Abstract
Breast cancer progression involves genetic changes and changes in the extracellular matrix (ECM). To test the importance of the ECM in tumor cell dissemination, we cultured epithelium from primary human breast carcinomas in different ECM gels. We used basement membrane gels to model the normal microenvironment and collagen I to model the stromal ECM. In basement membrane gels, malignant epithelium either was indolent or grew collectively, without protrusions. In collagen I, epithelium from the same tumor invaded with protrusions and disseminated cells. Importantly, collagen I induced a similar initial response of protrusions and dissemination in both normal and malignant mammary epithelium. However, dissemination of normal cells into collagen I was transient and ceased as laminin 111 localized to the basal surface, whereas dissemination of carcinoma cells was sustained throughout culture, and laminin 111 was not detected. Despite the large impact of ECM on migration strategy, transcriptome analysis of our 3D cultures revealed few ECM-dependent changes in RNA expression. However, we observed many differences between normal and malignant epithelium, including reduced expression of cell-adhesion genes in tumors. Therefore, we tested whether deletion of an adhesion gene could induce sustained dissemination of nontransformed cells into collagen I. We found that deletion of P-cadherin was sufficient for sustained dissemination, but exclusively into collagen I. Our data reveal that metastatic tumors preferentially disseminate in specific ECM microenvironments. Furthermore, these data suggest that breaks in the basement membrane could induce invasion and dissemination via the resulting direct contact between cancer cells and collagen I.
Collective cell migration is an important mechanism for both normal epithelial development and cancer invasion (1). During collective cell migration, cells move in coordinated groups and maintain cell–cell adhesion. In the normal mammary gland, ducts transition from a polarized bilayer into a proliferative, motile, multilayered epithelium and then migrate collectively through the stromal tissue (2, 3). Mammary carcinomas also originate from a polarized adult epithelium, transition from a simple to multilayered organization, and migrate collectively (4, 5). Despite these similarities, normal ductal morphogenesis in vivo does not involve local dissemination of cells and eventually results in restoration of polarized simple epithelial architecture. In contrast, breast carcinomas continue to grow, disseminate cells locally, and frequently metastasize to distant sites (6). These observations raise the fundamental question: What features of tumor progression can regulate the transition from a collective to a disseminative phenotype?
Cancer is a genetic disease, and sequencing has revealed that genes encoding cell–cell and cell–matrix adhesion proteins frequently are mutated (7, 8). However, breast cancer also involves characteristic changes in the ECM and the tumor microenvironment (9–12). For example, collagen I is enriched and aligned at the stromal border in breast tumors (10, 13), changes in collagen I organization are independent negative prognostic indicators (14), and increased collagen I crosslinking accelerates progression in experimental cancer models (15). Additionally, basement membrane proteins and their integrin receptors have been shown to regulate carcinoma cell behavior (16–18). A major challenge today is to distinguish the relative contributions of specific genetic and microenvironmental changes to the migration and local dissemination of carcinoma cells.
In vivo, there are vast differences in the soluble signals, the stromal cells, and the ECM microenvironments surrounding carcinomas and normal ducts (9). It is difficult to manipulate these signals independently in an intact tumor and even more challenging to assess the acute cell behavioral consequences of experimental manipulations. The relative optical inaccessibility of mammalian tissues led our laboratory and others to establish 3D ex vivo models of both normal and malignant mammary epithelial growth (5, 19–24). We have applied these techniques to test the relative importance of genetic and microenvironmental changes in regulating the pattern of collective cell migration and the likelihood of local dissemination.
Results
An epithelial cell in a mammary duct exists in a highly structured 3D environment and receives extensive inputs from cell–cell, cell–matrix, and soluble signals. We previously identified the critical conditions that enable primary mammary epithelium to undergo an organotypic program of branching morphogenesis (2). We found that, despite extensive cell migration, normal mammary morphogenesis in 3D Matrigel cultures and in vivo occurs without ECM-directed protrusions (2, 3). In contrast, carcinomas in vivo can migrate with protrusions and can disseminate cells locally and to distant sites (6, 25). Because the tumor microenvironment changes in parallel with genetic changes in the cancer cells (10), it is unclear whether the protrusive migration and dissemination of carcinoma cells are the result of cell-intrinsic motility differences or of interactions of the cancer cells with their microenvironment. Therefore, we exploited organotypic culture techniques to isolate and culture fragments from individual primary human mammary carcinomas in different ECM microenvironments (Fig. 1A and SI Materials and Methods). We first optimized the medium conditions to yield consistent branching morphogenesis in samples of normal human breast epithelium (Fig. S1 A–D). We then focused on two ECM environments: a gel composed of basement membrane proteins (Matrigel) to model the normal breast epithelial microenvironment and 3 mg/mL collagen I to model the stromal matrix encountered by invading mammary carcinomas (10). Although fibrillar collagen I is present near normal mammary ducts, it remains outside an intact basement membrane even during branching morphogenesis, limiting contact with normal epithelial cells (26).
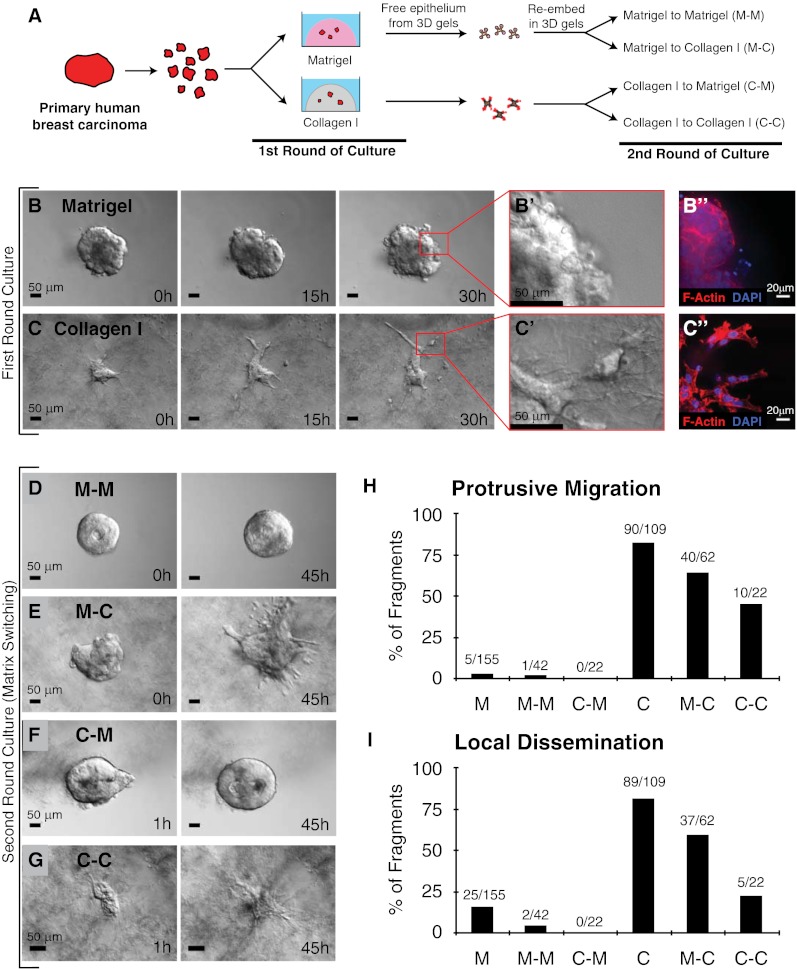
Fig. 1.
