fig. 2A). Fusion quantified for PIV5 HN and PIV5-NDV HN at 33 °C showed that the energy requirement of the PIV5-NDV HN chimera was similar to that of the PIV5 HN protein (Fig. 5E), unlike the PIV5 1–117 HN stalk, which was unaffected by temperature (Fig. 2A). Additionally, like PIV5 HN-induced fusion (Fig. 4B), fusion induced by the PIV5-NDV HN chimera could be blocked by 0.1 U/mL C. perfringens neuraminidase (Fig. S1B).
Structurally Conserved Loops in the Paramyxovirus HN Globular Head Domains Have Important Regulatory Roles in Fusion and Receptor Binding.
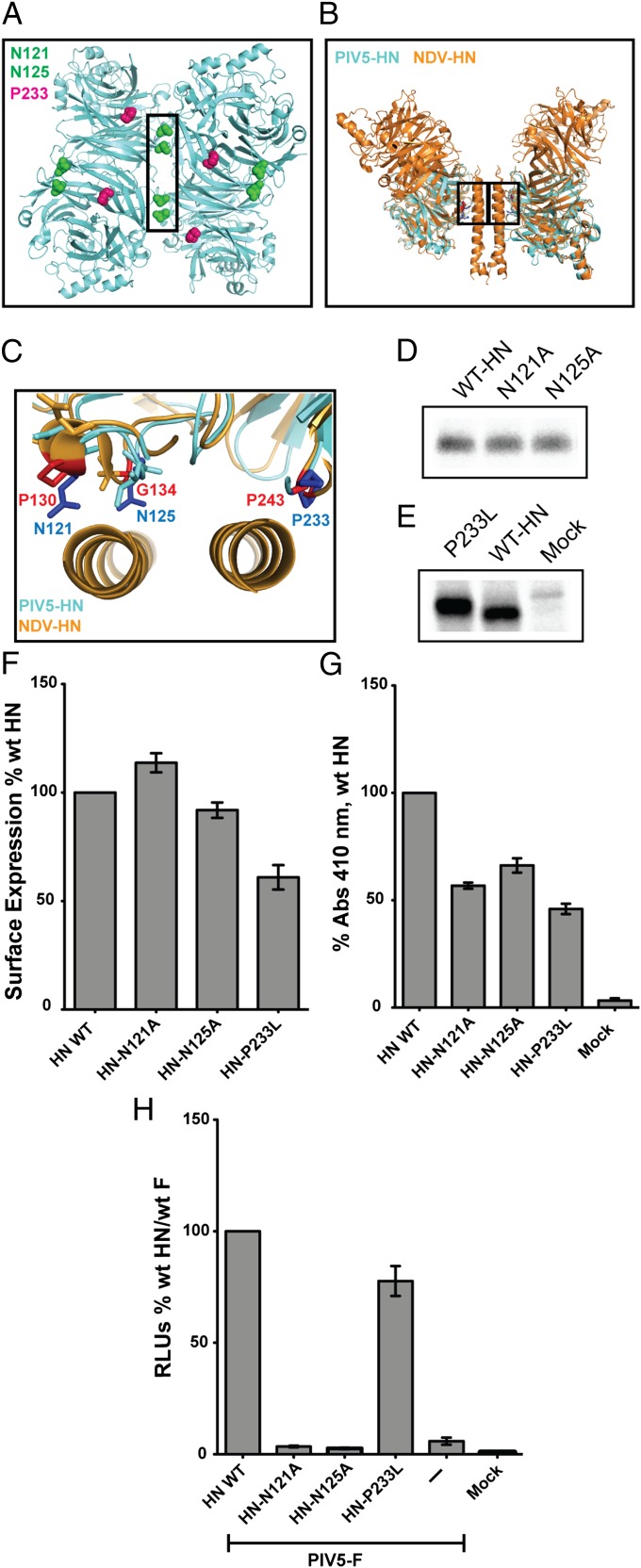
The two known positions of the HN head domains create different surfaces for protein–protein interactions and make unambiguous mutagenesis studies of function more challenging. The four-heads-up position in the PIV5 HN atomic structure (16) creates a dimer-of-dimer interface that buries a surface area of 657 Å2 (Fig. 6A). The four-heads-down conformation of the NDV HN atomic structure (15) revealed an interface of interaction between two globular NA head domains and the 4HB of the stalk, burying a significant area (928 Å2) of protein–protein interaction. We aligned the structures of PIV5 HN head domain (16) with the NDV HN head domain (15), highlighting a region conserved structurally in the NA domains of both viruses and that form a part of the stalk–head interface in NDV HN (Fig. 6B). Side chains of residues P243, P130, and G134 in NDV HN and their corresponding residues P233, N121, and N125 in PIV5 HN are in positions that potentially form contacts between the globular head and the 4HB stalks in the four-heads-down arrangement (Fig. 6C). Interestingly, residues N121 and N125 (PIV5 HN), in addition to their putative contacts in the head–stalk interface of the ‘four-heads-down form (Fig. 6C), also form contacts within the dimer-of-dimer interface of the PIV5 HN four-heads-up form (Fig. 6A). Unlike N121 and N125, P233 (PIV5 HN) is not involved in this dimer-of-dimer interface. To test the role of these interfaces observed in the two different structural arrangements obtained for PIV5 HN (16) and NDV HN (15), mutations were made in the PIV5 HN residues N121A, N125A, and P233L.
