Abstract
Chaperonin GroEL mediates the folding of protein encapsulated in a GroES-sealed cavity (cage). Recently, a critical role of negative charge clusters on the cage wall in folding acceleration was proposed based on experiments using GroEL single-ring (SR) mutants SR1 and SRKKK2 [Tang YC, et al. (2006) Cell 125:903–914; Chakraborty K, et al. (2010) Cell 142:112–122]. Here, we revisited these experiments and discovered several inconsistencies. (i) SR1 was assumed to bind to GroES stably and to mediate single-round folding in the cage. However, we show that SR1 repeats multiple turnovers of GroES release/binding coupled with ATP hydrolysis. (ii) Although the slow folding observed for a double-mutant of maltose binding protein (DMMBP) by SRKKK2 was attributed to mutations that neutralize negative charges on the cage wall, we found that the majority of DMMBP escape from SRKKK2 and undergo spontaneous folding in the bulk medium. (iii) An osmolyte, trimethylamine N-oxide, was reported to accelerate SRKKK2-mediated folding of DMMBP by mimicking the effect of cage-wall negative charges of WT GroEL and ordering the water structure to promote protein compaction. However, we demonstrate that in-cage folding by SRKKK2 is unaffected by trimethylamine N-oxide. (iv) Although it was reported that SRKKK2 lost the ability to assist the folding of ribulose-1,5-bisphosphate carboxylase/oxygenase, we found that SRKKK2 retains this ability. Our results argue against the role of the negative charges on the cage wall of GroEL in protein folding. Thus, in chaperonin studies, folding kinetics need to be determined from the fraction of the real in-cage folding.
Keywords: guanidium chloride, urea, fluorescence, anisotropy, apyrase
The bacterial GroEL/GroES chaperonin is an essential molecular chaperone that mediates the folding of various proteins (1, 2). GroEL consists of two rings stacked back to back, and each ring, made up of seven 57-kDa subunits, possesses a large central cavity. GroEL binds to a wide range of denatured proteins at the hydrophobic apical end of the central cavity to make a binary complex of GroEL/substrate protein. On binding of ATP to GroEL, GroES attaches to the apical end of the GroEL ring as a lid, generating a GroEL/GroES/substrate protein ternary complex, in which the substrate protein in the sealed cage starts folding. Hydrolysis of bound ATP triggers the detachment of the GroES lid to allow the substrate protein in the cage, whether folded or denatured, to be free. The next denatured protein is captured by GroEL, ATP binds to GroEL, and the cycle repeats. Single-round folding can be observed without the complication of coordinated ATP hydrolysis turnover of the two rings in GroEL by the use of a single-ring (SR) mutant of GroEL (SR1) that holds the GroES lid long enough for the substrate protein to finish folding in the cage (3).
Two model mechanisms for the function of chaperonin in mediating protein folding have been proposed. The passive Anfinsen cage model explains that proteins fold in a spontaneous manner in the cage without the risk for aggregation (4). The confinement model proposes that proteins folding in the narrow space of the cage destabilize off-pathway (or slow) folding intermediates and can be more rapid than the spontaneous folding (5–8). These models assume that a substrate protein starts folding essentially as a free polypeptide in the cage on GroES binding. However, we recently found that polypeptide chains were loosely tethered to GroEL/GroES interface regions and were subsequently released either into the cage, in which the protein completed folding (in-cage folding), or into the outside bulk medium, in which it folded in a spontaneous manner (out-of-cage folding) (9).
Recently, important aspects of chaperonin function were reported by Tang et al. (6) and Chakraborty et al. (8). Using SR variants of GroEL, they extensively studied chaperonin-mediated folding of maltose binding protein, especially its mutant with two amino acid replacements (DMMBP; V8G/Y283D). They generated a unique mutant derived from SR1 in which three negatively charged residues (D359, D361, and E363) were replaced with lysines (SRKKK2), and they found that SRKKK2 had largely impaired chaperonin activity. In other words, it encapsulated DMMBP but was unable to accelerate folding and failed in assisting the folding of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco). Folding by SRKKK2 was accelerated by the addition of an osmolyte, which was assumed to compensate for the loss of the negative charge effect by limiting structural flexibility of polypeptides. Based on these observations, these investigators proposed that clusters of negative charges exposed on the cage wall play a critical role in folding acceleration by chaperonin and that their removal resulted in the conversion of an active chaperonin cage to a largely passive folding environment. Taking out-of-cage folding into account, we revisited the studies mentioned above. Surprisingly, we found that the majority of DMMBP was folded via out-of-cage folding; in other words, spontaneous folding. This finding, in addition to others discussed here, argues against a critical role of negative charges on the GroEL cage wall in the accelerated folding of protein substrates.
Results
Iterative Turnover of SR1 in Guanidium Chloride.