ECM microenvironments modulate the pattern of collective migration and local dissemination in human mammary carcinomas. (A) Schematic description of isolation and 3D culture of human mammary carcinoma fragments. In the first round of culture, tumor fragments were embedded in either 3D Matrigel or collagen I. In the second round of culture, the same tumor fragments were freed from the 3D gels and were re-embedded in the same or were swapped to the other microenvironment. (B–C′′) Representative DIC time-lapse sequences of human mammary carcinomas in Matrigel (B) or collagen I (C). (B′ and C′) Enlarged views of B and C at 30 h showing the smooth and protrusive leading fronts, respectively. (B′′ and C′′) Micrographs of the border of the same mammary carcinoma embedded in Matrigel or collagen and stained with phalloidin–F-actin and DAPI. (D–G) Representative frames of DIC time-lapse movies of human mammary carcinomas switched from Matrigel to Matrigel (M–M) (D), Matrigel to collagen I (M–C) (E), collagen I to Matrigel (C–M) (F), or collagen I to collagen I (C–C) (G) at 0 or 1 h in culture (Left) or 45 h in culture (Right). (H and I) Bar graphs showing the number of tumor fragments in each ECM condition with protrusive migration (H) or local dissemination (E) relative to the number of primary human tumor fragments analyzed in each condition.
Human Mammary Carcinomas Invade and Disseminate Preferentially into Collagen I.
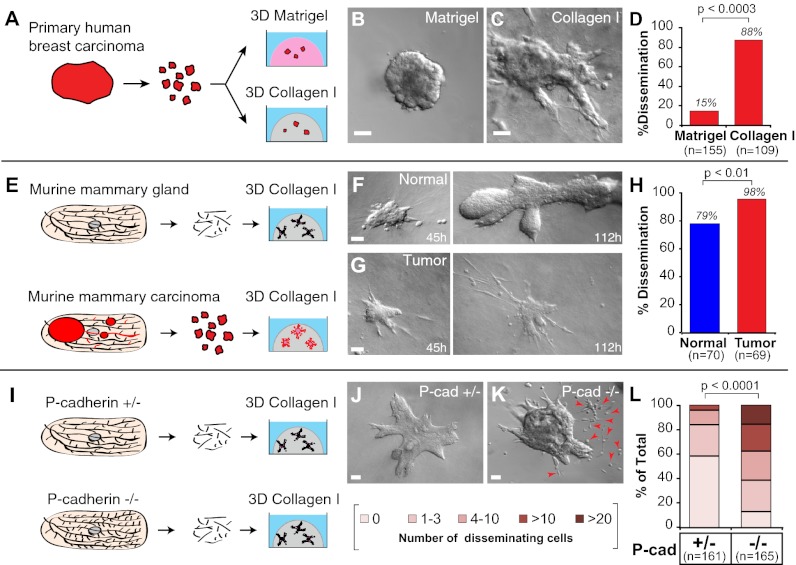
We explanted fragments from primary human mammary carcinomas (n = 7 tumors) (Fig. S1E and SI Materials and Methods) into 3D ECM cultures (Fig. 1A). The starting point for culture was epithelial fragments of a few hundred to a few thousand cells. We allocated fragments of the same tumor to different 3D ECM microenvironments and observed ECM-dependent carcinoma migration strategies (Fig. 1 B–C′′). In 3D Matrigel, we observed both indolent behavior and collective epithelial migration (Fig. 1 B–B′′). We observed single-cell protrusions from the epithelium in Matrigel only rarely and did not observe robust collective protrusive migration. In contrast, fragments from the same primary human mammary carcinoma exhibited protrusive migration and disseminated cells extensively into 3D gels of 3 mg/mL collagen I (Fig. 1 C–C′′, H, and I). Although the extent of invasion and dissemination varied among tumor fragments (Fig. S2 A–C), the borders of carcinoma fragments cultured in Matrigel maintained an epithelial appearance without protrusions (150/155 fragments from five human tumors) (Fig. 1H), whereas the borders of carcinoma fragments in collagen I were protrusive (90/109 fragments from five human tumors) and exhibited extensive local dissemination (89/109 fragments) (Fig. 1 H and I).
Current, Local ECM Microenvironment Determines the Cellular Strategy of Invasion for Primary Human Mammary Carcinomas.
We next sought to determine whether protrusive migration and dissemination could be reversed if the ECM composition returned to basement membrane-like composition. To test this concept, we first cultured primary human tumor fragments in either Matrigel or collagen I until the pattern of migration was clear and then digested the ECM and transferred the tumor fragments to another ECM environment (Fig. 1 D–G). We tested all reciprocal combinations, including transfer from Matrigel to Matrigel, from Matrigel to collagen I, from collagen I to Matrigel, and from collagen I to collagen I. Transfer between Matrigel and Matrigel resulted in a restarting of collective migration (Fig. 1D). Transfer from Matrigel to collagen I resulted in protrusive migration in the new environment (Fig. 1E), whereas transfer from collagen I to Matrigel resulted in a retraction of protrusions and confined, collective growth (Fig. 1F). Carcinoma fragments transferred from collagen I to collagen I were protrusive but on average were less disseminative (Fig. 1 G–I), suggesting there may be a limited subpopulation of highly invasive cells in a tumor. However, we cannot exclude the possibility that the more extensive enzymatic digestion required to free epithelial fragments from a collagen I gel might have reduced their invasive behavior. We therefore conclude that the current, local ECM environment determines the migration strategy and likelihood of dissemination.
Collagen I Induces Protrusive Migration and Local Dissemination of Murine Carcinoma Cells.
Our investigations with human breast carcinomas suggested that the local ECM microenvironment is sufficient to induce or repress protrusive and disseminative behavior. However, live primary human carcinoma tissue is scarce, and the details of tumor pathology vary widely from one available sample to the next (Fig. S1E). Therefore we modeled this regulatory interaction using mouse mammary carcinomas. We selected a mammary carcinoma model in which the mouse mammary tumor virus long terminal repeat drives the expression of the polyomavirus middle T oncogene (MMTV-PyMT), because it exhibits progressive cellular and molecular changes that parallel those observed in human breast cancer (27, 28). Gene expression in this model clusters with the highly aggressive luminal B subtype of human breast cancer (29).
