Abstract
Fluorescence live-cell imaging has temporally resolved the conserved choreography of more than 30 proteins involved in clathrin and actin-mediated endocytic budding from the plasma membrane. However, the resolution of these studies is insufficient to unveil how the endocytic machinery actually drives membrane deformation in vivo. In this study, we use quantitative immuno-EM to introduce the temporal dimension to the ultrastructural analysis of membrane budding and define changes in the topography of the lipid bilayer coupled to the dynamics of endocytic proteins with unprecedented spatiotemporal resolution. Using this approach, we frame the emergence of membrane curvature with respect to the recruitment of endocytic factors and show that constriction of the invaginations correlates with translocation of membrane-sculpting proteins. Furthermore, we show that initial bending of the plasma membrane is independent of actin and clathrin polymerization and precedes building of an actin cap branched by the Arp2/3 complex. Finally, our data indicate that constriction and additional elongation of the endocytic profiles require the mechanochemical activity of the myosins-I. Altogether, this work provides major insights into the molecular mechanisms driving membrane deformation in a cellular context.
Keywords: Bin/Amphiphysin/Rvs proteins, Wiskott–Aldrich syndrome protein-interacting protein, amphiphysin, huntingtin interacting protein
Budding of transport intermediates from cellular membranes is an energetically unfavorable process that involves a variety of molecular machines. Protein coats, membrane-sculpting domains such as ENTHs (epsin N-terminal homology) and BARs (Bin/Amphiphysin/Rvs), enzymes that translocate or modify lipids, actin nucleating promoting factors (NPFs), and molecular motors participate in membrane deformation (1). Much information has been learned about how these proteins function in vitro. However, little is known about how they effectively bend the lipid bilayer in vivo.
Clathrin and actin-dependent endocytic budding from the plasma membrane (PM) is one of the best-characterized membrane traffic events, which requires more than 30 proteins (ref. 2 and references therein and ref. 3). A comprehensive description of the sequence of events leading to the generation of endocytic vesicles was first elucidated in Saccharomyces cerevisiae (Fig. 1A). Live-cell fluorescence microscopy in yeast has shown that clathrin and the early module composed of the scaffolding protein Ede1/Eps15 and the F-BAR (FCH-BAR) protein Syp1/FCHO1/2 (Fer/Cip4 homology domain only proteins 1 and 2) are the first to arrive at the endocytic sites (4–10). Up to 90 s later, the intermediate coat module formed by the yeast epsins Ent1/2 and the HIP1R (huntingtin interacting protein-related) homolog Sla2 is recruited to the patch (4, 9, 11, 12). The late coat components, such as the clathrin adaptor Sla1, and the NPFs Las17/WASP (Wiskott–Aldrich syndrome protein) and Pan1 immediately follow (9, 11, 13). Actin polymerization is initiated a few seconds later (11) on arrival of Bzz1/syndapin and Vrp1/WIP (WASP-interacting protein), two proteins that modulate the activity of the NPFs (13). After an initial stage of actin-dependent corralled motility, the endocytic coat initiates a slow movement into the cytosol, which reflects the elongation of a tubular invagination capped by a hemispherical clathrin coat (11, 14–16). Initiation of the inward movement coincides with the recruitment of the third NPF, the myosin-I Myo5, and initiation of massive actin polymerization (11, 13, 14, 17). Arrival of the amphiphysins Rvs161/167 precedes scission of the vesicles, which travel rapidly into the cytosol while still attached to actin (4, 18, 19). The myosin-I ATPase and actin nucleating promoting activity (NPA) are essential to power the slow inward movement of the coat, which also requires the NPA of Las17. In addition, Las17 and Pan1 play a function earlier in the process (13, 14). The need for different NPFs and their exact role in the process are not well-understood.
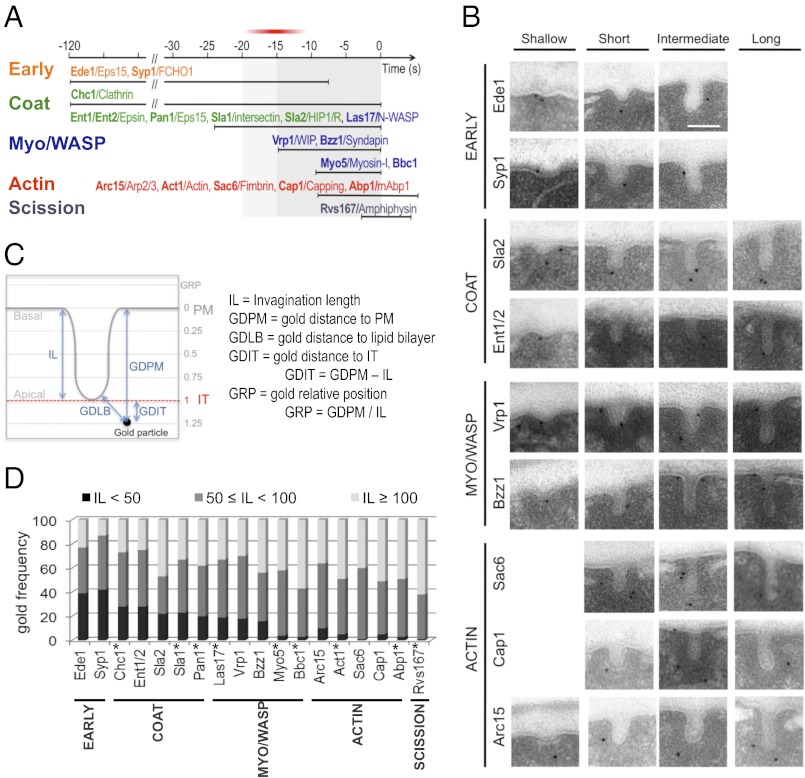
Fig. 1.
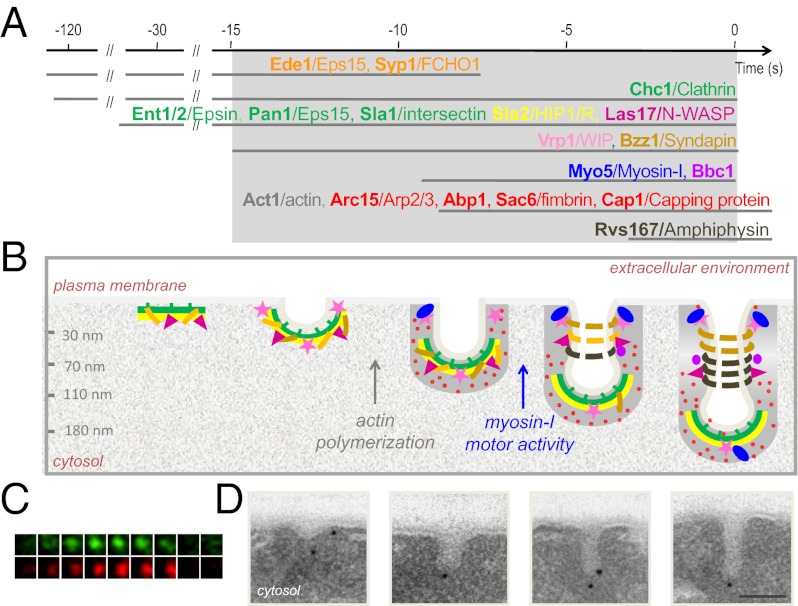
The invagination length can be used as a parameter to define the age of the endocytic profiles. (A) Proposed cortical dynamics of endocytic proteins included in the current statistical analysis based on or inferred from live-cell imaging (ref. 2 and references therein). The proteins are grouped in modules (4, 7) represented in different colors. The timing of clathrin recruitment with respect to the early module is not well-defined (4–10). The exact time at which the Arp2/3 complex is recruited with respect to other endocytic proteins is controversial (13, 17). Yeast proteins and their respective mammalian homologs are shown in bold and regular types, respectively. Time 0 corresponds to scission (18). The area highlighted in gray shows the broadest time window analyzed in this study. The red bar indicates when initial curvature may appear. (B) Representative immuno-EM micrographs of ultrathin sections from yeast showing endocytic invaginations of different sizes labeled with immunogolds against the HA tag versions of the indicated proteins. (Scale bar: 100 nm.) (C) Diagram illustrating the parameters used to analyze the position of the immunogolds. The levels of the PM and the invagination tip (IT) are indicated. (D) Relative frequency of immunogolds labeling the indicated proteins binned according to the IL. The number of immunogolds recorded for each protein (ng) was 120. SI Appendix, Fig. S1B shows the GLM analysis of the IL. Proteins labeled with an asterisk were analyzed in ref. 16 and are included as references.