Fig. 6.
Structurally conserved loops in the paramyxovirus HN globular head domains have important regulatory roles in fusion and receptor binding. (A) Bottom view of the PIV5 HN four-heads-up form showing the positions of N121 (green), N125 (green), and P233 (red) residues on all four monomers. The dimer-of-dimer interface is indicated by a black box. (B) Side view of the four-heads-down form as a structural alignment of PIV5 HN globular heads (aquamarine) with the NDV HN structure (orange) showing the head/stalk interface (black boxes). (C) Magnified top view of B. Corresponding PIV5 HN residues (blue) and NDV HN residues (red) are highlighted in the head–stalk interface. (D and E) Expression of PIV5 HN, N121A, and N125A (D) and P233L mutant polypeptides (E) immunoprecipitated from radiolabeled cell lysates with PIV5 HN pAb R471. (F) Surface expression of the PIV5 HN globular head mutants expressed as a percentage of WT PIV5 HN surface expression using PIV5 HN pAb R471 and flow cytometry. (G) Receptor-binding activity of mutant proteins expressed as a percentage of WT PIV5 HN bound to chicken RBCs at 4 °C. (H) Luciferase reporter assay for fusion showing fusion promotion by the N121A, N125A, and P233L mutants cotransfected with PIV5-F, expressed as a percentage of WT PIV5-F and WT PIV5 HN fusion.
To analyze expression of the mutant HN proteins, transfected cells were radiolabeled metabolically with [35S], and proteins were immunoprecipitated. The expression levels of the mutants were comparable to those of WT HN protein (Fig. 6 D and E). The cell-surface expression levels of N121A, N125A, and P233L mutants were comparable to WT HN, ranging between 70–110% of WT PIV5 HN (Fig. 6F). The receptor-binding ability of these mutants, determined by their ability to bind chicken RBCs (hemadsorption), was variable. When normalized to surface expression, P233L has a normal receptor-binding capability, whereas N121A and N125A have somewhat reduced receptor-binding ability (Fig. 6G). However, N121A and N125A (part of both head–stalk and dimer-of-dimer interfaces) were severely impaired for fusion activation, whereas P233L (part of the head–stalk interface only) showed WT levels of fusion with PIV5 F (Fig. 6H). There are two explanations for these data. One is that altering the stalk–head interaction by N121A, N125A, or P233L in the four-heads-down form does not affect fusion activation. In this case, we assume that in the four-heads-up form, the N121A and N125A mutations alter the dimer-of-dimer interface and fusion activation. In contrast P233L does not disrupt an interface in the four-heads up form, and fusion activation is observed. The alternative explanation is that the mutations N121A and N125A stabilize the head–stalk interaction and thus inhibit fusion.
Discussion
The F protein of paramyxoviruses causes membrane fusion through the concerted action of the F protein and the receptor-binding protein HN, H, or G. The F protein folds initially to form a trimeric metastable prefusion form that is triggered to undergo large-scale irreversible conformational changes to form the trimeric postfusion conformation. It is thought that F-protein refolding couples the energy released with membrane fusion. The irreversible nature of F-protein refolding requires a highly specific mechanism so that triggering occurs only on receptor binding by an attachment protein, i.e., HN, H, or G. The precise mechanism by which HN, H, or G activates F is unclear.