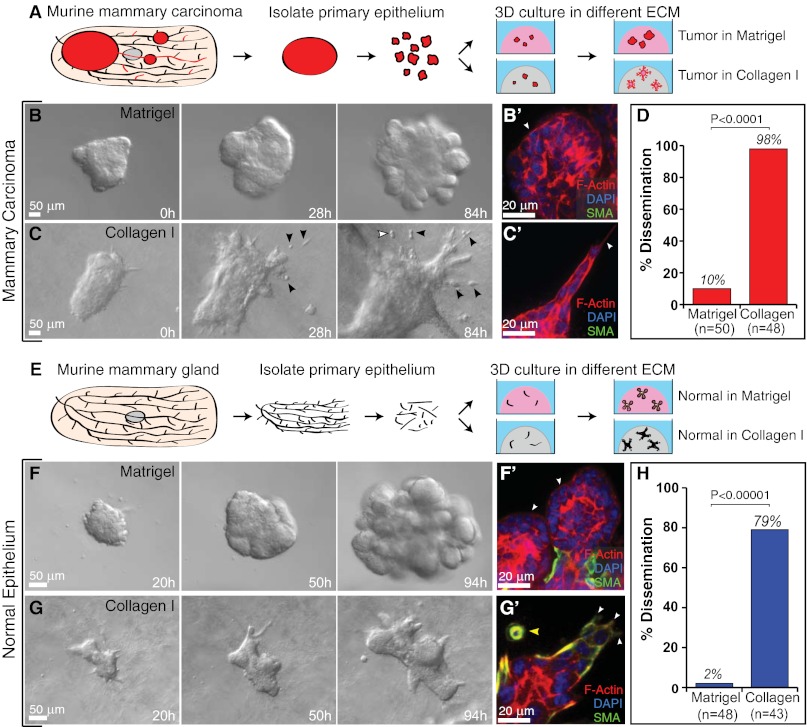
We isolated epithelial fragments of 200–1,000 cells from advanced murine mammary carcinomas (12–15 wk, 1.5- to 2-cm tumors) and embedded them into Matrigel or collagen I (Fig. 2A). Carcinoma fragments in Matrigel culture developed into budded structures with high efficiency (Fig. 2B). Cells within these fragments remained in a stratified organization, without lumens, throughout their time in Matrigel culture, and cells at the ECM border maintained an epithelial appearance, without protrusions (Fig. 2 B and B′). Although the MMTV-PyMT model metastasizes to the lungs with high efficiency in vivo, 90% of the carcinoma fragments did not disseminate cells into Matrigel (Fig. 2D and 45/50 movies). In contrast, carcinoma fragments embedded in 3 mg/mL collagen I developed extensive protrusions and frequently (98%) disseminated cells into collagen I (Fig. 2 C–D and 47/48 movies). Both the protrusions and dissemination were detectable by transmitted light microscopy (Fig. 2 C and C′). We conclude that the ECM microenvironment determines the collective cell migratory strategy and the likelihood of local dissemination in both human breast cancer cells and murine mammary carcinoma models.
Fig. 2.
The ECM governs the migratory pattern and disseminative behavior of both tumor and normal murine mammary epithelium. (A) Schematic description of isolation and 3D culture of murine tumor fragments. (B and C) Representative frames of DIC time-lapse movies of tumor fragments in Matrigel (B) and collagen I (C). Black arrowheads indicate disseminated cells, some of which are observed to proliferate (white arrowhead). (B′ and C′) Localization of actin, SMA, and DAPI in tumor fragments in Matrigel (B′) and collagen I (C′). White arrowheads mark the leading fronts. (D) Percent of tumor fragments showing cell dissemination in Matrigel and collagen I. n, total number of movies (four biological replicates, Student's t test, two-tailed, unequal variance). (E) Schematic description of isolation and 3D culture of normal mammary organoids. (F and G) Representative frames from DIC time-lapse movies of normal organoids in Matrigel (F) and collagen I (G). (F′ and G′) Localization of actin, SMA, and DAPI in normal organoids in Matrigel (F′) and collagen I (G′). White arrowheads mark the leading fronts. Yellow arrowhead indicates myoepithelial cell dissemination. (H) Percent of normal organoids showing dissemination in Matrigel and collagen I. n, total number of movies (four biological replicates, Student's t test, two-tailed, unequal variance).
Normal Mammary Epithelial Cells Exhibit Protrusive Migration and Local Dissemination in a Collagen I Microenvironment.
We next asked whether a protrusive, disseminative response to collagen I was tumor specific. We isolated epithelial fragments (organoids) from the mammary glands of FVB mice and cultured them in Matrigel or 3 mg/mL collagen I (Fig. 2E). Again, the cell-migratory strategy and likelihood of local dissemination depended on the ECM microenvironment. In Matrigel, normal organoids migrated collectively to accomplish branching morphogenesis without protrusions into the ECM (Fig. 2 F and F′), as we previously reported (2). In collagen I, organoids isolated from the same mouse migrated with extensive protrusions, and cells disseminated locally into the ECM (Fig. 2 G and G′). To test the reversibility of the ECM-induced changes in migratory program, we next cultured normal epithelial fragments for 90 h in Matrigel or collagen I (Fig. S3 A–C) and then recovered and re-embedded the epithelium in either the same or the opposite matrix (Fig. S3D). Similar to the results observed in human carcinoma fragments (Fig. 1 G–H), the current, local ECM microenvironment dictated the migratory pattern, with collective epithelial migration in Matrigel (Fig. S3 E and G) and collective protrusive migration in collagen I (Fig. S3 F and H).
Mammary Carcinomas Exhibit Sustained Local Dissemination in Collagen I.
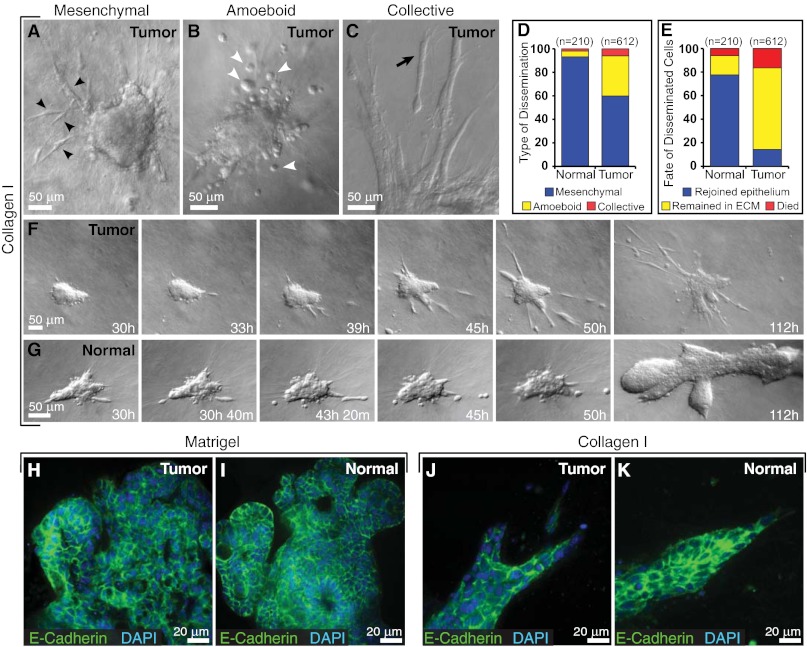
To understand better how epithelia transition from collective migration to individual cell dissemination, we quantified specific dissemination behaviors. Carcinoma fragments disseminated cells into collagen I throughout culture, with an average of 13 cells observed to leave each tumor mass (612 disseminating cells observed in 48 movies; see SI Materials and Methods). We classified the disseminating tumor cells, based on previous morphological definitions (1, 30), as mesenchymal, amoeboid, or collective (Fig. 3 A–D). Most carcinoma cells disseminated with a mesenchymal morphology (60%) as they protruded into the ECM and maintained an elongated morphology while migrating through the collagen I matrix (Fig. 3A). Other carcinoma cells (34%) disseminated in an amoeboid fashion as rounded cells that rolled or squeezed through the collagen I matrix (Fig. 3B). In a minority of cases (6%), we also observed collective dissemination of groups of cells (Fig. 3C). Individual carcinoma fragments typically exhibited both mesenchymal and amoeboid dissemination (Fig. 3F). Most disseminated carcinoma cells remained motile in the ECM during the entire period of observation (69%), but 17% of the disseminated cells died, and 14% rejoined the tumor fragment (Fig. 3E). Once in the matrix, individual cancer cells were observed to convert between elongated and rounded morphologies, consistent with a previous report on melanoma cells (31). Despite local dissemination, carcinoma cells localized E-cadherin to intercellular borders in both Matrigel and collagen I (Fig. 3 H and J).