Fluorescence live-cell imaging has established the basis for the molecular models explaining clathrin and actin-dependent endocytosis (20–22). However, the resolution offered by the most sophisticated versions of this technique is insufficient to detect changes in the morphology of the lipid bilayer coupled to the dynamics of endocytic proteins (23, 24). Thus, for instance, it is unknown when membrane curvature emerges with respect to the recruitment of endocytic factors.
Quantitative immuno-EM (QIEM) has shown that the primary endocytic profiles in yeast are tubular invaginations of up to 180 nm in length (16). Statistical analysis of the length of the invaginations labeled for different proteins shows that early endocytic factors are significantly more represented in the shortest invaginations compared with the late components, indicating that the length of the invagination can be used as a parameter to describe its age (16). In this work, we use QIEM to introduce the temporal dimension to the ultrastructural study of endocytic budding. Combined statistical analysis of parameters defining the shape of the endocytic profiles and the localization of immunogolds labeling 18 key endocytic proteins builds up a comprehensive and dynamic map of the endocytic machinery coupled to membrane deformation, with unprecedented spatiotemporal resolution. In addition, we investigate the morphology of endocytic profiles after interfering with key pieces of the endocytic machinery. Altogether, our data define the time window when membrane curvature first appears with respect to the recruitment of endocytic proteins and show that actin is required to elongate the incipient invaginations but not initially bend the lipid bilayer. Strikingly, we also find that clathrin does not play an essential structural role to maintain the cohesion of the endocytic coat in yeast or shape the endocytic invaginations. Finally, we show that constriction and growth of the endocytic profiles from 70 nm on correlate with the assembly of BAR proteins at the invagination neck and require the myosin-I ATPase.
Results and Discussion
Initial Bending of the PM Does Not Require Actin or Clathrin Polymerization and Coincides with the Recruitment of Vrp1 and Bzz1.
We have previously used QIEM to describe the localization of immunogolds labeling nine endocytic proteins along PM invaginations of increasing length (16). In that study, images of immunogolds labeling each endocytic protein were acquired under transmission electron microscope, and the length of the associated invaginations (IL) was recorded (16). Statistical comparison of the average IL for each protein showed that this parameter can be used to describe the age of the endocytic profile (16). Here, we used this approach to define when membrane curvature emerges with respect to the recruitment of the endocytic factors. We reasoned that proteins with similar disassembly dynamics recruited at different time points before or at emergence of membrane curvature should have similar IL distributions (SI Appendix, Fig. S1A). However, this parameter should significantly increase for proteins sequentially recruited during the elongation of the profiles (SI Appendix, Fig. S1A). We extended our analysis to a total of 18 proteins to cover the complete time window of endocytic budding (Fig. 1A). The factors analyzed in the current work included the components of the early module Ede1 and Syp1 (6–8, 10); Ent1/2 and Sla2, which are recruited after clathrin and slightly before Sla1, Pan1, and Las17 (4, 9, 11, 12); Bzz1 and Vrp1, which arrive about 10 s later (13); and finally, the Arp2/3 complex, Sac6, and Cap1, which are recruited with actin and disassembled after the endocytic patch has moved inside the cell (4, 11, 17, 19) (Fig. 1A).
Ultrathin sections from chemically fixed yeast expressing HA-tagged versions of these proteins were prepared and labeled with a monoclonal anti-HA and gold-labeled secondary antibodies (immunogolds). Images from 120 immunogolds in association with PM invaginations were collected for each protein (Fig. 1B), and the IL was measured for every immunogold (Fig. 1C). In addition, a number of parameters describing the position of the immunogold with respect to the invagination were recorded for ulterior analysis (Fig. 1C). Statistically significant differences among the IL distribution for different proteins were detected using a general linear model (GLM) (SI Appendix, Fig. S1B). All previously examined proteins (16) were included in the analysis as references after assessing the reproducibility of the methodology (Material and Methods).
The overall comparison of the IL distributions showed a good correlation between the dynamics of the proteins at the endocytic patch (Fig. 1A) and the presence of labeling on invaginations of increasing length (Fig. 1D and SI Appendix, Fig. S1 B and C). Only small deviations from the expected behavior were detected for Sla2 and Bbc1 (Fig. 1D and SI Appendix, Fig. S1 B and C) (16), which could reflect slight differences in the dynamics of the HA and GFP-tagged proteins. Interestingly, Vrp1, which is recruited ∼20 s before scission and halfway between the latest coat components and the actin module (13) (Fig. 1A), did not show significant differences with Sla1, Pan1, Ent1/2, Las17, or clathrin (Chc1), whereas it significantly differed from actin (Act1), Sac6, Abp1, and Cap1 (SI Appendix, Fig. S1B). These data indicated that the recruitment of Vrp1 either immediately follows or prompts membrane curvature. Consistent with Bzz1 arriving immediately after Vrp1 (13), Bzz1 had an IL distribution closest to the IL distribution of Vrp1 (Fig. 1D and SI Appendix, Fig. S1B), but it showed more similarities to the actin module (SI Appendix, Fig. S1B). These data framed our study within a time window of maximum 20 s before scission (Fig. 1A), and they indicate that the clathrin coat remains flat for up to 90 s after its recruitment to the PM. Because we did not analyze immunogolds associated with flat PM, events occurring before the recruitment of Vrp1 were necessarily invisible to our analysis. In this context, the differences observed for the IL distribution for the early module components Ede1 and Syp1 most likely reflected their early departure from the endocytic sites rather than their early arrival (6–9) (Fig. 1 A and D and SI Appendix, Fig. S1 A and B).