It is well established for many paramyxoviruses that F and HN, H, or G interact physically, as shown by coimmunoprecipitation and cocapping studies (3, 25, 32, 41, 42). However, coimmunoprecipitation of PIV5 F and HN has been difficult to demonstrate; it may be a weak and/or transient interaction. The available data suggest that the sites of F interaction reside within the stalk domains of HN, H, or G (21–23, 25–34). The commonly adopted hypothesis for F activation is that, on receptor binding, the receptor-binding protein HN, H, or G undergoes structural changes in the globular head that are transmitted through the connecting loops to the stalks, where F interacts with the stalk (20, 24, 35, 43). However, our finding of fusion promotion by a headless PIV5 HN 1–117 stalk suggests that the F-activating region residing within the HN 1–117 stalk is sufficient to activate PIV5 fusion. Addition of carbohydrate chains to residues on the outer surface of the PIV5 HN 1–117 stalk 4HB ablate fusion, a finding indicative of an F protein–HN 1–117 stalk interaction. Furthermore, the PIV5 HN 1–117 stalk does not activate other, noncognate F proteins.
Two lines of evidence indicate that the expression of PIV5 HN 1–117 stalk overcomes an energy barrier more readily than WT HN. First, at 33 °C HN 1–117 stalk triggers fusion to a greater extent than does WT HN. Second, HN 1–117 stalk triggers fusion of the hypofusogenic mutant F-P22L orders of magnitude more readily than WT HN. The explanation best supported by the data suggests a distinct energy requirement by the globular heads of PIV5 HN, possibly to undergo structural rearrangements, whereas the HN 1–117 stalk may be able to activate F-P22L because of an unlimited period of F–HN stalk interaction. Similarly, in the hybrid PIV5-NDV HN construct, fusion was greater at 37 °C than at 33 °C.
To expand the observation that PIV5 F can be activated by its cognate HN stalk, we expressed various HN stalk constructs of NDV and hPIV3 both with and without HA-epitope tags at their N and C termini (Table S1). None of these HN stalk constructs activated the cognate F protein for fusion. The addition of a HA-tag to the C terminus of PIV5 HN 1–117 stalk reduced fusion activity; expression of a PIV5 HN stalk that was shorter in length (residues 1–110) and lacked the interchain disulfide bond also reduced fusion. Because of the apparent loss of fusion activity upon epitope-tagging the PIV5 HN 1–117 stalk and because of the inability to detect hPIV3 and NDV HN protein stalks on the cell surface using available antibodies, the F-triggering capabilities of these proteins cannot be interpreted. As the mechanism of F activation by the PIV5 HN 1–117 stalk, we propose that the PIV5 HN 1–117 stalk in absence of the head domains folds into a receptor-bound F-triggering 4HB conformation, similar in concept to the structural rearrangements that occur on the CDV H stalk with receptor binding (23). This F-triggering–competent PIV5 HN stalk encounters F only on reaching the cell surface (44) and triggers F in the absence of receptor binding. Appending tags to or removing N-terminal residues from the PIV5 HN 1–117 4HB may disrupt the F-triggering conformation, causing the PIV5 HN 1–117 stalk to lose its F-triggering capability. For other paramyxoviruses the attachment protein heads may be essential to stabilize the receptor-bound F-triggering conformation, which does not occur with expression of the stalk alone.
The major question that arises is whether activation of fusion by PIV5 HN 1–117 stalk is a mechanism that is unique to PIV5 or is related to processes occurring in other paramyxoviruses. The atomic structure of NDV HN (15) indicates there is a flexible linker between the globular head domains and the 4HB of the stalk, suggesting that the head domains can move. Different positions of the PIV5 HN head domains are observed by electron microscopy (Fig. 2E). The energy requirements for moving HN, H, or G heads may vary among paramyxoviruses and may depend on receptor engagement. Similarly, different paramyxoviruses may differ in the flexibility and movement of the top of the stalk. A recent study using engineered disulfide bonds between head domains of the measles H dimer proposed possible conformational changes between monomers of this dimer that could influence fusion promotion by measles virus H (19). However, different results have been obtained in studies with NDV HN, in which minor or no effects were observed on fusion using mutants in which the globular head dimers were linked through disulfide bridges (45). Additionally, the atomic structures of NDV HN (15), PIV5 HN (16), and measles virus H bound to its cellular receptor SLAM (20) support the idea that the dimer interfaces in these attachment proteins remain intact; instead, the dimer-of-dimer interfaces appear more prone to disruptions or changes. Differences in fusion activation (Fig. 6H) and receptor-binding abilities (Fig. 6G) in the dimer-of-dimer interface of PIV5 HN N121A and N121A mutants suggest that maintaining a head dimer in the upright position is essential for triggering fusion, and the dimer-of-dimer interface in the PIV5 HN structure creates an arrangement whereby both the dimers can attain a heads-up conformation. On the other hand, P233L did not affect fusion in the putative head–stalk interface of PIV5 HN, suggesting that this contact may not be essential in the F-triggering process. However, we cannot discount the possibility that N121A and N125A mutations influence interactions in the head–stalk interface.