Fig. 3.
Cell dissemination into collagen I is persistent in tumor and transient in normal epithelium. (A–C) Tumor cells disseminate with mesenchymal (black arrowheads) (A), amoeboid (white arrowheads) (B), and collective (black arrow) (C) morphologies. (D and E) Distribution of morphological types of dissemination (D) and fate of disseminated cells in normal and tumor organoids (E) in collagen I. n, total number of disseminated cells observed in each condition. (F and G) Representative frames from DIC time-lapse movies of tumor (F) and normal organoids (G) in collagen I. (H and I) Localization of E-cadherin and DAPI in tumor (H) and normal organoids (I) cultured in Matrigel. (J and K) Localization of E-cadherin and DAPI in tumor (J) and normal organoids (K) cultured in collagen I.
Protrusive Migration and Local Dissemination Are Transient Responses of Normal Myoepithelial Cells to Collagen I.
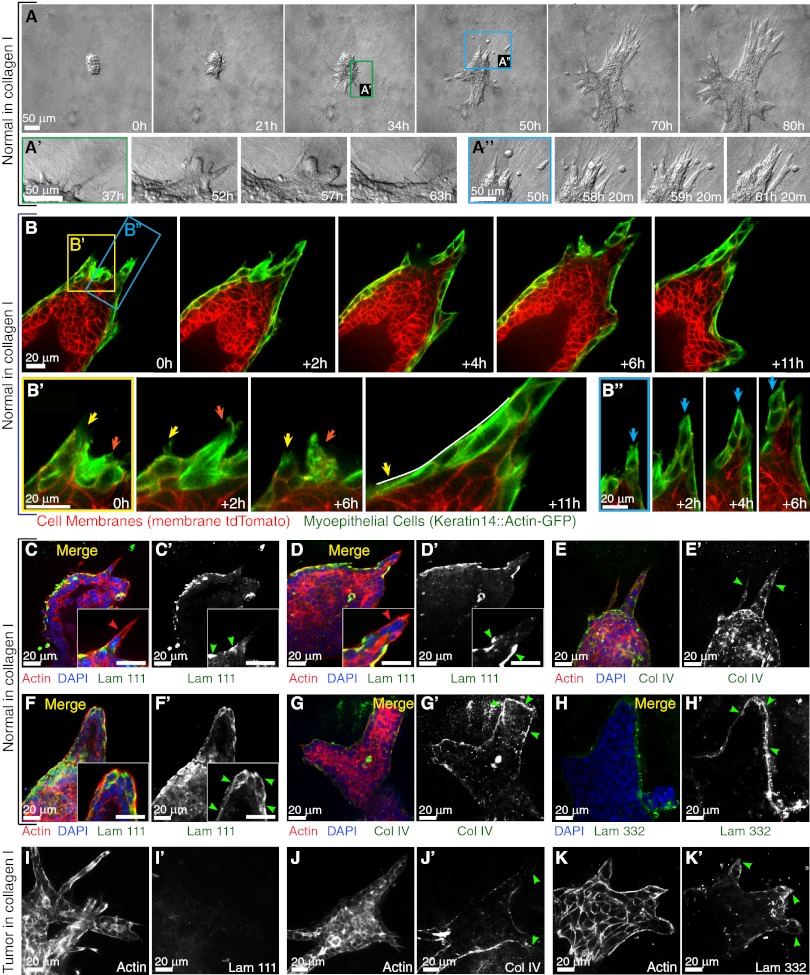
We previously observed no ECM-directed protrusions at the front of elongating mammary ducts in Matrigel or in vivo (2). In contrast, collagen I induced acute protrusive and disseminative behaviors in both normal and malignant mammary epithelium (Fig. 3 F and G, 1–50 h). We observed dissemination from 79% (34/43 movies) of normal epithelial organoids (Fig. 2H), with an average of five cells leaving each epithelial group (210 disseminating cells observed in 43 movies), typically at the protrusive borders (Fig. 4 A and A′′). Despite local dissemination, cells in the main epithelial group remained E-cadherin+ in both Matrigel and collagen I (Fig. 3 I and K). The main difference in cell behavior between normal and carcinoma cells was that both protrusions and dissemination were transient in normal epithelial cells (Fig. 3G), because normal epithelial organoids ceased protrusive activity and reverted to a program of branching morphogenesis. The protrusive normal cells stained positive for the myoepithelial marker smooth muscle α-actin (SMA) (Fig. 2G′) in 67/69 samples. Using a transgenic myoepithelial cell reporter to visualize the protrusive behavior in real time (Fig. 4B, keratin-14::actin-GFP, and ref. 32), we observed subcellular protrusions extending and retracting from single myoepithelial cells (Fig. 4B′) as well as multicellular extensions of myoepithelial cells (Fig. 4B′′). Live imaging revealed that the transition from protrusive to epithelial organization at the ECM interface (Fig. 4A′) represented a change in cell behavior in individual myoepithelial cells (Fig. 4B′). In contrast, in Matrigel, myoepithelial cells remained closely adherent to the luminal epithelial cells and did not extend protrusions into the ECM (2).
Fig. 4.
Normal epithelium transiently protrudes and disseminates into collagen I but reestablishes a complete basement membrane. (A) Representative frames of a DIC time-lapse movie of a normal organoid grown in collagen I. (A′) Higher magnification of a transition from a protrusive to a smooth border with ECM. (A′′) Higher magnification of the reintegration of disseminated cells with the epithelial group. (B) Representative frames from a confocal time-lapse movie of a normal organoid grown in collagen I. (B′) Higher magnification of transient protrusions within single myoepithelial cells. Arrows indicate individual protrusions, retractions, and epithelial reorganization. (B′′) Higher magnification of a multicellular extension of myoepithelial cells (blue arrows) at the leading front. (C–G) Normal epithelia in collagen reform a multicomponent basement membrane. (C–D′ and F and F′) Localization of actin, DAPI, and laminin 111 in a merge of all channels (C, D, and F) and in a single channel of laminin 111 (C′, D′, and F′) in a normal organoid with a single-cell protrusion (C′), a multicellular extension (D′), and a normal organoid after reorganization (F′). All the Insets highlight negative correlation of cell protrusion (C, D, and F) and laminin 111 (C′, D′, and F′) at the leading front. (E and E′ and G and G′) Localization of actin, DAPI, and collagen IV in a merge of all channels (E and G) and in a single channel of collagen IV in a normal organoid (E′ and G′) with multicellular extensions (E′) and after reorganization (G′). (H and H′) Localization of DAPI and laminin 332 in a merge of two channels (H) and a single channel of laminin 332 (H′). (I–K′) Tumor epithelia display incomplete basement membrane coverage. Single channels show the localization of actin (I, J, and K), laminin 111(I′), collagen IV (J′), and laminin 332 (K′) in tumor organoids in collagen I. Red and green arrowheads indicate actin-based protrusions and signals of basement membrane components, respectively.
Normal Mammary Organoids Progressively Organize a Basement Membrane in Collagen I.