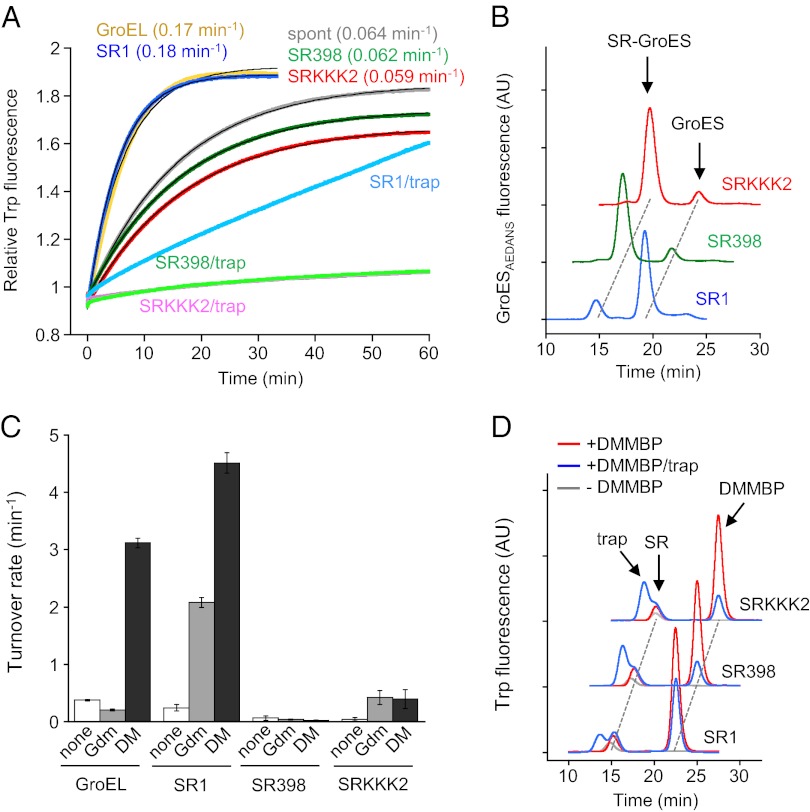
Chakraborty et al. (8) reported that the folding of DMMBP in the SR1 cage occurs much more rapidly than spontaneous folding. Indeed, when we monitored the folding of DMMBP by an increase in Trp fluorescence under the reported conditions [“buffer B” (20 mM Tris⋅HCl [pH 7.5], 20 mM KCl, 5 mM Mg(OAc)2, 1 mM DTT) in the report by Chakraborty et al. (8) was used], the apparent folding rate constants of DMMBP by SR1 (0.18 min−1) and by GroEL (0.17 min−1) were about threefold larger than that of spontaneous folding (0.064 min−1) (Fig. 1A). However, in these experiments, DMMBP was denatured by guanidium chloride (GdmCl), which has been known to destabilize the association of GroES with GroEL (10). In fact, the final GdmCl concentration in the folding solution derived from GdmCl-denatured DMMBP was 60 mM, a concentration high enough to induce the detachment of GroES from SR1 in the presence of ATP (9). Another version of the SR mutant of GroEL, SR398 [SR1(D398A)] is known to form a stable complex with GroES in the presence of GdmCl (9, 11). Using SR398 as a control, we thoroughly checked the stability of GroES association with SR1. Fluorescently labeled GroESAEDANS in the SR1/DMMBP/GroESAEDANS ternary complex was exchanged in the presence of ATP with nonlabeled GroES added in excess and appeared as free GroESAEDANS in gel-filtration analysis (Fig. 1B). The fluorescence anisotropy change showed that GroESAEDANS was detached gradually (0.34 min−1) from SR1 and rapidly (9.6 min−1) from the SR1/DMMBP complex (Fig. S1A). SR398, on the contrary, retained GroESAEDANS in the ternary complex in these tests. These results indicate that the folding of GdmCl-denatured DMMBP by SR1 occurs through iterative cycles of chaperonin reactions coupled with ATP hydrolysis; SR1 detaches GroES and releases DMMBP, and denatured DMMBP binds to SR1 again to start the next cycle. In fact, increased acceleration of ATP hydrolysis in the presence of 60 mM GdmCl and denatured DMMBP was observed for SR1 but not at all for SR398 (Fig. 1C). The fact that the majority of folded DMMBP after SR1-mediated folding reactions was found as free molecules (Fig. 1D, red trace) further supports the above contention. Thus, what was observed for SR1-mediated DMMBP folding by Chakraborty et al. (8) turned out to be folding through iterative chaperonin cycles and not single-round in-cage folding. The rate constant of DMMBP folding by SR1 (0.18 min−1) was nearly the same as that of iterative folding by GroEL (0.17 min−1) (Fig. 1A). The in-cage folding rate of GdmCl-denatured DMMBP by SR1 was difficult to measure, but the rate by SR398 was obtainable (0.20 min−1) (Fig. S1H), which reasonably resembles the iterative folding rate of GdmCl-denatured DMMBP by SR1 (0.18 min−1). It is worth mentioning that careful interpretation is needed for other reports that also assumed a single-round folding reaction when SR1-mediated folding of GdmCl-denatured substrate protein was observed (5, 6, 12).