Syncytia formation by PIV5 F protein expressed alone has been observed repeatedly at 37 °C, but it is greatly enhanced by coexpression of PIV5 HN (Fig. 1C) (2, 46) or by incubation of cells expressing F at 42 °C (Fig. 4 B and C). Hyperfusogenic mutants of PIV5 such as F-S443P expressed alone can cause syncytia formation at 25 °C (38). Taken together, these data suggest that a major role of HN is to lower the energy barrier for triggering metastable F. Syncytia formation without HN expression suggests either that another receptor molecule interacts with F or that the close proximity of confluent tissue-culture cells permits F to fuse without receptor engagement. An atomistic model of pre-hairpin F predicts that the distance F can span is 210 Å (47). Syncytia formation mediated by respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) that lack G has been well documented (48, 49), and both RSV and hMPV F proteins have been shown to interact with glycosaminoglycans and heparin sulfate, respectively (50, 51). Also, mutants of NDV F and Sendai virus F have been made that are hyperfusogenic and have a much lower need for HN (52, 53). Thus, it seems possible that mutants of PIV5, NDV, and Sendai virus F that lower the energy barrier do not require a receptor molecule for syncytia formation in tissue-culture cells.
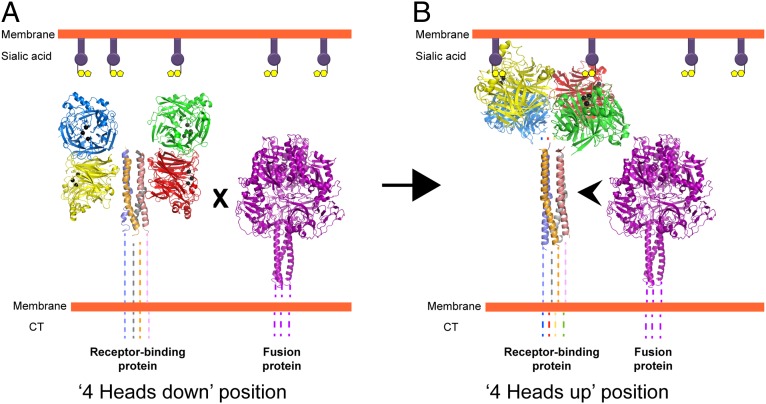
We propose a simple model for activation of fusion in paramyxovirus (Fig. 7 A and B). The HN, H, or G attachment protein stalk harbors the region for fusion activation, which remains covered by the globular heads in the four-heads-down form (Fig. 7A). In this conformation, F is unable to interact with the F-activating region of the attachment protein stalk. When receptor is bound via the HN globular head regardless of whether there is one sialic acid-binding site or two (NDV HN), energy is required to move the heads to attain the four-heads-up form (Fig. 7B). Because the heads can bind receptor at 4 °C, this energy probably is required to expose the region of F activation on the stalk and to allow F to interact with the stalk of the attachment protein to trigger fusion.
Fig. 7.
Schematic model of paramyxovirus F activation based on putative structural rearrangements in the receptor-binding protein. (A) The receptor-binding protein in the four-heads-down position. The formation of the contacts in the head–stalk interface prevents physical interaction of F with the stalk region of the receptor-binding protein. (B) Creation of the contacts in the dimer-of-dimer interface of the receptor-binding protein in the four-heads-up position moves the heads upwards, exposing the stalk. This exposure allows F to interact with the HN stalk, causing F triggering. Black balls indicate the sialic acid binding site residues in the four neuraminidase head domains (colored red, green, blue, and yellow) of paramyxovirus HN. Prefusion F is represented by the PIV5 prefusion F structure (purple) (6). Dotted lines represent regions of paramyxovirus F and HN proteins for which no structural data are available. CT, cytoplasmic tail.