We observed a shift from protrusive to smooth, organized basal surfaces in normal mammary organoids cultured in collagen I (Fig. 4 A and B). To test whether this shift might relate to reestablishment of a basement membrane, we used antibodies to stain for laminin 111, laminin 332, and collagen IV. We observed a negative correlation between protrusions and basement membrane organization. We observed single-cell protrusions (Fig. 4 C and C′) and multicellular protrusive groups (Fig. 4 D and D′) that extended through gaps in the laminin 111. Collagen IV staining was diffuse and incomplete in protrusive areas of normal epithelium (Fig. 4 E and E′). Late in culture in collagen I, normal epithelium typically became covered by a complete basement membrane that stained positive for all three markers (Fig. 4 F–H′). In contrast, at the ECM border of carcinoma fragments, we observed no laminin 111 and only scattered laminin 332 and incomplete collagen IV coverage (Fig. 4 I–K′). The most striking difference between tumor and normal epithelium in collagen I was the lack of laminin 111 along tumor borders, even late in culture (Fig. 4 I and I′ vs. F and F′).
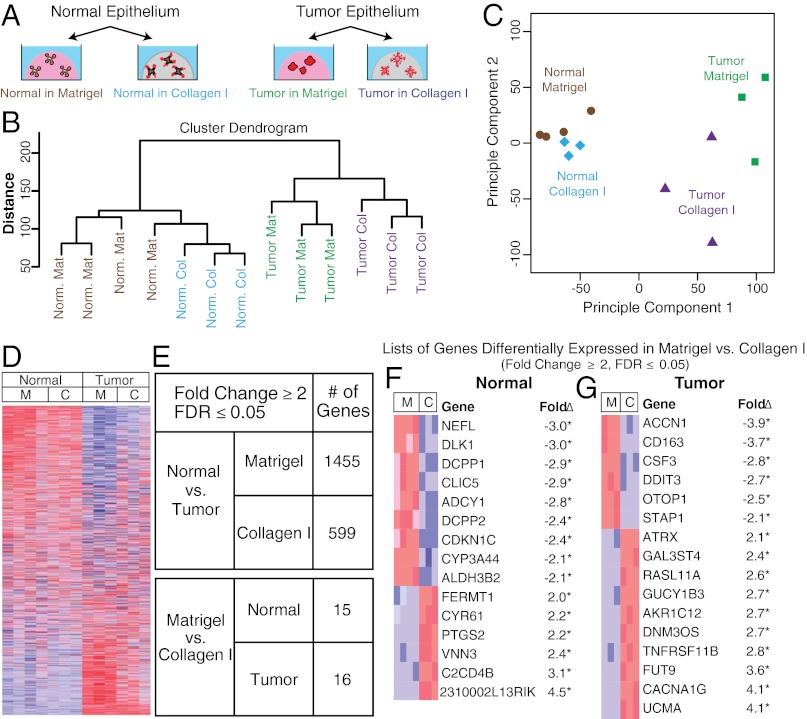
ECM Microenvironment Has Minor Effects on Average Gene Expression.
Taken together, our results suggest that the pattern of epithelial migration and local dissemination are constrained by the local ECM microenvironment. We next sought to identify changes in RNA expression that could regulate these changes in cell behavior. Accordingly, we collected RNA from normal and malignant epithelium during active growth at day 4 of culture in either Matrigel or collagen I. Our goal was to compare average gene expression, so we isolated RNA from whole cultures. We hybridized the resulting RNA to Agilent single-color microarrays (Fig. 5A), with a minimum of three biologically independent microarray replicates per condition (SI Materials and Methods). To test the relative importance of the ECM to gene expression, we performed complete-linkage hierarchical clustering. Normal samples clustered together regardless of their ECM microenvironment, distinct from all tumor samples (Fig. 5B). A principal component analysis confirmed that the first principal component was whether the epithelium was normal or tumor, and the second principal component was the ECM condition (Fig. 5C). Using a fold change ≥2 and a false discovery rate (FDR) ≤0.05 as criteria for significance we found only 15 or 16 genes were differentially expressed based on ECM condition in normal or tumor epithelium, respectively (Fig. 5 E–G). However, these genes did not have obvious mechanistic connections to dissemination (Fig. 5 F and G). Thus, the ECM microenvironment had a relatively small impact on average RNA expression, despite its large effects on migratory strategy and local dissemination. Our experimental design cannot exclude the possibility of changing gene expression within subpopulations of the epithelium.
Fig. 5.
Tumor and normal epithelium remain transcriptionally distinct despite morphological similarities induced by the ECM. (A) Schematic description of 3D cultures of normal and tumor murine epithelial fragments in Matrigel and collagen I for mRNA expression analyses. n = at least 3 biological replicates. (B) Complete-linkage hierarchical clustering of the experimental conditions. (C) Principal component analysis of the experimental conditions. (D) Heatmap representation of the 19,693 genes included in the microarray (blue and red indicate lower and higher expression, respectively). (E) Summary of differentially expressed genes with fold changes ≥2 and FDR ≤0.05. (F and G) Genes differentially expressed based on ECM condition in normal (F) and tumor (G). C, collagen I; M, Matrigel.
Normal and Malignant Epithelia Differ in Their Expression of Cell-Adhesion Genes and Modifiers of the Extracellular Microenvironment.
In contrast to the modest differences observed between ECM environments, we found significant differences in gene expression between normal and malignant epithelia, even when cultured in the same ECM: 1,455 genes were differentially expressed between normal and tumor samples in Matrigel, and 599 genes were differentially expressed between normal and tumor samples in collagen I (Fig. 5 D and E). These data suggest that normal epithelium and tumors accomplish morphologically similar migration processes despite widely different gene expression.
We next sought gene-expression signatures that might explain the sustained dissemination of carcinoma cells in collagen I (Fig. S4A). The epithelial-to-mesenchymal transition (EMT) has been proposed as a mechanism for cancer metastasis (33). Conceptual models of EMT center on decreased expression of E-cadherin and increased expression of genes such as N-cadherin and vimentin (33). However, the local dissemination we observed in collagen I was achieved with membrane-localized E-cadherin (Fig. 3 J and K). Surprisingly, E-cadherin RNA expression was not statistically significantly different between any two conditions (Fig. S4C). Furthermore, carcinoma cells displayed reduced levels of both N-cadherin and vimentin RNA, and the EMT regulators Snail-1, Snail-2, and Twist-1 also were not differentially expressed (defined as a greater-than-twofold change, FDR <0.05) in any condition (Fig. S4C). Our results suggest that a classic molecular EMT program is not activated in the tumor or in response to a collagen microenvironment, despite vigorous dissemination of cells into the ECM.
Importantly, our enrichment analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) functional annotation revealed large and statistically significant differences between normal tissue and tumors in gene sets for cell adhesion and for proteins that function in the extracellular space (Fig. S4 A and B and SI Materials and Methods). Analysis of mRNA expression of genes defined by structural motifs related to adhesion (e.g., cadherins and integrins) and of ECM and ECM-remodeling genes [e.g., collagens and matrix metalloproteinases (MMPs)] (Figs. S4B and S5) revealed widespread changes across gene families. Moreover, adhesion gene sets (cadherins, immunoglobulin cell-adhesion molecules, integrins, lectins, and other cell-adhesion molecules) were down-regulated in tumor relative to normal epithelium (∼60% of differentially expressed genes higher in normal epithelium; P ≤ 0.0003, Wilcoxon rank sum test). We specifically observed strong down-regulation in the cadherin gene family, with 75% of differentially expressed genes down-regulated in tumor relative to normal tissue (P ≤ 0.0001, Wilcoxon rank sum test) (Fig. S4B). These data suggest that down-regulation of cell-adhesion genes may contribute to the sustained dissemination of carcinoma cells in collagen I.