Fig. 1.
Folding of GdmCl-denatured DMMBP. (A) Folding of GdmCl-denatured DMMBP diluted into buffer B containing 0.5 μM GroES (spont, gray) or buffer B containing 0.5 μM GroES and 0.2 μM SRs (or 0.1 μM GroEL) (GroEL, yellow; SR1, blue; SR398, green; SRKKK2, red) at 25 °C. ATP was added at time 0. Trap(D87K) was added before ATP when indicated (trap, final = 0.1 μM). Folding was monitored by the increase in Trp fluorescence of DMMBP. SDs of rate constants from three independent experiments are shown in Table S1. The folding rate constant is shown in parentheses. The baseline fluorescence of chaperonin was subtracted from the data. (B) Gel-filtration analysis of fluorescently labeled GroESAEDANS associated with SRs in buffer B containing diluted GdmCl-denatured DMMBP (final GdmCl = 60 mM). Excess nonlabeled GroES (1 μM) was added 10 s after the start of the reaction. The folding solutions were analyzed 1 h after the start of the folding reaction. (C) ATP hydrolysis activity of GroEL and SRs in buffer B containing none, 60 mM GdmCl (Gdm), or GdmCl-denatured DMMBP (DM) (final GdmCl = 60 mM). The averaged turnover rates for a ring (7 subunits of GroEL) and SDs from three independent experiments are shown. (D) Gel-filtration analysis of the folding solution of SR-mediated folding of GdmCl-denatured DMMBP. Elution was monitored by Trp fluorescence of DMMBP. The folding solutions in A were analyzed 1 h after the start of the folding reaction. Red, folded DMMBP; blue, folded DMMBP in the presence of trap(D87K); gray baseline without DMMBP. AU, arbitrary units.
Dominance of Out-of-Cage Folding in SRKKK2-Mediated Folding.
Similar to SR398, SRKKK2 retains the associated GroESAEDANS in 60 mM GdmCl (Fig. 1B and Fig. S1A). Apparent folding rate constants of DMMBP by SRKKK2 (0.059 min−1) and by SR398 (0.062 min−1) were similar to the rate constant of spontaneous folding (0.064 min−1) (Fig. 1A). The rate constants of spontaneous folding and SRKKK2-mediated folding agree with reported values (8), but an unnoticed aspect of the folding by SRKKK2 was revealed when trap(D87K) was included in the folding solutions. Trap(D87K) is a mutant of GroEL that captures denatured protein tightly and does not release it even in the presence of ATP. As shown in Fig. 1A, trap(D87K) mostly abolished the folding of DMMBP by SRKKK2 and SR398, indicating that denatured DMMBP escaped out of the cage and was captured by trap(D87K) in the bulk medium. The slow folding of DMMBP by SR1 in the presence of trap(D87K) was assumed to be caused by the SR1-mediated folding of denatured DMMBP occasionally released from trap(D87K) during iterative folding cycles by SR1 (Fig. 1A). When the folding solutions of SRKKK2 and SR398 were analyzed by gel filtration after reactions, the majority of DMMBP was found in the form of free molecules folded via out-of-cage folding in the absence of trap(D87K), but they were found in trap(D87K)-denatured DMMBP complexes in the presence of trap(D87K) (Fig. 1D, red and blue traces). Folded DMMBP in the SR fraction (∼5%) was unaffected in the presence of trap(D87K), confirming the absence of “forced escape” (9). When proteinase K was added immediately after the addition of adenylyl imidodiphosphate (AMPPNP), a large fraction of DMMBP corresponding to the escaped, denatured DMMBP was digested (Fig. S1G). Out-of-cage folding consists of two events: escape and spontaneous folding. The escape of DMMBP from the cage of SRKKK2 and SR398 was monitored directly by FRET between donor-labeled DMMBP and acceptor-labeled trap(D87K) (Fig. S1C). The final FRET values indicated that ∼90% of DMMBP escaped out of the cage and was trapped by trap(D87K). Major rate constants of the escape of DMMBP were 1.4 min−1 for SRKKK2 and 5.6 min−1 for SR398, which were much faster than the rate constant of SR398- and SRKKK2-mediated folding (∼0.06 min−1). This rapid escape contributed little to the overall folding rate, and the out-of-cage folding of DMMBP apparently occurred at a rate very similar to a spontaneous folding rate. Thus, SRKKK2 mediated DMMBP folding mostly by out-of-cage folding under the reported conditions (8).
In-Cage Folding of Urea-Denatured DMMBP.