Bzz1 releases the inhibition imposed on Las17 by Sla1 (13), and binding of Vrp1 to Myo5 is essential for the myosin to develop its NPA (13, 25, 26). To test if actin polymerization triggered by Vrp1 and/or Bzz1 might prompt initial membrane curvature, the endocytic profiles were examined in cells expressing Sla1-HA treated with either the actin monomer sequestering drug Latrunculin A (LatA) (27) or DMSO. QIEM showed striking differences for the IL distributions of Sla1-HA in LatA and DMSO-treated cells (Fig. 2 A–C and SI Appendix, Table S1). Consistent with actin polymerization sustaining the slow inward movement of the coat (11), less than 20% of the Sla1-HA labeling was associated with invaginations longer than 40 nm in LatA-treated cells compared with 80% in mock-treated cells (Fig. 2C). LatA does not preclude assembly of the coat at the PM, which was shown by fluorescence microscopy (4, 5, 15), but the resolution of this technique is insufficient to discern the bending state of the underlying membrane. Our analysis showed that significant labeling for Sla1-HA was associated with shallow invaginations in actin-poisoned cells. They were bona fide endocytic structures, because they were also labeled with immunogolds against clathrin (Fig. 2A). These results showed that actin polymerization was critical for growth of the endocytic invaginations, but it was not strictly required to initiate membrane curvature. In agreement, invaginations shorter than 40 nm were poorly labeled for actin and actin-binding proteins compared with the endocytic coat components (16) (Fig. 1D and SI Appendix, Fig. S2).
Fig. 2.
Ultrastructure of the yeast endocytic profiles after interfering with actin and clathrin. (A and D) Immuno-EM micrographs showing representative PM invaginations labeled with immunogolds against Sla1-HA (α-Sla1-HA) or Clathrin (α-Chc1) on (A) WT cells treated with 200 μM LatA or DMSO or (D) chc1Δ cells and the isogenic WT (CHC1). (B and E) Scatter graphs representing the GDPM (gold distance to the plasma membrane) of 120 immunogolds against Sla1-HA vs. the corresponding IL (parameter definition in Fig. 1C) for each of the indicated strains and the experimental conditions described in A and D, respectively. The x axis corresponds to the level of the basal PM. The red dotted line indicates the position of the IT. The gray lines converging at the origin define constant GRP values. Blue and red circles indicate the control strains and the LatA-treated or mutant strains, respectively. (C and F) Histograms showing the relative frequency of Sla1-HA immunogolds from the scatter graphs shown in B and E, respectively, binned according to the length of the corresponding invaginations. Student t test P values comparing the length of Sla1-HA–labeled invaginations or the positional parameters of the corresponding immunogolds are described in SI Appendix, Table S1.
Clathrin polymerization was classically considered a driving force bending the lipid bilayer (28, 29), and its role as a scaffold for cargo adaptors is widely accepted (30). We have previously shown that the endocytic coat components, including clathrin, form a 40-nm hemisphere around the tip of the endocytic invaginations (16). QIEM analysis of other components of the endocytic coat module, such as the epsins Ent1/2 or the HIP1R homolog Sla2, also confirmed this view. More than 85% of the immunogolds labeling the coat components were at less than 40 nm from the invagination tip [GDIT (gold distance to the invagination tip) < 40 nm] and had GRPs (gold relative position) close to one (Fig. 1C, parameter definition, Fig. 3, and SI Appendix, Fig. S3 and Tables S2 and S3, GDIT). Therefore, we hypothesized that assembly and subsequent bending of the clathrin coat might be sufficient to generate curvature. To test this hypothesis, we analyzed the morphology of endocytic invaginations labeled for Sla1-HA in chc1∆ and WT yeast (Fig. 2 D–F). Surprisingly, depletion of clathrin did not significantly alter the length of the Sla1-HA–labeled invaginations or their morphology in any other obvious respect (Fig. 2 D–F). Interestingly, the analysis of the different positional parameters (parameter definition in Fig. 1C) for Sla1-HA did not reveal major changes in its localization on depletion of clathrin (SI Appendix, Table S1). This finding suggested that clathrin does not play an essential structural role in yeast to define the morphology or size of the endocytic coat, but rather, it might have a regulatory function (4, 31, 32). Sufficient concentration of membrane-sculpting BAR and ENTH domains held by scaffolding proteins, such as Ede1, Pan1, or Sla1, might be sufficient to initiate membrane curvature by steric crowding mechanism (33).
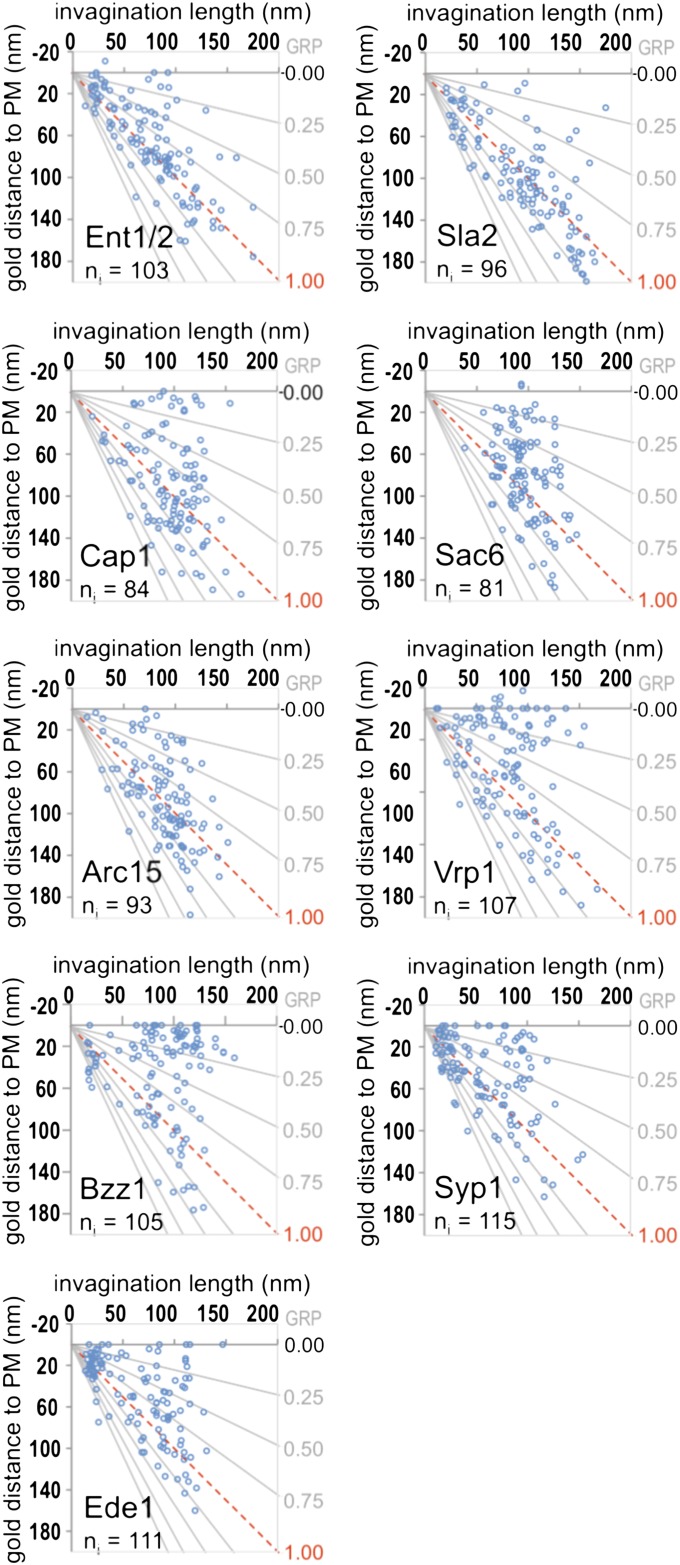
Fig. 3.