The model we propose suggests a core mechanism of paramyxovirus F activation by HN, H, or G proteins. However, certain additional requirements and variations among paramyxovirus subfamilies exist (43). The atomic structures of measles virus H and henipavirus G proteins (9, 10, 12, 14, 20) indicate that the arrangement of the globular heads is somewhat different from that of HN proteins, and the proteinaceous receptor is bound by a lateral surface of the β-propeller, not at the top, where HN binds sialic acid. Nonetheless, a degree of functional conservation in F activation among viruses binding different receptors is apparent, especially in the stalk domain as found for the CDV H protein (23). However, the morbillivirus H and henipavirus G proteins require structural changes (20, 23) or a unique domain (22) in their putative stalk 4HBs to activate their cognate F. No effect on fusion was seen in experiments in which the length of the measles H stalk was extended above the F-interacting region, but insertions below this region abrogated fusion (25). These data also argue against any F-triggering interaction with the HN, H, or G head.
The site in the cell where F associates with the attachment protein differs among paramyxoviruses. F–HN proteins do not interact in the endoplasmic reticulum (ER) (44, 54), whereas F–H proteins associate as complexes in the ER (55, 56). For this reason the PIV5 HN 1–117 stalk, even though it lacks the regulatory head domains, can traffic independently to the surface without triggering F prematurely. Also measles virus H mutants have been found to associate with F although unable to trigger fusion (28), and a stronger F–G or F–H association leads to lower fusion activity in henipaviruses and morbilliviruses (41, 58–59), suggesting that F–G or F–H association, by itself, may not trigger F.
Given the positions of the heads in the measles tetramer, the differences in stalk length between HN, H, or G proteins, the different positions of receptor-binding sites on the globular heads of H and G proteins, and conformational changes in the morbillivirus H stalk for F triggering, the prereceptor-bound forms of morbillivirus or henipavirus stalks may differ significantly from the four-heads-down form observed in NDV-HN. In addition, the prereceptor-bound form of H or G may associate with F in a complex before F triggering. However, we suggest that triggering F upon receptor binding would require both structural rearrangements between the HN/H/G globular head dimers to expose the F-activating region and flexibility within the stalk 4HB of the attachment protein to allow F interactions with the central region of the HN, H, or G stalk domain, thus providing the core trigger at the heart of the F-activation mechanism.
Experimental Procedures
Cells and Antibodies.
Vero, 293T, BHK-21F, and BSR-T7/5 (a BHK clone expressing T7 RNA polymerase) cells were grown as described (38). Antibodies specific for PIV5 HN globular head included a mix of mAb HN-1b ascites fluid and mAb HN-4b hybridoma supernatant (37). The PIV5 HN full-length and stalk were detected using HN pAb R471, as described previously (21). PIV5 F antibodies included mAb F1a ascites fluid (37), specific to prefusion cleaved PIV5 F, and mAb 6-7, specific to the postfusion form of PIV5 F (60). NDV HN mAbs 4a and 2b were cell-culture supernatants (40).
Cloning and Mutagenesis.
pCAGGS-HN, pCAGGS-F, and pCAGGS-F P22L expression constructs harboring the PIV5 (W3A) HN, F, and F-P22L mutant genes, respectively, were used as described previously (38). The PIV5 HN stalk construct (1–117) was created by PCR amplification of the stalk region (residues 1–117) from the full-length PIV5 HN (residues 1–565). The PCR fragment then was cloned into the pCAGGS vector. Mutagenesis of pCAGGS-HN or pCAGGS-HN 1–117 stalk was done as described (21). The PIV5-NDV-HN chimeric protein construct was amplified using four-primer PCR from pCAGGS-NDV-HN (Australia-Victoria strain) and pCAGGS-PIV5 HN (strain W3A) and was cloned into the pCAGGS vector. The nucleotide sequences were verified using an Applied Biosystems 3100-Avant automated DNA sequencer (Life Technologies Corp.).
Expression of HN and F Glycoproteins in Mammalian Cells.
PIV5 WT F and HN (W3A strain) and their mutant proteins were expressed by transient transfection using pCAGGS constructs in Vero, BHK-21F, and 293T cells.
Immunoprecipitation and SDS/PAGE.