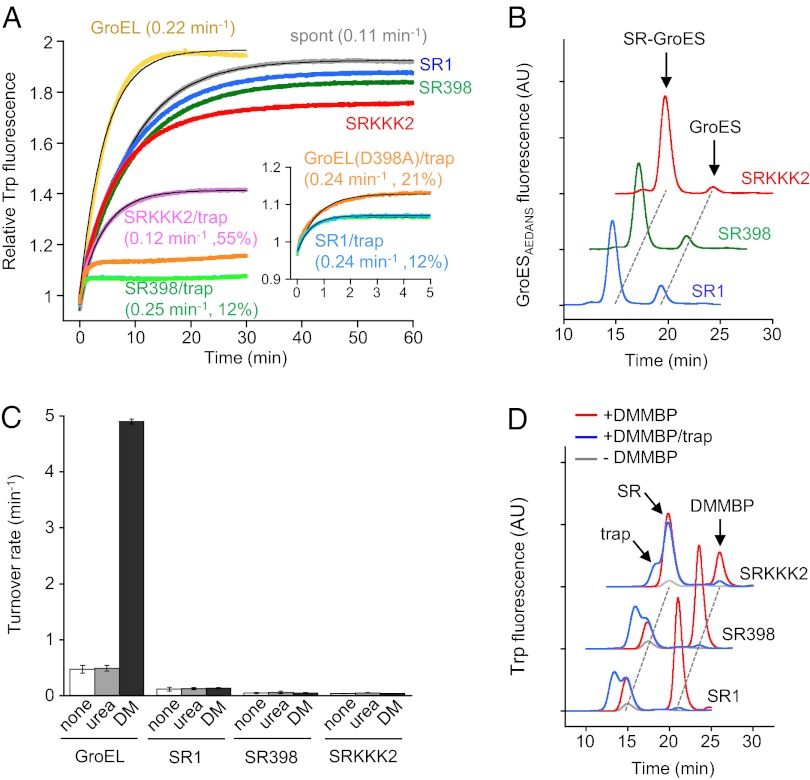
In-depth examinations demonstrated that the association of SR1 with GroES is unstable in GdmCl at >20 mM and in urea at >0.5 M (Fig. S2A). Also, Mg2+ (5 mM) in buffer B is insufficient to stabilize the association (Fig. S2B). Using 8 M urea for denaturation of DMMBP, we carried out the same series of experiments as performed in Fig. 1 in buffer containing 10 mM Mg2+ [HKM buffer (50 mM Hepes⋅NaOH [pH 7.5], 50 mM KCl, 10 mM MgCl2, 1 mM DTT)]. Spontaneous folding and GroEL-mediated folding of urea-denatured DMMBP (0.11 min−1 and 0.22 min−1, respectively) were more rapid than those of GdmCl-denatured DMMBP (0.064 min−1 and 0.17 min−1, respectively). Under these conditions, not only SR398 and SRKKK2 but SR1 did not exchange GroESAEDANS with free GroES (Fig. 2B and Figs. S1B and S2C) and ATP hydrolysis activities were not stimulated by 80 mM urea and denatured DMMBP (Fig. 2C). Gel-filtration analysis of the folding solution after the reaction showed that a small (SR1 and SR398) or large (SRKKK2) amount of folded DMMBP was associated with the SR fraction, representing the in-cage folding (Fig. 2D, red traces). Out-of-cage folding produced free DMMBP, which was abolished when trap(D87A) was present (Fig. 2D, blue traces). Time courses of folding were measured in the presence of trap(D87A) (in-cage folding) and in the absence of trap(D87A) (in-cage folding + out-of-cage folding) (Fig. 2A). The rate constant of in-cage folding was obtained as the product of the apparent rate constant and the fraction of the in-cage folding yield (SI Text). The exact value of the in-cage yield was obtained after taking into account the observation that the folded DMMBP(4C) with two internal disulfide cross-links was confined in the cage at ∼100% yield and its magnitude of Trp fluorescence was 78% that of the folded DMMBP in the bulk medium (Fig. S3). The rate constant of in-cage folding of DMMBP by SR1 (0.24 min−1, yield of 12%) was almost the same as that by SR398 (0.25 min−1, yield of 12%). SRKKK2, however, underwent in-cage folding at a slower rate constant (0.12 min−1) and a higher yield (55%). These in-cage yields were consistent with the fraction of DMMBP protected from proteinase K added immediately after addition of AMPPNP (Fig. S1G). Thus, compared with spontaneous folding (0.11 min−1), the in-cage folding of DMMBP by SR1 and by SR398 was about twofold more rapid, whereas that by SRKKK2 was at a similar level. We confirmed that GroEL(D398A), a double-ring version of SR398, was also capable of in-cage folding of urea-denatured DMMBP (0.24 min−1, yield of 21%). The timing of the trap(D87K) addition did not affect the folding and escape of DMMBP (Fig. S1 D and F).
Fig. 2.