Localization of immunogolds labeling endocytic proteins on PM invaginations of increasing length. Scatter graphs showing the GDPM vs. the corresponding IL parameters (parameter definition in Fig. 1C) for each recorded immunogold labeling the indicated proteins. ng = 120 for each protein. The number of invaginations (ni) associated with the recorded immunogolds is indicated. The x axis corresponds to the level of the basal PM. The red dotted line indicates the position of the IT. The gray lines converging at the origin define constant GRP values.
Initial Actin Polymerization Builds a Branched Actin Network That Caps the Endocytic Coat and Moves with It as the Invagination Elongates.
We showed that actin polymerization was not required to sustain initial PM bending, but it was critical for elongation of the endocytic invaginations. Accordingly, invaginations longer than 40 nm show significant labeling for actin (16). This labeling is initially scattered over a dark ribosome-free area about 50-nm thick that surrounds the endocytic coat. Then, it extends to the neck as the profile elongates, and it finally splits into basal and apical pools on invaginations longer than 110 nm (16) (Fig. 4A).
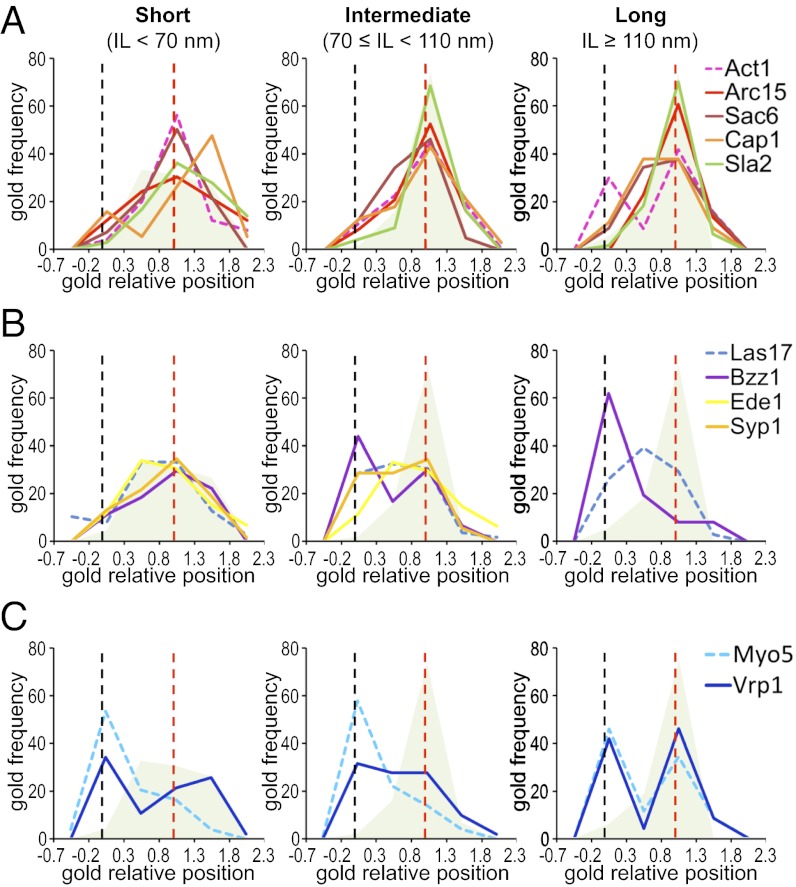
Fig. 4.
Comparative ultrastructural dynamics of endocytic proteins. (A–C) Graphs representing the frequency of immunogolds labeling the indicated proteins binned according to their GRP (parameter definition in Fig. 1C) associated with short (IL < 70 nm), intermediate (70 ≤ IL < 110 nm), and long (IL ≥ 110 nm) invaginations. ng = 120 for each protein. Dashed graphs correspond to proteins analyzed in ref. 16. The gray/green shadow depicts the distribution of Sla1, which indicates the position of the endocytic coat. The black and red dashed vertical lines indicate the positions of the basal PM and the IT, respectively. SI Appendix, Fig. S3 shows analogous GRP graphs, including all components of the endocytic modules analyzed so far by QIEM, and SI Appendix, Fig. S4 shows GRP, GDPM, and GDIT graphs for each of the newly analyzed endocytic factors. SI Appendix, Table S2 shows the homogeneity analysis of the GRP, GDIT, and GDPM parameters as a function of the IL, SI Appendix, Table S3 shows descriptive statistics, and SI Appendix, Table S4 shows the GLM analysis of the 18 endocytic factors.
To gain insight into the architecture of the endocytic actin structures, we analyzed the localization of proteins controlling its organization. These proteins included Sac6/fimbrin, which bundles actin filaments (34), the Arp2/3 complex, which nucleates and branches actin (35, 36), and Cap1/capping protein, which binds to the barbed, fast-growing ends of the filaments and prevents their growth (37, 38). Similar to actin, labeling for Sac6 and Cap1 was scattered around the invagination tip and spread to the base as the profile elongated (Figs. 3 and 4A and SI Appendix, Fig. S4). However, no obvious bimodal distribution could be observed for Sac6 or Cap1 in the longest profiles (Fig. 4A). Interestingly, a closer inspection of the labeling pattern for Cap1 indicated that the immunogolds were not randomly distributed, but rather, they seemed to be associated with the rims of the ribosome-free area, the coat, and the base of the invaginations (SI Appendix, Fig. S5). This Cap1 distribution suggested the presence of actin filaments with multiple polarities.
Similar to other members of the actin module, labeling for Arc15 initially appeared covering the endocytic coat (Figs. 3 and 4A). However, as the profiles elongated, Arc15 did not spread to the base, but it remained associated with the invagination tip (Figs. 3 and 4A and SI Appendix, Fig. S4 and Table S2). Thus, on invaginations longer than 110 nm, no labeling for Arc15 was detected close to the basal PM (GRP < 0.3), whereas more than 30% of the actin labeling was found within this area (Figs. 3 and 4A and SI Appendix, Table S3, GRP) (16). Over 90% of the Arc15 labeling always appeared at less than 70 nm from the invagination tip (GDIT < 70 nm) (Fig. 3 and SI Appendix, Fig. S4C and Table S3, GDIT), suggesting that a cap of branched actin filaments might initially form, which then moves together with the endocytic coat as the profile elongates. Interestingly, the endocytic coat component Sla2 showed no significant differences with Arc15 for any of the positional parameters analyzed, including the distance to the lipid bilayer [SI Appendix, Table S4, GDLB (gold distance to the lipid bilayer)]. The immunogolds against Sla2-HA were, on average, at 22.74 ± 17.85 nm from the lipid bilayer, similar to the actin-binding proteins and significantly different from the coat components, which had a GDLB close to 10 nm (SI Appendix, Tables S3 and S4). This localization was consistent with the proposed role of Sla2/HIP1R working as a linker between the endocytic coat and actin (32, 39, 40). It was proposed that a strong actin network stabilized by the Arp2/3 complex and Sac6 and cross-linked to the coat through Sla2 is important for force transduction during membrane invagination (20). Consistent with this view, deletion of Sla2 caused a very significant decrease in the IL (SI Appendix, Fig. S6). Compared with the WT, sla2Δ cells accumulated shallow and short invaginations surrounded by a thick electron-dense area that was heavily labeled for actin (SI Appendix, Fig. S6).
Translocation of F-BAR Proteins to the Invagination Neck Correlates with Its Constriction.