To examine expressed proteins in transfected 293T cells, proteins were labeled metabolically with [35S] at 18 h posttransfection, and proteins were immunoprecipitated as described previously (21). Polypeptides were analyzed on 10% (wt/vol) or 15% (wt/vol) acrylamide gels.
Flow Cytometry.
The cell-surface expression of PIV5 HN and its mutants was quantified by flow cytometry using a FACSCaliber flow cytometer (Becton Dickinson) as described previously (21). The PIV5 HN 1–117 stalk was detected using the PIV5 HN pAb R471 at 1:100 dilution (21). To detect WT F or F-P22L conversion to the postfusion form on the cell surface, 1:200 dilutions of mAb F1a or mAb 6-7 were used. NDV HN monoclonal antibodies 2b and 4a were used at 1:10 dilution to detect chimeric proteins with NDV HN globular heads.
Hemadsorption Assay and Neuraminidase Assay.
Transfected 293T cell monolayers were allowed to bind 1% chicken RBCs as described previously (38). The neuraminidase activity of the PIV5 HN globular head mutants was determined as described previously (21).
Syncytia Formation.
Transfected BHK-21 cells were placed at 37 °C, 33 °C, or 42 °C to allow fusion. Eighteen hours posttransfection, the cells were washed with PBS, fixed, and stained using a Hema3 staining protocol (Fisher Scientific) according to the manufacturer’s instructions. For analyzing sialic acid receptor engagement by WT HN and HN 1–117 during fusion, DMEM containing C. perfringens neuraminidase (Sigma Scientific) or zanamivir was added just after transfection, and cells were fixed and stained 18 h posttransfection. A 50-mM stock solution of zanamivir was prepared from Relenza Rotadisks (GlaxoSmithKline) (5 mg zanamivir with lactose) (61).
Luciferase Reporter Assay.
To quantitate fusion observed in the syncytia assay, Vero cell monolayers were transfected with 1 μg of pCAGGS-F, pCAGGS-HN, or the HN mutant DNA and pT7-luciferase, a plasmid that expresses firefly luciferase under T7 polymerase control. BSR-T7/5 cells were overlaid on the Vero cell monolayer at 15 h posttransfection and were incubated further for 6–7 h at 37 °C or 33 °C. Cells (0.5 mL) were lysed with 2× Reporter Lysis Buffer (Promega). Subsequently, the cell lysates were frozen overnight at −80 °C to facilitate lysis and release of luciferase. Cell debris was pelleted from the samples by centrifugation (2,000 × g for 10 min at room temperature), and 150 μL of the cleared lysates was added to a 96-well dish together with 150 μL of the luciferase assay substrate (Promega). The luciferase activity in relative light units then was determined using a SpectraMax M5 plate reader (Molecular Devices).
Electron Microscopy.
Solutions of PIV5 HN ectodomain (residues 56–565) (5 μg/mL) were absorbed onto 300-mesh copper grids covered with a carbon film that had been freshly glow discharged. Grids were stained with a 1% aqueous solution of uranyl formate, and protein was observed in a JEOL 1230 electron microscope operated at 100 kV. Images were acquired with a Gatan 831 CCD camera.
Supplementary Material
Acknowledgments
The mAbs specific for Newcastle disease virus HN protein were a generous gift from Ronald Iorio (University of Massachusetts Medical School). This research was supported in part by National Institutes of Health Research Grants AI-23173 (to R.A.L.) and GM-61050 (to T.S.J.). B.D.W. was an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
See Author Summary on page 15549 (volume 109, number 39).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213813109/-/DCSupplemental.