Folding of urea-denatured DMMBP. Experimental procedures were the same as in Fig. 1, except that urea-denatured DMMBP and HKM buffer were used. Colors of data and labels in figures are the same as in Fig. 1. (A) Folding of urea-denatured DMMBP in HKM buffer. (Inset) First 5 min of folding by SR1, SR398, and GroEL(D398A) in the presence of trap(D87K). The folding rate constant and in-cage yield are shown in parentheses. SDs of rate constants from three independent experiments are shown in Table S1. The residuals of curve fitting are shown in Fig. S1E. (B) Gel-filtration analysis of GroESAEDANS associated with SRs in HKM buffer containing diluted urea-denatured DMMBP. Excess nonlabeled GroES (1 μM) was added at 10 s, and the folding solutions were analyzed 1 h after the start of the folding reaction. AU, arbitrary units. (C) ATP hydrolysis activity of GroEL and SRs in HKM buffer containing none, 80 mM urea (urea), or urea-denatured DMMBP (DM). (D) Gel-filtration analysis of the folding solution of SR-mediated folding of urea-denatured DMMBP. The folding solutions in A were analyzed 1 h after the start of the folding reaction.
Effect of Trimethylamine N-Oxide on SRKKK2-Mediated Folding.
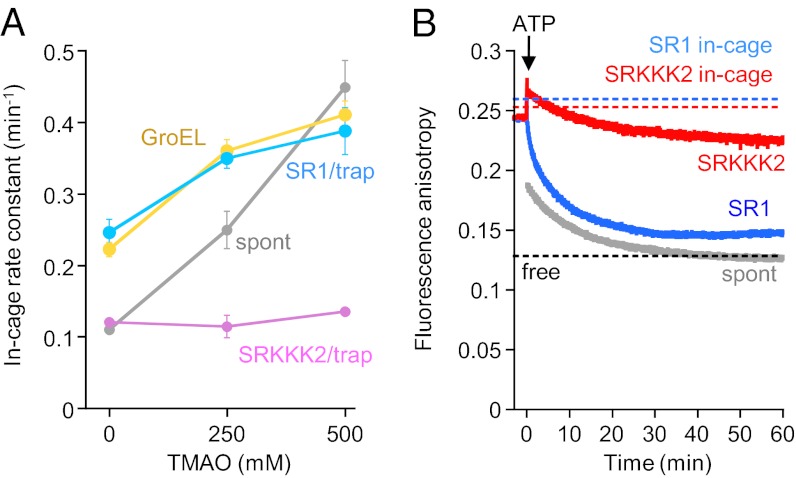
Chakraborty et al. (8) observed that trimethylamine N-oxide (TMAO) accelerated SRKKK2-mediated folding of DMMBP, as well as spontaneous folding. TMAO is an osmolyte suggested to reduce structural flexibility of proteins (13), and these investigators interpreted the results to mean that WT chaperonin mimics the effect of TMAO, consistent with their proposal that the negative charge lining of the cage may promote protein compaction by an ordering effect on water structure. However, because DMMBP mostly escapes from the cage of SRKKK2 and folds spontaneously under the conditions in the study by Chakraborty et al. (8), their observation most likely reflects the acceleration of spontaneous folding by TMAO. We reexamined their experiments under the in-cage folding condition and found that TMAO had no effect on the in-cage folding rate constant of DMMBP by SRKKK2 (Fig. 3A). On the contrary, TMAO accelerated in-cage folding by SR1 and iterative folding by GroEL to the same extent. Spontaneous folding was accelerated the most by TMAO.
Fig. 3.
Effect of TMAO and anisotropy change of urea-denatured DMMBP. (A) In-cage folding rate constant of urea-denatured DMMBP in the presence of trap(D87K) at various concentrations of TMAO. The concentration of DMMBP is 0.05 μM. Other folding conditions were the same as described in Fig. 2A. (B) Fluorescence anisotropy change during folding of urea-denatured DMMBP(A52C)Alexa in HKM buffer (final = 0.02 μM). Spontaneous folding (spont, gray), SR1-mediated folding (blue), and SRKKK2-mediated folding (red) are shown. Dashed lines represent anisotropy values of folded DMMBP(A52C)Alexa in the medium (black) and in the cages of SR1 (blue) and SRKKK2 (red).
Mobility of DMMBP in the Cage of SRKKK2.