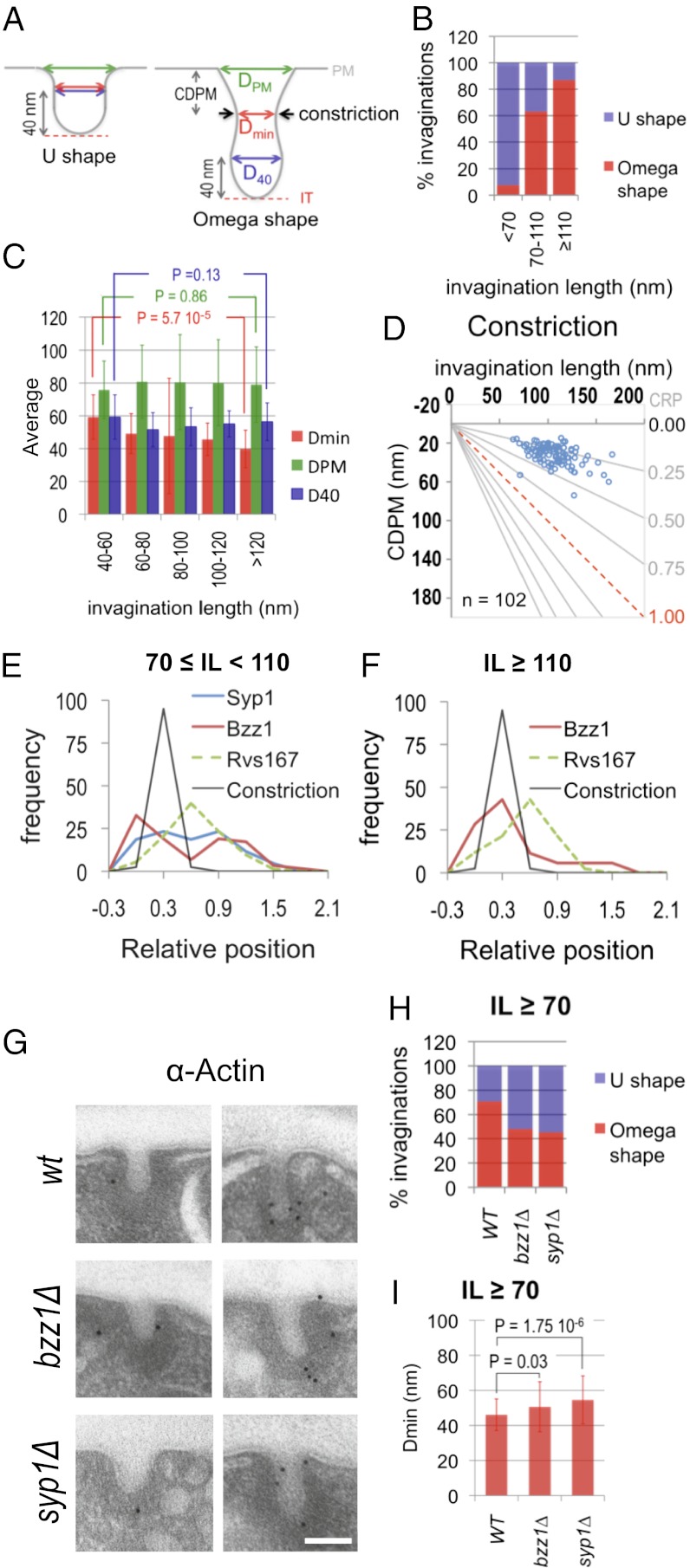
The observation that the Arp2/3 complex only occupies the tip of endocytic invaginations longer than 70 nm suggested a possible change in the mechanism of elongation of the profiles at this point. Accordingly, we detected important changes in the relative position (GRP) of several endocytic proteins at IL 70 nm using the Mann–Whitney Wilcoxon test (MWW) test. The most significant change in the GRP was observed for Bzz1 (SI Appendix, Table S2). Labeling for this protein initially occupied the invagination tip, where it colocalized with the endocytic coat and Las17 (Figs. 3 and 4B and SI Appendix, Table S3, GRP). In longer profiles, Bzz1 progressively moved to meet Myo5 at the invagination base (Figs. 3 and 4B and SI Appendix, Figs. S3 and S4 and Table S3, GRP). Interestingly, the MWW test also evidenced the displacement of the other F-BAR protein Syp1 at this point (SI Appendix, Table S2). In short profiles (IL < 70 nm), the GRP for Syp1 was close to one, identical to the GRP of the coat protein Sla1 (Figs. 3 and 4B and SI Appendix, Table S3, GRP). In longer profiles (IL ≥ 70 nm), Syp1 spread to the intermediate area of the invagination, where it colocalized with Las17 (16) (Figs. 3 and 4B and SI Appendix, Fig. S4C and Table S3, GRP). Although the variation in GRP for Syp1 was milder than the variation observed for Bzz1, its physiological relevance was corroborated by the observation that Ede1, a protein that physically interacts with Syp1 and shows identical dynamics at the endocytic patch (7, 10), behaved similarly (Figs. 3 and 4B and SI Appendix, Fig. S4C and Tables S2–S4). In agreement with the membrane-sculpting activity of F-BAR proteins (10, 41), we observed that their assembly around the invagination neck correlated with the constriction of the profiles. Whereas more than 90% of the invaginations shorter than 70 nm were U-shaped, most of the longest profiles exhibited an ω-contour (Figs. 1B and 5 A and B). Thus, the narrowest diameter along the invagination neck (Dmin) progressively decreased from 59.24 ± 13.52 nm (IL < 60 nm) to 39.79 ± 11.44 nm (IL > 120 nm) (Fig. 5C), whereas the diameter at the base of the endocytic coat (D40) or the level of the basal PM (DPM) remained constant (Fig. 5C).
Fig. 5.
Constriction of the endocytic invaginations correlates with the translocation of F-BAR proteins to the profile neck. (A) Diagram illustrating the two main shapes adopted by the yeast endocytic profiles. The invagination geometry is defined by Dmin, DPM, and D40. Invaginations were defined as ω-shaped when Dmin was smaller than D40. Black arrows indicate the position of the constriction. CDPM is the distance from the constriction to the PM. (B) Percentages of ω- vs. U-shaped invaginations as a function of their length. The number of invaginations analyzed was 26, 95, and 46 for the IL bins <70, 70–110, and ≥110 nm, respectively. Data were randomly collected from immuno-EM micrographs of invaginations of WT cells labeled with immunogolds against different HA-tagged endocytic proteins (n = 167). (C) Averages and SDs for the Dmin, DPM, and D40 of endocytic invaginations binned according to their length. Student t test P < 0.005 indicates a very significant difference. Invaginations analyzed were the same as in B. (D) Graph representing the CDPM plotted against the IL. n represents the number of the constricted invaginations analyzed. The x axis represents the level of the basal PM. The red dotted line indicates the positions of the IT. The gray lines converging at the origin define constant CRP values (CRP = CDPM/IL). (E and F) Graphs showing the relative frequency of immunogolds labeling Syp1, Bzz1, and Rvs167 binned according to their GRP and the relative frequency of the constriction according to its relative position (CRP) corresponding to invaginations with 70 ≤ IL < 110 nm (E) and IL ≥ 110 nm (F). Dashed graphs indicate proteins previously analyzed in ref. 16. SI Appendix, Table S5 shows the statistical analysis of the colocalization of the constriction with the BAR proteins. (G) Representative immuno-EM micrographs showing endocytic invaginations from WT, syp1∆, and bzz1∆ strains labeled with immunogolds against actin. (Scale bar: 100 nm.) (H) Percentages of ω- vs. U-shaped invaginations longer than 70 nm for the indicated strains. The number of the analyzed invaginations was 84, 100, and 106 for the WT, bzz1∆, and syp1∆ strains, respectively. (I) Average and SD for the Dmin of endocytic invaginations longer than 70 nm for the indicated strains. Invaginations analyzed were the same as in H. MWW test P < 0.005 indicates very significant differences.