Reduced Intercellular Adhesion Cooperates with a Collagen I Microenvironment to Permit Sustained Dissemination of Nontransformed Cells.
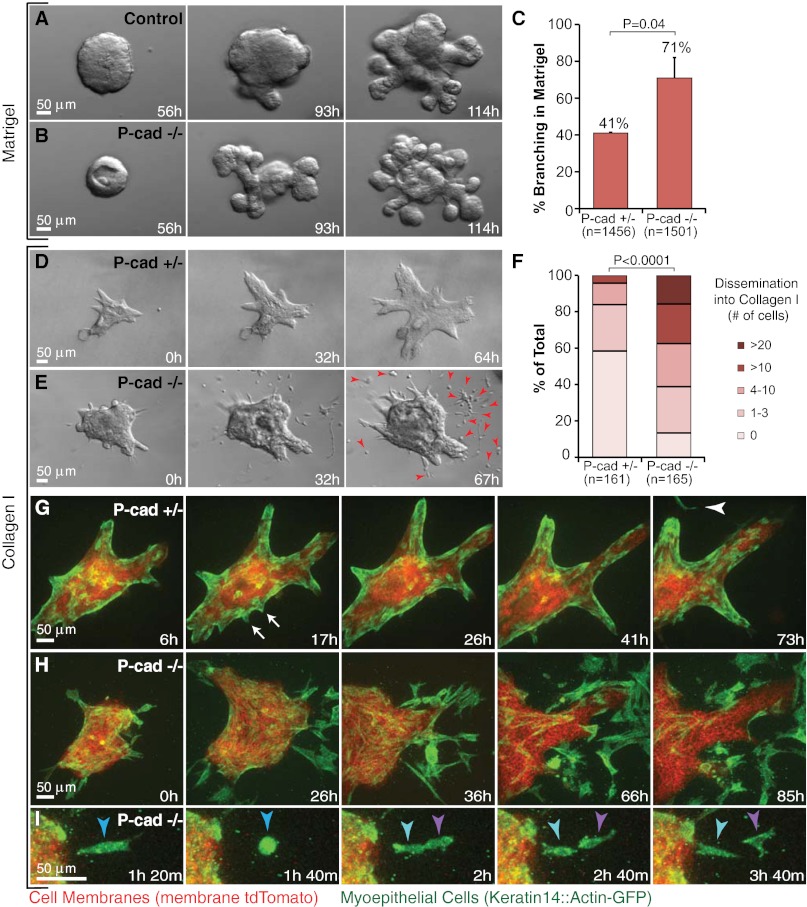
Recent breast cancer genome-sequencing efforts have revealed mutations in multiple families of cell-adhesion genes, including both classical cadherins and protocadherins (34). More than 70% of breast tumors in a recent study had mutations in a cell-adhesion gene, but few of these mutations occurred in more than one tumor (35). We chose to test genetically whether altering cell adhesion is sufficient to enable sustained dissemination of nontransformed cells into collagen I. Because myoepithelial cells were the only protrusive or disseminative cells in our normal epithelial cultures, we focused on P-cadherin (Cdh3), a classical cadherin specifically expressed in myoepithelial but not luminal epithelial cells (36). Deletion of P-cadherin in vivo results in precocious alveolar differentiation and luminal epithelial hyperplasia (37). We hypothesized that loss of P-cadherin might synergize with a collagen I-rich microenvironment to induce sustained myoepithelial dissemination. In Matrigel, we observed precocious branching (Fig. 6 A and B) and increased branching efficiency (Fig. 6C) in P-cadherin–null epithelial fragments but no protrusions or dissemination. In contrast, P-cadherin–null epithelial fragments explanted into collagen I disseminated more cells relative to controls, and dissemination was sustained throughout culture (Fig. 6 D–F). Disseminating cells were myoepithelial in nature (K14+) (Fig. 6H), and they survived and proliferated in collagen I (Fig. 6I). Indeed, we frequently observed nearly complete depletion of myoepithelial cells from the surface of P-cadherin–null organoids (Fig. 6H). We conclude that deletion of a cell-adhesion gene is sufficient to induce sustained myoepithelial dissemination in specific ECM microenvironments.
Fig. 6.
Loss of P-cadherin causes precocious branching morphogenesis in Matrigel and enhanced, sustained dissemination into collagen I. (A and B) Representative frames from DIC time-lapse movies of (A) control [P-cadherin+/+ (P-cad+/+)] and (B) P-cadherin−/− (P-cad−/−) epithelium grown in parallel in Matrigel. (C) Percent of P-cad+/− and P-cad−/− organoids branching in Matrigel on day 7. n, total number of organoids counted (three biological replicates; *P = 0.04; Student’s t test, two-tailed, unequal variance). (D and E) Representative frames from DIC time-lapse movies of P-cad+/− (D) and P-cad−/− (E) epithelium grown in parallel in collagen I. Arrowheads indicate persistent cell dissemination. (F) Distribution of number of disseminated cells per organoid in P-cad+/− and P-cad−/− epithelia. n, total number of movies (three biological replicates; *P < 0.0001; upper one-sided χ2 test). (G) Representative frames from a confocal time-lapse movie of P-cad+/−, mT/mG, K14::Actin-GFP epithelium in collagen I. Arrows indicate transient, myoepithelial-led protrusions. Arrowhead indicates a single disseminated myoepithelial cell. (H) Representative frames from a confocal time-lapse movie of enhanced myoepithelial dissemination into collagen I by P-cad−/−, mT/mG, K14::Actin-GFP epithelium. (I) Proliferation of a disseminated P-cad−/− myoepithelial cell.
Discussion
In the present study we sought to isolate the specific role of the ECM in regulating collective epithelial migration and local dissemination by explanting fragments from the same epithelium into different ECM microenvironments. We found that murine and human mammary carcinomas cultured in 3D Matrigel were indolent or migrated collectively as a multilayered epithelium. Surprisingly, we observed local dissemination only rarely in Matrigel, even from metastatic human and murine mammary carcinomas. This result demonstrates that a metastatic genotype is not sufficient for local dissemination in all ECM microenvironments. In contrast, both normal and malignant mammary epithelium disseminated vigorously into collagen I.
Epithelial Cells Have ECM-Specific Migration Programs.
We observed large, ECM-specific differences in the pattern of collective migration and frequency of local dissemination with fragments from the same epithelium and identical culture medium. Moreover, these large differences in migratory pattern had few corresponding ECM-specific differences in gene expression. Despite the local dissemination of epithelial cells into collagen I, we did not detect a classic molecular EMT response in either normal or tumor tissue. However, all our transcriptome experiments compared RNA extracted from whole cultures, so our experimental design cannot exclude gene-expression or signaling changes in the cells directly in contact with the ECM. Comparable molecular profiling studies comparing in situ and invasive breast cancer at the tissue level also have failed to define a gene signature predictive of invasion (38).