References
- 1.Heminway BR, Yu Y, Galinski MS. Paramyxovirus mediated cell fusion requires co-expression of both the fusion and hemagglutinin-neuraminidase glycoproteins. Virus Res. 1994;31:1–16. doi: 10.1016/0168-1702(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 2.Horvath CM, Paterson RG, Shaughnessy MA, Wood R, Lamb RA. Biological activity of paramyxovirus fusion proteins: Factors influencing formation of syncytia. J Virol. 1992;66:4564–4569. doi: 10.1128/jvi.66.7.4564-4569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu XL, Ray R, Compans RW. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol. 1992;66:1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison T, McQuain C, McGinnes L. Complementation between avirulent Newcastle disease virus and a fusion protein gene expressed from a retrovirus vector: Requirements for membrane fusion. J Virol. 1991;65:813–822. doi: 10.1128/jvi.65.2.813-822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao Q, Hu X, Compans RW. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J Virol. 1997;71:650–656. doi: 10.1128/jvi.71.1.650-656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin H-S, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb RA, Jardetzky TS. Structural basis of viral invasion: Lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowden TA, et al. Crystal structure and carbohydrate analysis of Nipah virus attachment glycoprotein: A template for antiviral and vaccine design. J Virol. 2008;82:11628–11636. doi: 10.1128/JVI.01344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 11.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 12.Hashiguchi T, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci USA. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence MC, et al. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 14.Xu K, et al. Host cell recognition by the henipaviruses: Crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci USA. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan P, et al. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci USA. 2011;108:14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan P, et al. Structural studies of the parainfluenza virius 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:1–13. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 17.Ng DT, Hiebert SW, Lamb RA. Different roles of individual N-linked oligosaccharide chains in folding, assembly, and transport of the simian virus 5 hemagglutinin-neuraminidase. Mol Cell Biol. 1990;10:1989–2001. doi: 10.1128/mcb.10.5.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan P, Leser GP, Demeler B, Lamb RA, Jardetzky TS. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology. 2008;378:282–291. doi: 10.1016/j.virol.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navaratnarajah CK, et al. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat Struct Mol Biol. 2011;18:128–134. doi: 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashiguchi T, et al. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol. 2011;18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 21.Bose S, et al. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol. 2011;85:12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maar D, et al. Cysteines in the stalk of the Nipah virus G glycoprotein are located in a distinct subdomain critical for fusion activation. J Virol. 2012;86:6632–6642. doi: 10.1128/JVI.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ader N, et al. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J Biol Chem. 2012;287:16324–16334. doi: 10.1074/jbc.M112.342493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porotto M, et al. The second receptor binding site of the globular head of the Newcastle disease virus hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J Virol. 2012;86:5730–5741. doi: 10.1128/JVI.06793-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paal T, et al. Probing the spatial organization of measles virus fusion complexes. J Virol. 2009;83:10480–10493. doi: 10.1128/JVI.01195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishop KA, et al. Residues in the stalk domain of the Hendra virus G glycoprotein modulate conformational changes associated with receptor binding. J Virol. 2008;82:11398–11409. doi: 10.1128/JVI.02654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bousse T, Takimoto T, Gorman WL, Takahashi T, Portner A. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology. 1994;204:506–514. doi: 10.1006/viro.1994.1564. [DOI] [PubMed] [Google Scholar]
- 28.Corey EA, Iorio RM. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J Virol. 2007;81:9900–9910. doi: 10.1128/JVI.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng R, et al. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology. 1999;253:43–54. doi: 10.1006/viro.1998.9501. [DOI] [PubMed] [Google Scholar]
- 30.Deng R, Wang Z, Mirza AM, Iorio RM. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology. 1995;209:457–469. doi: 10.1006/viro.1995.1278. [DOI] [PubMed] [Google Scholar]
- 31.Melanson VR, Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the Newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol. 2004;78:13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melanson VR, Iorio RM. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol. 2006;80:623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stone-Hulslander J, Morrison TG. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J Virol. 1999;73:3630–3637. doi: 10.1128/jvi.73.5.3630-3637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanabayashi K, Compans RW. Functional interaction of paramyxovirus glycoproteins: Identification of a domain in Sendai virus HN which promotes cell fusion. J Virol. 1996;70:6112–6118. doi: 10.1128/jvi.70.9.6112-6118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porotto M, et al. Spring-loaded model revisited: Paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J Virol. 2011;85:12867–12880. doi: 10.1128/JVI.05873-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iorio RM, Melanson VR, Mahon PJ. Glycoprotein interactions in paramyxovirus fusion. Future Virol. 2009;4:335–351. doi: 10.2217/fvl.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randall RE, Young DF, Goswami KKA, Russell WC. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J Gen Virol. 1987;68:2769–2780. doi: 10.1099/0022-1317-68-11-2769. [DOI] [PubMed] [Google Scholar]