To investigate which BAR protein might promote constriction, we compared their localization. We found that the constriction relative position [CRP; CRP = constriction distance to the PM (CDPM)/IL] was 0.30 ± 0.09 and did not significantly change as the profile elongated (Fig. 5D). Comparison of the CRP with the GRP of the BAR proteins indicated maximum coincidence with Bzz1, especially in the longest profiles (Fig. 5 E and F and SI Appendix, Table S5). However, a significant pool of Syp1 labeling also concentrated at this position when invaginations were shorter than 110 nm (Fig. 5E). No obvious overlap with Rvs167 could be detected (Fig. 5 E and F and SI Appendix, Table S5). Consistent with a role of Syp1 and Bzz1 in the constriction of the invagination neck, we observed that the percentage of ω-shaped profiles longer than 70 nm dropped from 71% in WT cells to 48% and 45% in bzz1Δ and syp1Δ strains, respectively (Fig. 5 G and H). Furthermore, depletion of Syp1 caused a very significant increase in the Dmin of invaginations longer than 70 nm (from 46 ± 12 nm in the WT to 55 ± 15 nm in syp1Δ cells) (Fig. 5 G and I). Depletion of Bzz1 had a milder effect on this parameter (Fig. 5 G and I).
Myosins-I Are Required for Elongation and Constriction of Endocytic Profiles.
The rearrangement of endocytic factors observed at IL 70 nm suggested that the Las17 NPA might be inhibited at this point. On invaginations longer than 70 nm, Las17 lost contact with its activator Bzz1 and spread to the neck to meet two inhibitors, Syp1 and Bbc1 (8, 13, 16) (Fig. 4B and SI Appendix, Fig. S3) (16). However, we have previously shown that Myo5 associates with the base of invaginations longer than 50 nm and the tip of profiles longer than 110 nm (16). To define which pool of myosin provided the predominant NPA in long profiles (IL > 70 nm), we analyzed the position of its coactivator Vrp1 (13). Interestingly, Vrp1 exhibited a bimodal distribution, which did not change as the profiles elongated (Figs. 3 and 4C and SI Appendix, Table S2, GRP). On short invaginations, a significant fraction of the Vrp1 labeling colocalized with the basal Myo5 (Fig. 4C). On profiles longer than 110 nm, Vrp1 and Myo5 showed no significant difference for any of the positional parameters (Fig. 4C and SI Appendix, Table S6). Together with previous observations (4, 13, 14, 42), our data strongly suggested that, after an initial burst of actin polymerization induced by Las17 and Pan1 on the endocytic coat, Myo5/Vrp1 take over to localize the predominant NPA at the base of the profiles. In the longest invaginations, a new burst of actin polymerization ignited by Myo5/Vrp1 on the tip might participate in vesicle fission (SI Appendix, Fig. S7).
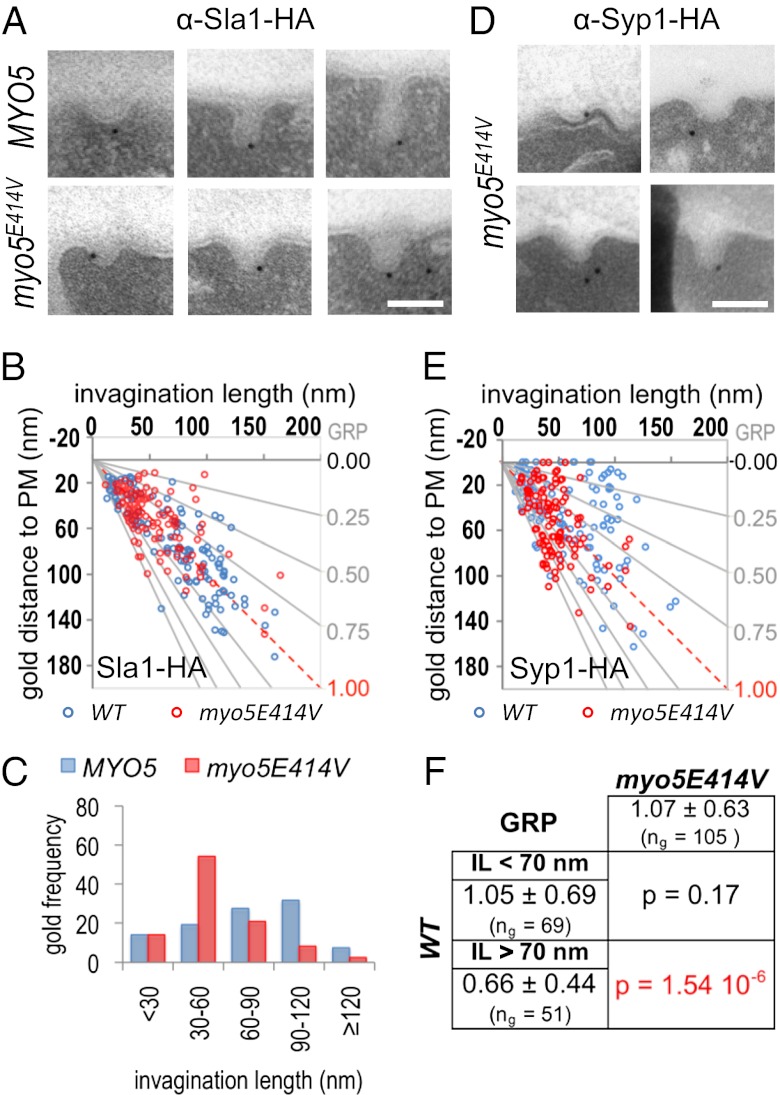
In addition to actin polymerization, the myosin-I ATPase is essential to sustain the slow movement of the coat into the cytosol (13). However, unlike actin polymerization, the molecular motor is most likely not required to sustain the corralled movement of the coat observed before the arrival of the myosin (13–15, 17). To characterize the corralled vs. slow inward movement and the function of the myosin ATPase at the ultrastructural level, we analyzed the morphology of Sla1-HA–labeled endocytic profiles in a double myosin-I deleted strain (myo3∆ myo5∆) expressing either the WT MYO5 or a mutant bearing the E414V substitution in a conserved residue of the ATPase active site (43). QIEM showed a significant difference in the IL of the myosin mutant compared with the WT (Fig. 6 A–C). The myo3∆ myo5-E414V strain accumulated endocytic invaginations of about 70 nm covered by the ribosome-free area (Fig. 6 A–C). The data indicated that building of the actin cap branched by the Arp2/3 complex might cause the corralled movement and promote growth of invaginations shorter than 70 nm, whereas the myosin ATPase would be required for farther directed elongation of the profiles. Interestingly, we also noticed that the invaginations in the myosin-I mutant were predominantly U-shaped (Fig. 6A), indicating that either the mechanochemical activity of Myo5 was directly required for the constriction of the invaginations and/or the elongation of the profiles powered by the molecular motor was necessary for the F-BAR proteins to translocate to the invagination neck. Consistent with the first hypothesis, we found that the basal pool of Myo5 colocalized with the position of the constriction (MWW, P > 0.02). However, consistent with the second possibility, QIEM showed that, unlike Syp1 in the WT strain (Figs. 1B and 3, Syp1), the F-BAR protein failed to move from the tip to the neck of the invaginations in the myosin ATPase mutant (Fig. 6 D–F).