Taken together, our data suggest that cancer cells could possess all the gene expression required for sustained local dissemination but remain indolent while the basement membrane remains intact. However, if the basement membrane were disrupted, the resulting direct contact between cancer cells and the stromal collagen I matrix could induce protrusive and disseminative cell behaviors rapidly. Breach of the basement membrane can be accomplished by MMP-based proteolysis by the cancer cells (10, 39), by immune cells recruited during inflammatory processes, or by the actions of carcinoma-associated macrophages (40) or fibroblasts (41). Consistent with this model, correlative studies in human breast tumors show that even microscopic breaks in the myoepithelium correlate with poor patient prognosis (42).
Stromal ECM Is Not Sufficient for Sustained Dissemination.
The acute reaction of normal and carcinoma-derived epithelium to collagen I was very similar, because both exhibited protrusive migration and a mixture of amoeboid and mesenchymal dissemination. All the individual cell behaviors observed in the tumor fragments were observed in the normal fragments also. However, this similarity was transient: Normal epithelium reestablished basement membrane coverage and underwent branching morphogenesis. In contrast, dissemination from carcinoma fragments was sustained throughout culture, and polarized epithelial architecture was not restored. Taken together, our data support a requirement for coordinate changes in both the cancer cell and the microenvironment to enable sustained dissemination. Our data also suggest that the final signal triggering invasion and local dissemination can be provided by changes in the ECM microenvironment rather than by genetic changes in the cancer cell. This suggestion is consistent with recent sequencing efforts that identified similar gene expression and mutations within in situ and invasive breast tumors (38, 43). Additionally, central fibrosis, which is characterized by high levels of collagen I, independently correlates negatively with patient outcome even among the most aggressive types of breast cancers (44).
Cell–Cell Adhesion and the ECM Microenvironment Coordinately Regulate Dissemination.
Cell-adhesion genes frequently are down-regulated or mutated in metastatic human tumors (7, 8) and in our mouse carcinoma model. Our data support the hypothesis that deletion of a cell-adhesion gene can enable sustained dissemination of otherwise normal cells. Because the transiently disseminating normal cells were myoepithelial, we focused on P-cadherin (37), the major classical cadherin in myoepithelial cells (36). The phenotype of P-cadherin deletion in Matrigel and in vivo was luminal epithelial hyperplasia. However, the phenotype of P-cadherin deletion in collagen I was excess, sustained myoepithelial dissemination. The consequences of molecular perturbations therefore can be qualitatively different in different ECM microenvironments with respect to clinically important variables such as local dissemination. Our data are consistent with recent reports that myoepithelial cells are structurally and molecularly abnormal in nonmalignant regions adjacent to primary human breast tumors (45). Our data suggest that observed changes in the ECM composition of the tumor microenvironment in these regions may help explain these abnormalities (45).
Translational Implications for Breast Cancer.
Our data indicate that the cellular migratory strategy and the likelihood of local dissemination depend not only on the genetic state of the cancer cells but also on the ECM in the tumor microenvironment. Importantly, we demonstrated through matrix-switching experiments that, even after an ECM-induced transition to protrusive migration and local dissemination, human malignant carcinomas can revert to confined, nonprotrusive growth in response to basement membrane signals. Our data are consistent with past work on the normalization of tumor architecture by basement membrane signals such as laminin 111 (17, 46, 47) and on the invasion-associated behavior of cells in collagen I (10, 13, 48).
Future Directions.
It is now necessary to isolate the molecular basis for the differential effects of different ECM microenvironments on collective cell migration and dissemination. Collagen I and Matrigel have distinct rigidity, protein composition, and supramolecular organization. It remains unclear which of these variables is most important to dissemination, although past studies suggest a role for increased matrix rigidity in cancer progression (10). Our data also suggest that deletion of a single adhesion gene is sufficient to induce sustained dissemination of nontransformed cells into a stromal matrix such as collagen I. It will be important to determine whether deletion of other adhesion genes will promote local dissemination similarly. Because the collective migration strategy of epithelial cells differs in different ECMs, it also is possible that specific genetic perturbations contribute to invasion and dissemination only in specific microenvironmental contexts.
Materials and Methods
Isolation of Primary Murine Mammary Organoids.
We isolated mouse mammary organoids from normal mice using previously described techniques (2, 49). Briefly, we dissected the no. 3 and no. 4 mammary glands and digested this tissue into epithelial fragments by a combination of mechanical disruption and collagenase/trypsin digestion. We then separated these fragments from single cells by differential centrifugation. The final pellet was composed of epithelial fragments, each containing several hundred cells; we term these fragments “organoids.” Tumors were harvested from mice at 12–16 wk of age, when carcinomas were poorly differentiated. We surgically isolated the largest tumor in each mouse and processed it as above. Any incompletely digested large tumor fragments were removed before differential centrifugation. We also added additional rounds of differential centrifugation as needed to remove single cells. This protocol was adapted further for isolation of organoids from primary human mammary tumor (see SI Materials and Methods for a complete description of isolation methods for human tissue).
Primary Murine Mammary Organoid Culture.
We embedded organoids derived from normal and tumor epithelium in 3D Matrigel (354230; BD Biosciences) or rat-tail collagen I (354236; BD Biosciences). Cultures were set up in 24-well coverslip-bottomed plates (EK-42892; E&K Scientific) or in two-well or four-well coverslip-bottomed chambers (155383; Nunc). Acid-solubilized rat-tail collagen I gels (3 mg/mL collagen I, pH 7–7.5) were prepared as described in SI Materials and Methods. For each matrix, organoids were mixed to yield a suspension of two or three organoids/μL. A 100-μL suspension of organoids was plated in each well on a 37-°C heating block, followed by incubation at 37 °C for 45 min to allow polymerization. Epithelial fragments in collagen I were plated on top of an underlay of cell-free collagen I of the same concentration. Murine samples were cultured in 1 mL of 2.5-nM FGF2 in murine organoid medium (2).
Time-Lapse Differential Interference Contrast Microscopy.
Live imaging of normal and tumor murine organoids was conducted using a Zeiss Cell Observer system with a Zeiss AxioObserver Z1 and an AxioCam MRM camera. In general, images were collected at 20-min intervals with exposure times of ∼250 ms. Some of the movies of human tumor were collected on a Zeiss Axiovert S-100 microscope and a Cohu CCD camera, as previously reported (2). Temperature was held at 37 °C and CO2 at 5%.
Gene-Expression Analysis of Normal and Tumor Fragments in Parallel ECM Conditions.
In total, four different conditions were profiled (tumor vs. normal; collagen I vs. Matrigel); each was replicated at least three times in biologically independent experiments. Sample preparation, labeling, and array hybridizations were performed according to standard protocols from the University of California, San Francisco Shared Microarray Core Facilities and Agilent Technologies (http://www.arrays.ucsf.edu and http://www.agilent.com). Equal amounts of Cy3-labeled target were hybridized to Agilent whole mouse genome 4 × 44K Ink-jet arrays. Arrays were scanned using the Agilent microarray scanner, and raw signal intensities were extracted with Feature Extraction v. 9.1 software (Agilent). Samples were confirmed to be of good quality and were quantile normalized using R/Bioconductor packages. Pairwise differentially expressed genes were detected using the limma package in R. q-Values ≤0.05 were deemed statistically significant. A stand-alone program written in Java was developed to interface with the R program via the command-line to generate heatmaps for publication (available upon request). Genes changed by twofold or more and with q-values ≤0.05 were used as input for DAVID Gene Set Analysis (50). Gene sets associated with structurally similar gene families (including cell–cell adhesion, cytoskeletal networks, and actin–myosin contractility) were curated manually from Mouse Genome Informatics and Interpro (available upon request). Lists of cell-adhesion genes were cross-referenced further with OKCAM, an online database of cell-adhesion molecules (51). Microarray data have been made available on the National Center for Biotechnology Information’s GEO database (accession no. GSE39173).