- 38.Paterson RG, Russell CJ, Lamb RA. Fusion protein of the paramyxovirus SV5: Destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- 39.Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc Natl Acad Sci USA. 2006;103:17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iorio RM, Bratt MA. Monoclonal antibodies to Newcastle disease virus: Delineation of four epitopes on the HN glycoprotein. J Virol. 1983;48:440–450. doi: 10.1128/jvi.48.2.440-450.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aguilar HC, et al. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol. 2006;80:4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JK, et al. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem. 2008;283:16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plemper RK, Brindley MA, Iorio RM. Structural and mechanistic studies of measles virus illuminate paramyxovirus entry. PLoS Pathog. 2011;7:e1002058. doi: 10.1371/journal.ppat.1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paterson RG, Johnson ML, Lamb RA. Paramyxovirus fusion (F) protein and hemagglutinin-neuraminidase (HN) protein interactions: Intracellular retention of F and HN does not affect transport of the homotypic HN or F protein. Virology. 1997;237:1–9. doi: 10.1006/viro.1997.8759. [DOI] [PubMed] [Google Scholar]
- 45.Mahon PJ, Mirza AM, Musich TA, Iorio RM. Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein:Iimplications for the mechanism of fusion promotion. J Virol. 2008;82:10386–10396. doi: 10.1128/JVI.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paterson RG, Hiebert SW, Lamb RA. Expression at the cell surface of biologically active fusion and hemagglutinin/neuraminidase proteins of the paramyxovirus simian virus 5 from cloned cDNA. Proc Natl Acad Sci USA. 1985;82:7520–7524. doi: 10.1073/pnas.82.22.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim YH, et al. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc Natl Acad Sci USA. 2011;108:20992–20997. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biacchesi S, et al. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: Deletion of G yields a promising vaccine candidate. J Virol. 2004;78:12877–12887. doi: 10.1128/JVI.78.23.12877-12887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Techaarpornkul S, Collins PL, Peeples ME. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology. 2002;294:296–304. doi: 10.1006/viro.2001.1340. [DOI] [PubMed] [Google Scholar]
- 50.Feldman SA, Audet S, Beeler JA. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. 2000;74:6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang A, Masante C, Buchholz UJ, Dutch RE. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J Virol. 2012;86:3230–3243. doi: 10.1128/JVI.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sergel TA, McGinnes LW, Morrison TG. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J Virol. 2000;74:5101–5107. doi: 10.1128/jvi.74.11.5101-5107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rawling J, Cano O, Garcin D, Kolakofsky D, Melero JA. Recombinant Sendai viruses expressing fusion proteins with two furin cleavage sites mimic the syncytial and receptor-independent infection properties of respiratory syncytial virus. J Virol. 2011;85:2771–2780. doi: 10.1128/JVI.02065-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J, Quinlan E, Mirza A, Iorio RM. Mutated form of the Newcastle disease virus hemagglutinin-neuraminidase interacts with the homologous fusion protein despite deficiencies in both receptor recognition and fusion promotion. J Virol. 2004;78:5299–5310. doi: 10.1128/JVI.78.10.5299-5310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plemper RK, Hammond AL, Cattaneo R. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem. 2001;276:44239–44246. doi: 10.1074/jbc.M105967200. [DOI] [PubMed] [Google Scholar]
- 56.Tong S, Compans RW. Alternative mechanisms of interaction between homotypic and heterotypic parainfluenza virus HN and F proteins. J Gen Virol. 1999;80:107–115. doi: 10.1099/0022-1317-80-1-107. [DOI] [PubMed] [Google Scholar]
- 57.Aguilar HC, et al. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol. 2007;81:4520–4532. doi: 10.1128/JVI.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bishop KA, et al. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J Virol. 2007;81:5893–5901. doi: 10.1128/JVI.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plemper RK, Hammond AL, Gerlier D, Fielding AK, Cattaneo R. Strength of envelope protein interaction modulates cytopathicity of measles virus. J Virol. 2002;76:5051–5061. doi: 10.1128/JVI.76.10.5051-5061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsurudome M, et al. Hemagglutinin-neuraminidase-independent fusion activity of simian virus 5 fusion (F) proteinDdifference in conformation between fusogenic and nonfusogenic F proteins on the cell surface. J Virol. 2001;75:8999–9009. doi: 10.1128/JVI.75.19.8999-9009.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Porotto M, et al. Paramyxovirus receptor-binding molecules: Engagement of one site on the hemagglutinin-neuraminidase protein modulates activity at the second site. J Virol. 2006;80:1204–1213. doi: 10.1128/JVI.80.3.1204-1213.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]