Based on the decrease in anisotropy of fluorescently labeled DMMBP as a probe of increasing mobility of DMMBP during SR1- and SRKKK2-mediated folding, Tang et al. (6) suggested that reducing the negative net charge of the cage wall strongly impaired the mobility of DMMBP in the cage. We measured anisotropy change under the conditions of single-round folding in HKM buffer using urea-denatured DMMBP(A52C)Alexa (Fig. 3B). Native DMMBP(A52C)Alexa, as well as spontaneously folded DMMBP(A52C)Alexa, in the medium shows a low anisotropy value (0.13), reflecting high mobility. On the contrary, anisotropy values of folded DMMBP(A52C)Alexa in the isolated SR1/GroES complex and in the isolated SRKKK2/GroES complex are high (0.25), almost the same as the value of unfolded DMMBP(A52C)Alexa bound to SRs before the start of the folding reaction (0.24). Thus, mobility of folded DMMBP(A52C)Alexa in the cage is very restricted regardless of the presence or absence of negative charges in the cage. Therefore, time courses of anisotropy decrease solely reflect the progress of out-of-cage folding, and final values (SR1 = 0.15, SRKKK2 = 0.22) are parallel to the yields of the out-of-cage folding (SR1 = ∼90%, SRKKK2 = ∼40%). These results do not support the suggestion that the negative net charge on the cage wall contributes to the mobility of DMMBP in the cage.
Folding of Rubisco by SRKKK2.
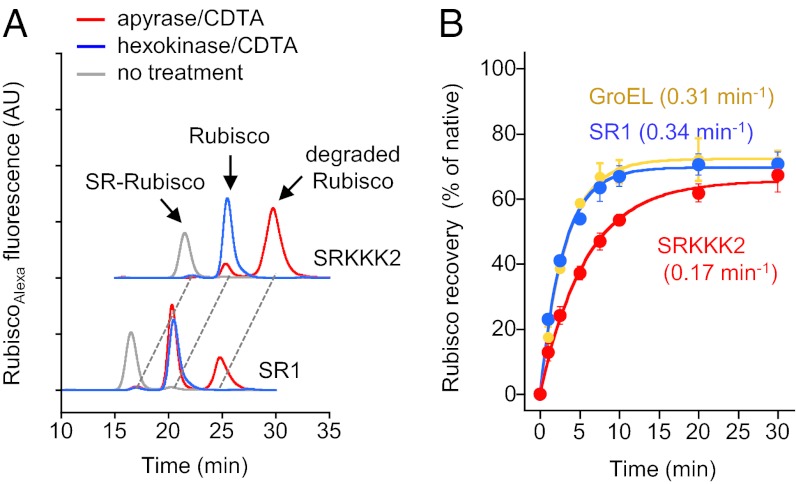
It was reported that SRKKK2 was unable to promote the folding of Rhodospirillum rubrum Rubisco (6), a stringent substrate protein in which folding is dependent on chaperonin (14). This observation has been proposed to be further evidence to support the critical role of three negatively charged residues in the cage, which are replaced by lysines in the SRKKK2 mutant. We examined folding of urea-denatured Rubisco by SRKKK2. To assess the amount of folded Rubisco monomer in the cage, the GroES lid of the cage must be removed from SRs to allow the folded Rubisco monomer to be free to form an enzymatically active dimer in the medium. Tang et al. (6) used apyrase/trans-1,2-cyclohexanediamine tetraacetic acid (CDTA) treatment for this purpose. However, when fluorescence-labeled Rubisco was used as a substrate and the folding solution of SRKKK2 was analyzed by gel filtration after treatment, the majority of Rubisco appeared as a large peak at a position equivalent to 18 kDa, which is much smaller than the Rubisco monomer (51 kDa) (Fig. 4A, red trace). SDS/PAGE analysis showed that ∼70% of Rubisco was degraded during apyrase/CDTA treatment of the SRKKK2 solution (Fig. S4). Therefore, this 18-kDa peak was derived from digested Rubisco by contaminated proteinase activity in the commercial apyrase. In the case of SR1, a Rubisco dimer peak appeared as a major peak and as an 18-kDa minor peak. It is possible that the rapid lid-opening procedure and dimer formation in the case of SR1 would result in less degradation of Rubisco monomer. To avoid apyrase, we have developed another lid-opening procedure based on a previously reported procedure (11) in which hexokinase/CDTA treatment is followed by freezing in liquid nitrogen, and we found that the Rubisco dimer was formed from both SR1- and SRKKK2-mediated folding (Fig. 4A, blue traces). Contrary to the report by Tang et al. (6), the recovery time course of Rubisco activity showed that SRKKK2 can promote the folding of Rubisco, although at a slower rate constant (0.17 min−1) than SR1 (0.34 min−1) and GroEL (0.31 min−1) (Fig. 4B). SRKKK2 was also capable of mediating the folding of rhodanese, another stringent substrate protein, and GFP as rapidly as SR398 folds (Fig. S5).
Fig. 4.
Folding of Rubisco by SRKKK2. (A) Gel-filtration analysis of the SR1- and SRKKK2-mediated folding solutions of urea/acid-denatured fluorescently labeled RubiscoAlexa before (gray) or after GroES-detaching treatments. The folding solutions 30 min after the start of the reaction were treated with apylase/CDTA for 30 min at 25 °C (red) or with hexokinase/CDTA, followed by freezing with liquid nitrogen (blue). The positions of the SR/Rubisco complex, Rubisco dimer, and degraded Rubisco are shown by arrows. AU, arbitrary units. (B) Recovery time course of Rubisco activity at 25 °C by GroEL (yellow), SR1 (blue), and SRKKK2 (red) measured after hexokinase/CDTA treatment.