Fig. 6.
Elongation and constriction of the endocytic profiles require the myosin-I ATPase. (A) Representative immuno-EM micrographs showing endocytic invaginations from a myo3Δ myo5Δ strain expressing either the WT MYO5 or the myo5E414V mutant labeled with immunogolds against Sla1-HA. (B) Scatter graph representing the GDPM of immunogolds labeling Sla1-HA in the strains described in A vs. the corresponding IL (parameter definition in Fig. 1). ng = 120 for each strain. (C) Histogram representing the relative frequency of the Sla1-HA immunogolds from B binned according to the IL. The MWW test showed very significant differences (P < 0.001) for the Sla1-HA IL distribution of the indicated strains. (D) Representative micrographs showing endocytic invaginations from a myo3Δ myo5Δ strain expressing the myo5E414V mutant labeled with immunogolds against Syp1-HA. (E) Scatter graph representing the GDPM of 120 immunogolds labeling Syp1-HA for the strain described in D (red circles) vs. the corresponding IL (parameter definition in Fig. 1). For comparison, the graph was superimposed on the graph showing the position of Syp1-HA in WT cells (blue circles) (Fig. 3). (F) Statistical comparison of the Syp1-HA GRP in the myo3Δ myo5E414V mutant vs. the GRP of Syp1-HA in a WT strain for invaginations shorter (IL < 70 nm) or longer (IL > 70 nm) than 70 nm. The average GRP, SD, and number of immunogolds analyzed (ng) for each population are indicated. MWW tests (P < 0.005) show very significant differences and are highlighted in red.
Conclusions
Altogether, our data provide detailed positional information on 18 proteins coupled to membrane deformation during endocytic budding from the PM in a cellular context. Combining immunogold labeling, high-resolution transmission EM, and statistics allowed for description of changes in the membrane topography coupled to the dynamics of endocytic proteins, with a resolution down to 7 nm (SI Appendix, Tables S3, GDLB, and S4, GDLB). Such resolution has not yet been reached by emerging techniques using correlative EM and fluorescence microscopy, which were recently applied to the study of endocytic budding in yeast (18). Those EM approaches used high-pressure freezing and freeze substitution to fix yeast, and they probably preserve the ultrastructure of endocytic profiles better than chemical fixation (18, 41). However, immunolabeling of endocytic structures on these samples is inefficient (44), and therefore, it is not feasible at the moment to use this technique to apply a statistical analysis of positional parameters describing the dynamics of a comprehensive set of endocytic proteins at the ultrastructural level. Thus, our methodology offers a good compromise to describe changes in the topography of the lipid bilayer coupled to the dynamics of endocytic proteins. In any case, our fixation protocol may slightly distort the endocytic structures, and therefore, care should be taken not to conclude that our measurements exactly reflect the morphology of the endocytic invaginations and the position of the proteins involved in the budding process. In the future, the use of cryo-EM of vitreous sections will help define the exact morphology of the endocytic invaginations.
Live imaging studies have been the key to initially define the functional modules involved in the process, both in yeast and mammals (3, 4). Our ultrastructural analysis has now confirmed the existence of some modules at a higher resolution and unveiled higher levels of complexity for others (SI Appendix, Fig. S3). Thus, we have confirmed that the components of the early module or the endocytic coat (4, 12) show nearly indistinguishable behaviors for any of the parameters analyzed in our study (SI Appendix, Fig. S3 and Tables S2 and S4), and therefore, they can also be considered as functional modules at the ultrastructural level. Our analysis has also helped define their function by describing their dynamics at much higher resolution and coupling them to membrane deformation (Fig. 7 and SI Appendix, Fig. S3). However, we found that the components of the previously defined WASP/Myo and actin modules have a much more complex behavior, which could not be anticipated by other techniques (SI Appendix, Fig. S3). The data presented here provide valuable information regarding when and where physiological interactions occur, which significantly helps to refine the current models explaining the process (Fig. 7).
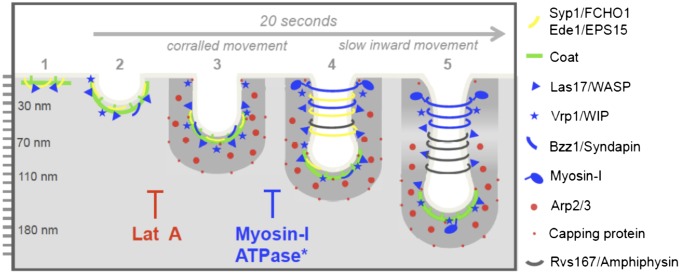
Fig. 7.
Scheme of ultrastructural dynamics of endocytic proteins during endocytic budding. The scheme shows the dynamic localization of yeast endocytic proteins along growing endocytic profiles based on ref. 16 and this study. Critical steps during the endocytic process, which are blocked by interfering with actin dynamics (LatA) or the ATPase activity of myosins-I, are shown.
Materials and Methods
Yeast and Plasmids.
Yeast strains and plasmids used in this study are listed in SI Appendix, Table S7.
Preparation of Yeast Cells for Immuno-EM.
Yeast cells were fixed using 4% formaldehyde and 0.4% glutaraldehyde and processed for immuno-EM as previously described (16). For LatA treatment, cells were concentrated by filtration to 2 × 108 cells/mL and then incubated with 200 μM LatA in DMSO or an equivalent amount of DMSO for 20 min at 26 °C before fixation. For immunolabeling, anti-HA rat monoclonal antibody 3F10 from Roche or antiactin rat monoclonal antibody MAC237 (Abcam) were used as primary antibodies, and 12-nm gold-conjugated anti-rat IgG antibody (Jackson ImmunoResearch Laboratories) was used as secondary antibody. Clathrin labeling was performed as described (16) using 20-nm gold-conjugated antibodies. Ultrathin sections were examined using a Tecnai Spirit transmission electron microscope (FEI Company) at 120 kV accelerating voltage. Micrographs were acquired at 100,000× magnification using a CCD camera (MegaView III; Olympus) and the image acquisition analySIS software (Olympus). The specificity of the anti-HA labeling on PM invaginations was assessed as described (16) for the least abundant protein at the endocytic patch (Las17) and the most transiently recruited protein (Rvs167) (16). The maximum estimated distance from the epitope to the gold particle is 20–30 nm (45).
Statistical Analysis of the Immunogold Distribution.
All statistical analyses were performed using SAS System v9.2 (SAS Institute). Comparison of proteins with respect to the different parameters describing the position of the immunogolds labeling them was performed using either a nonparametric MWW (46) or a GLM (47). To test for the reproducibility of the immunolabeling patterns, labeling for Abp1-HA and Sla1-HA was repeated and compared with previously published data (16). Student t tests showed no significant differences between these data and the published immunogold populations for any of the parameters analyzed (P > 0.02), indicating that the results are independent of the strain background or the experimental handling. Bins of equal lengths were used to depict the frequency of gold particles according to the invagination length. Cutoffs at 70 and 110 nm were used to depict the changes in the distribution of endocytic proteins along the PM invaginations. The most appropriate cutoffs were defined by comparing the population of immunogolds according to the GRP that was associated with invaginations of different lengths using the MWW test. The cutoffs showing the most significant differences were used.