P-Cadherin–Deletion Experiments.
The P-cadherin and mT/mG mouse lines were acquired from the Jackson Laboratory. The keratin-14::actin-GFP transgenic line (32, 37, 52) was a kind gift of Elaine Fuchs (The Rockefeller University, New York). P-cadherin−/− and P-cadherin+/−, mT/mG, K14-Actin-GFP mammary organoids were isolated as described above. Organoids were grown in Matrigel or collagen I with 2.5 nM FGF2. Branching in Matrigel was quantified on day 7 in three independent biological replicates and scored as three or more buds per organoid. Organoids were grown in 3 mg/mL collagen I for 4–5 d, and cell dissemination was quantified from differential interference contrast (DIC) and confocal time-lapse movies in three independent biological replicates.
Supplementary Material
Acknowledgments
We thank Lisa Jacobs, Pedram Argani, and the Sidney Kimmel Comprehensive Cancer Center’s Breast Cancer Program for assistance in acquiring breast tumor tissue from Johns Hopkins Hospital; Jen Beck for technical support; Peng Huang for biostatistics support; Michael Ochs for bioinformatic support; and Mikala Egeblad for comments on the manuscript. This work was supported by the University of California at San Francisco Sandler Asthma Basic Research Center Functional Genomics Core Facility. This study was funded by National Cancer Institute Grants R01 CA056721 and R01CA138818 (to Z.W.) and U01 CA155758, U54CA151838, and P50 CA88843 (to A.J.E.); by National Institute of Environmental Health Sciences Grant U01 ES019458 (to Z.W. and P.Y.); and by funds from the Safeway Foundation Award for Breast Cancer Research (to A.J.E.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE39173). Movies are available via the American Society for Cell Biology (ASCB) Cell: An Image Library, http://www.cellimagelibrary.org (accession nos. 42151–42168).
See Author Summary on page 15543 (volume 109, number 39).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212834109/-/DCSupplemental.
References
- 1.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 2.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewald AJ, et al. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J Cell Sci. 2012;125:2638–2654. doi: 10.1242/jcs.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Friedl P, et al. Migration of coordinated cell clusters in mesenchymal and epithelial cancer explants in vitro. Cancer Res. 1995;55:4557–4560. [PubMed] [Google Scholar]
- 6.Nguyen DX, Bos PD, Massagué J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 8.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 9.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: Complex tissues that interface with the entire organism. Dev Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egeblad M, Rasch MG, Weaver VM. Dynamic interplay between the collagen scaffold and tumor evolution. Curr Opin Cell Biol. 2010;22:697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends Genet. 2009;25:30–38. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Egeblad M, et al. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Model Mech. 2008;1:155–167, discussion 165. doi: 10.1242/dmm.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provenzano PP, et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conklin MW, et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen OW, Rønnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weaver VM, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagios C, Lochter A, Bissell MJ. Tissue architecture: The ultimate regulator of epithelial function? Philos Trans R Soc Lond B Biol Sci. 1998;353:857–870. doi: 10.1098/rstb.1998.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 20.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 21.Ewald AJ. In: Practical Considerations for Long-Term Time-Lapse Imaging of Epithelial Morphogenesis in Three-Dimensional Organotypic Cultures. Imaging in Developmental Biology. 2nd Ed, Vol 3. Wong R, Sharpe J, editors. New York: Cold Spring Harbor Laboratory; 2010. pp. 623–645. [DOI] [PubMed] [Google Scholar]
- 22.Simian M, et al. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development. 2001;128:3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudjonsson T, Rønnov-Jessen L, Villadsen R, Bissell MJ, Petersen OW. To create the correct microenvironment: Three-dimensional heterotypic collagen assays for human breast epithelial morphogenesis and neoplasia. Methods. 2003;30:247–255. doi: 10.1016/s1046-2023(03)00031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson CM, Bissell MJ. Modeling dynamic reciprocity: Engineering three-dimensional culture models of breast architecture, function, and neoplastic transformation. Semin Cancer Biol. 2005;15:342–352. doi: 10.1016/j.semcancer.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedl P, Hegerfeldt Y, Tusch M. Collective cell migration in morphogenesis and cancer. Int J Dev Biol. 2004;48:441–449. doi: 10.1387/ijdb.041821pf. [DOI] [PubMed] [Google Scholar]
- 26.Williams JM, Daniel CW. Mammary ductal elongation: Differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev Biol. 1983;97:274–290. doi: 10.1016/0012-1606(83)90086-6. [DOI] [PubMed] [Google Scholar]
- 27.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvo F, et al. RasGRF suppresses Cdc42-mediated tumour cell movement, cytoskeletal dynamics and transformation. Nat Cell Biol. 2011;13:819–826. doi: 10.1038/ncb2271. [DOI] [PubMed] [Google Scholar]
- 31.Sanz-Moreno V, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 32.Vaezi A, Bauer C, Vasioukhin V, Fuchs E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev Cell. 2002;3:367–381. doi: 10.1016/s1534-5807(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Leary RJ, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci USA. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velculescu VE. Defining the blueprint of the cancer genome. Carcinogenesis. 2008;29:1087–1091. doi: 10.1093/carcin/bgn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daniel CW, Strickland P, Friedmann Y. Expression and functional role of E- and P-cadherins in mouse mammary ductal morphogenesis and growth. Dev Biol. 1995;169:511–519. doi: 10.1006/dbio.1995.1165. [DOI] [PubMed] [Google Scholar]
- 37.Radice GL, et al. Precocious mammary gland development in P-cadherin-deficient mice. J Cell Biol. 1997;139:1025–1032. doi: 10.1083/jcb.139.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polyak K. Molecular markers for the diagnosis and management of ductal carcinoma in situ. J Natl Cancer Inst Monogr. 2010;2010:210–213. doi: 10.1093/jncimonographs/lgq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf K, et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 40.DeNardo DG, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 42.Man YG, Sang QX. The significance of focal myoepithelial cell layer disruptions in human breast tumor invasion: A paradigm shift from the “protease-centered” hypothesis. Exp Cell Res. 2004;301:103–118. doi: 10.1016/j.yexcr.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 43.Miron A, et al. PIK3CA mutations in in situ and invasive breast carcinomas. Cancer Res. 2010;70:5674–5678. doi: 10.1158/0008-5472.CAN-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreike B, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trujillo KA, et al. Markers of fibrosis and epithelial to mesenchymal transition demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int J Cancer. 2011;129:1310–1321. doi: 10.1002/ijc.25788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: Tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gudjonsson T, et al. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115:39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenburg G, Hay ED. Epithelia suspended in collagen gels can lose polarity and express characteristics of migrating mesenchymal cells. J Cell Biol. 1982;95:333–339. doi: 10.1083/jcb.95.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fata JE, et al. The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGFalpha and FGF7 in morphogenesis of mouse mammary epithelium. Dev Biol. 2007;306:193–207. doi: 10.1016/j.ydbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 51.Li CY, et al. OKCAM: An ontology-based, human-centered knowledgebase for cell adhesion molecules. Nucleic Acids Res. 2009;37(Database issue):D251–D260. doi: 10.1093/nar/gkn568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]