Discussion
Chaperonin-mediated folding could be a mixture of in-cage folding and out-of-cage folding. Thorough examination is needed for individual substrate proteins because the fraction of out-of-cage folding differs from one protein to another and is dependent on GroEL variants (9). We found here that the folding of GdmCl-denatured DMMBP mediated by SRKKK2 undergoes mostly (∼95%) out-of-cage folding and its folding kinetics resemble those of spontaneous folding. Therefore, the analysis of SRKKK2-mediated folding of GdmCl-denatured DMMBP previously reported (6, 8) does not reflect in-cage folding but, rather, out-of-cage folding.
Using urea denaturation and buffer containing 10 mM Mg2+, we observed exclusive in-cage folding of DMMBP and found that DMMBP folds in the cage of SR1 and SR398 about twofold faster than it does in the cage of SRKKK2 and in free solution. The previously reported slow folding of DMMBP by SRKKK2 (6, 8) is, coincidently, in agreement with our results. However, SRKKK2 is not always inefficient; it is as efficient as SR398 in rhodanese folding as well as in GFP folding, which is twofold faster than the spontaneous folding. Thus, it is reasonable to state that the effect of the cage wall charge differs depending on the substrate protein.
Folding rates (and the yield of in-cage folding) vary depending on the combination of chaperonin variants and substrate proteins (9, 15), and the mutational effect of chaperonin on protein folding is not straightforward. Hydrophobicity of the cage wall has been thought to be an important factor that affects folding, but both the less hydrophobic SR398(Y203C) and the more hydrophobic pyrene-labeled SR398(F44C) fold rhodanese more slowly than SR398. Furthermore, a chaperonin variant optimized for the folding of GFP loses the ability to fold other substrates efficiently (16). Even free chloride ions affect the apparent folding rate (4, 17). Therefore, generalizing the mechanism of chaperonin folding based on the analysis of a single GroEL variant and substrate protein is insufficient because the folding of individual proteins appears to be distinct.
Materials and Methods
Proteins.
DMMBP and DMMBP(A52C), prepared as inclusion bodies from expressing Escherichia coli, were solubilized by GdmCl, refolded in buffer, and purified with amylose resin (New England Biolabs). SR398, SRKKK2, trap(D87K), GroES, fluorescently labeled GroESAEDANS, and RubiscoAlexa were prepared as described (9). Protein concentrations were measured by a Bradford protein assay kit (Bio-Rad). The labeling of DMMBP(A52C) by Alexa 488-maleimide (Molecular Probes) was performed as previously described (9).
Folding Assay.
Folding of DMMBP was assayed as follows. The reaction mixture containing 0.2 μM SR (or 0.1 μM WT GroEL) and 0.5 μM GroES in buffer B (8) or HKM buffer was incubated with stirring at indicated temperatures. DMMBP (10 μM) was denatured in a buffer containing 6 M GdmCl, 20 mM Tris⋅HCl (pH 7.5), and 1 mM DTT or in a buffer containing 8 M urea, 20 mM Tris⋅HCl, and 1 mM DTT for more than 30 min. Denatured DMMBP was diluted 100-fold into the reaction mixture, and the chaperonin-mediated folding reaction was started by the addition of ATP (1 mM). When indicated, 0.1 μM trap(D87K) was mixed before addition of ATP to measure the in-cage folding. DMMBP folding was monitored by the recovery of intrinsic Trp fluorescence of native DMMBP at 340 nm (excitation at 295 nm). The gel-filtration analysis was carried out with a Superdex 200 10/300GL column (GE Healthcare) equilibrated with HKM buffer. The Rubisco folding reaction was performed at 25 °C as described (9) with modifications. Rubisco denatured in 100 mM glycine⋅HCl (pH 2.2) containing 8 M urea and 5 mM DTT was diluted into HKM buffer (final = 0.2 μM) containing 0.5 μM SR and 2 μM GroES. The folding reaction was started by the addition of ATP (final = 1 mM). Aliquots were subjected to hexokinase/CDTA treatment. Recovered Rubisco activity was measured by a coupling enzyme assay (9).
Gel-Filtration Analysis.