Supplementary Material
Acknowledgments
We thank M. Cid and V. Robles; M. Pons for technical assistance; M. Borras and V. Paradicci for strain construction; G. Martínez, Y. Muela, N. Hernandez, L. Delgado, and C. López for assistance with the EM; and J. Casanova, M. Llimargas, S. Lemmon, M. Aldea, J. Giblin, and I. Fernández for critical reading of the manuscript. F.-Z.I. had a Ramón y Cajal Contract until May, 2011. This work was supported by Grants MTM2009-10893 (to A.E.), and BFU2008-03500, BFU2011-30185, and CSD2009-00016 (to M.I.G.) from the Spanish Government.
Footnotes
Conflict of interest statement: Sandra Lemmon (University of Miami Miller School of Medicine) coauthored a paper with F.-Z.I. and M.I.G. during the last 24 months, and therefore, she should be excluded as a referee.
This article is a PNAS Direct Submission.
See Author Summary on page 15541 (volume 109, number 39).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202789109/-/DCSupplemental.
References
- 1.Farsad K, De Camilli P. Mechanisms of membrane deformation. Curr Opin Cell Biol. 2003;15:372–381. doi: 10.1016/s0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22:1–13. [Google Scholar]
- 3.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 5.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Toshima JY, et al. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc Natl Acad Sci USA. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stimpson HE, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boettner DR, et al. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol. 2009;19:1979–1987. doi: 10.1016/j.cub.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll SY, et al. Analysis of yeast endocytic site formation and maturation through a regulatory transition point. Mol Biol Cell. 2012;23:657–668. doi: 10.1091/mbc.E11-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reider A, et al. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 12.Toret CP, Lee L, Sekiya-Kawasaki M, Drubin DG. Multiple pathways regulate endocytic coat disassembly in Saccharomyces cerevisiae for optimal downstream trafficking. Traffic. 2008;9:848–859. doi: 10.1111/j.1600-0854.2008.00726.x. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Galletta BJ, Chuang DY, Cooper JA. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K, et al. Actin-based motility during endocytosis in budding yeast. Mol Biol Cell. 2006;17:1354–1363. doi: 10.1091/mbc.E05-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Idrissi FZ, et al. Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J Cell Biol. 2008;180:1219–1232. doi: 10.1083/jcb.200708060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonsdottir GA, Li R. Dynamics of yeast Myosin I: Evidence for a possible role in scission of endocytic vesicles. Curr Biol. 2004;14:1604–1609. doi: 10.1016/j.cub.2004.08.055. [DOI] [PubMed] [Google Scholar]
- 18.Kukulski W, et al. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huckaba TM, Gay AC, Pantalena LF, Yang HC, Pon LA. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 2004;167:519–530. doi: 10.1083/jcb.200404173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 21.Galletta BJ, Mooren OL, Cooper JA. Actin dynamics and endocytosis in yeast and mammals. Curr Opin Biotechnol. 2010;21:604–610. doi: 10.1016/j.copbio.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boettner DR, Chi RJ, Lemmon SK. Lessons from yeast for clathrin-mediated endocytosis. Nat Cell Biol. 2012;14:2–10. doi: 10.1038/ncb2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu M, et al. Coupling between clathrin-dependent endocytic budding and F-BAR-dependent tubulation in a cell-free system. Nat Cell Biol. 2010;12:902–908. doi: 10.1038/ncb2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geli MI, Lombardi R, Schmelzl B, Riezman H. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 2000;19:4281–4291. doi: 10.1093/emboj/19.16.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lechler T, Shevchenko A, Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayscough KR, et al. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinrichsen L, Meyerholz A, Groos S, Ungewickell EJ. Bending a membrane: How clathrin affects budding. Proc Natl Acad Sci USA. 2006;103:8715–8720. doi: 10.1073/pnas.0600312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dannhauser PN, Ungewickell EJ. Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat Cell Biol. 2012;14:634–639. doi: 10.1038/ncb2478. [DOI] [PubMed] [Google Scholar]
- 30.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 31.Newpher TM, Idrissi FZ, Geli MI, Lemmon SK. Novel function of clathrin light chain in promoting endocytic vesicle formation. Mol Biol Cell. 2006;17:4343–4352. doi: 10.1091/mbc.E06-07-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boettner DR, Friesen H, Andrews B, Lemmon SK. Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Mol Biol Cell. 2011;22:3699–3714. doi: 10.1091/mbc.E11-07-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stachowiak JC, Hayden CC, Sasaki DY. Steric confinement of proteins on lipid membranes can drive curvature and tubulation. Proc Natl Acad Sci USA. 2010;107:7781–7786. doi: 10.1073/pnas.0913306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skau CT, et al. Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J Biol Chem. 2011;286:26964–26977. doi: 10.1074/jbc.M111.239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goode BL, Rodal AA, Barnes G, Drubin DG. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J Cell Biol. 2001;153:627–634. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim K, Yamashita A, Wear MA, Maéda Y, Cooper JA. Capping protein binding to actin in yeast: Biochemical mechanism and physiological relevance. J Cell Biol. 2004;164:567–580. doi: 10.1083/jcb.200308061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim T, Cooper JA, Sept D. The interaction of capping protein with the barbed end of the actin filament. J Mol Biol. 2010;404:794–802. doi: 10.1016/j.jmb.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engqvist-Goldstein AE, et al. The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J Cell Biol. 2001;154:1209–1223. doi: 10.1083/jcb.200106089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, Kaksonen M, Madden DT, Schekman R, Drubin DG. Interaction of Sla2p’s ANTH domain with PtdIns(4,5)P2 is important for actin-dependent endocytic internalization. Mol Biol Cell. 2005;16:717–730. doi: 10.1091/mbc.E04-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kishimoto T, et al. Determinants of endocytic membrane geometry, stability, and scission. Proc Natl Acad Sci USA. 2011;108:E979–E988. doi: 10.1073/pnas.1113413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barker SL, et al. Interaction of the endocytic scaffold protein Pan1 with the type I myosins contributes to the late stages of endocytosis. Mol Biol Cell. 2007;18:2893–2903. doi: 10.1091/mbc.E07-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman AL, Geeves MA, Manstein DJ, Spudich JA. Kinetic characterization of myosin head fragments with long-lived myosin.ATP states. Biochemistry. 1998;37:9679–9687. doi: 10.1021/bi973143f. [DOI] [PubMed] [Google Scholar]
- 44.Buser C, McDonald K. Correlative GFP-immunoelectron microscopy in yeast. Methods Enzymol. 2010;470:603–618. doi: 10.1016/S0076-6879(10)70024-0. [DOI] [PubMed] [Google Scholar]
- 45.Hermann R, Walther P, Müller M. Immunogold labeling in scanning electron microscopy. Histochem Cell Biol. 1996;106:31–39. doi: 10.1007/BF02473200. [DOI] [PubMed] [Google Scholar]
- 46.Hollander M, Wolfe DA. Nonparametric Statistical Methods. New York: Wiley; 1973. [Google Scholar]
- 47.Draper N, Smith H. Applied Regression Analysis. New York: Wiley; 1981. [Google Scholar]