Stability of the association of GroESAEDANS with SRs was examined by gel-filtration column chromatography monitored with the fluorescence of AEDANS at 490 nm (excitation at 340 nm). Denatured substrate protein (0.15 μM) was mixed with buffer containing 0.1 μM SR and 0.1 μM GroESAEDANS to form the ternary complex. ATP (1 mM) was added to the solution, and 1 μM GroES was added subsequently. After incubation under the indicated conditions, the solution was applied to a gel-filtration column equilibrated with HKM buffer. Location of Rubisco after the folding reaction was examined by gel-filtration column chromatography monitored with the fluorescence of Alexa at 520 nm (excitation at 480 nm). Denatured RubiscoAlexa was diluted into HKM buffer containing 0.1 μM SR, 0.25 μM GroES (final = 0.075 μM). Folding was initiated by addition of 1 mM ATP, and the solution was incubated for 30 min. For apyrase/CDTA treatment, solution was incubated with apyrase (0.17 U/μL; Sigma–Aldrich, Grade I from potato) and 15 mM CDTA for 5 min, and it was subsequently incubated with 50 mM Mg(OAc)2 for 1 h as reported (5), except that Rubisco mutant K168E was not added. For hexokinase/CDTA treatment, solution was incubated with hexokinase (0.2 U/μL; Roche Diagnostics) and 20 mM glucose for 15 s, and 15 mM CDTA was subsequently added; solution was frozen by means of liquid nitrogen. After being thawed on ice, Mg(OAc)2 (50 mM) was added. After incubation for 1 h at 25 °C, solution was analyzed by means of a gel-filtration column equilibrated with HKM buffer containing 10 mM NaHCO3. Alexa fluorescence of RubiscoAlexa was monitored.
ATP Hydrolysis Activity.
The reaction was started by the addition of ATP (1 mM) to buffer B or HKM buffer containing 0.5 μM SR, 2 μM GroES, and indicated components (60 mM GdmCl, 60 mM GdmCl and 1 μM denatured DMMBP, 80 mM urea, or 80 mM urea and 1 μM denatured DMMBP). Aliquots were mixed with 6% (vol/vol) perchloric acid to stop ATP hydrolysis, and produced Pi was assayed by the malachite green method. The steady state turnover rate of ATP hydrolysis was calculated from a linear increase in the amount of Pi.
Supplementary Material
Acknowledgments
We thank Yoko Ishizaki for technical assistance and Daniel Xu and Peter Gee for critical reading of the manuscript. This work was supported by Grant-in-Aid for Scientific Research on Priority Areas 19058004 (to M.Y.) and Grant-in-Aid for Scientific Research on Innovative Areas 23107728 (to F.M.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204547109/-/DCSupplemental.
References
- 1.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 2.Horwich AL, Fenton WA. Chaperonin-mediated protein folding: Using a central cavity to kinetically assist polypeptide chain folding. Q Rev Biophys. 2009;42(2):83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 3.Weissman JS, et al. Mechanism of GroEL action: Productive release of polypeptide from a sequestered position under GroES. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 4.Apetri AC, Horwich AL. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc Natl Acad Sci USA. 2008;105:17351–17355. doi: 10.1073/pnas.0809794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinker A, et al. Dual function of protein confinement in chaperonin-assisted protein folding. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- 6.Tang YC, et al. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 7.Tang YC, Chang HC, Chakraborty K, Hartl FU, Hayer-Hartl M. Essential role of the chaperonin folding compartment in vivo. EMBO J. 2008;27:1458–1468. doi: 10.1038/emboj.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty K, et al. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 2010;142:112–122. doi: 10.1016/j.cell.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Motojima F, Yoshida M. Polypeptide in the chaperonin cage partly protrudes out and then folds inside or escapes outside. EMBO J. 2010;29:4008–4019. doi: 10.1038/emboj.2010.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd MJ, Lorimer GH. Stability of the asymmetric Escherichia coli chaperonin complex. Guanidine chloride causes rapid dissociation. J Biol Chem. 1995;270:5388–5394. doi: 10.1074/jbc.270.10.5388. [DOI] [PubMed] [Google Scholar]
- 11.Rye HS, et al. Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature. 1997;388:792–798. doi: 10.1038/42047. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, et al. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell. 2008;133:142–153. doi: 10.1016/j.cell.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 13.Qu Y, Bolen DW. Hydrogen exchange kinetics of RNase A and the urea:TMAO paradigm. Biochemistry. 2003;42:5837–5849. doi: 10.1021/bi0206457. [DOI] [PubMed] [Google Scholar]
- 14.Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and Mg-ATP. Nature. 1989;342:884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- 15.Madan D, Lin Z, Rye HS. Triggering protein folding within the GroEL-GroES complex. J Biol Chem. 2008;283:32003–32013. doi: 10.1074/jbc.M802898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JD, Herman C, Tipton KA, Gross CA, Weissman JS. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell. 2002;111:1027–1039. doi: 10.1016/s0092-8674(02)01198-4. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi NK, Fenton WA, Deniz AA, Horwich AL. Double mutant MBP refolds at same rate in free solution as inside the GroEL/GroES chaperonin chamber when aggregation in free solution is prevented. FEBS Lett. 2011;585:1969–1972. doi: 10.1016/j.febslet.2011.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.