Abstract
Rab4A is a master regulator of receptor recycling from endocytic compartments to the plasma membrane. The protein TBC1D16 is up-regulated in melanoma, and TBC1D16-overexpressing melanoma cells are dependent on TBC1D16. We show here that TBC1D16 enhances the intrinsic rate of GTP hydrolysis by Rab4A. TBC1D16 is both cytosolic and membrane associated; the membrane-associated pool colocalizes with transferrin and EGF receptors (EGFRs) and early endosome antigen 1, but not with LAMP1 protein. Expression of two TBC1D16 isoforms, but not the inactive R494A mutant, reduces transferrin receptor recycling but has no effect on transferrin receptor internalization. Expression of TBC1D16 alters GFP-Rab4A membrane localization. In HeLa cells, overexpression of TBC1D16 enhances EGF-stimulated EGFR degradation, concomitant with decreased EGFR levels and signaling. Thus, TBC1D16 is a GTPase activating protein for Rab4A that regulates transferrin receptor recycling and EGFR trafficking and signaling.
Keywords: endocytosis, membrane traffic
Rab GTPases are master regulators of membrane trafficking (1). They exist in an active, GTP-bound state and an inactive, GDP-bound state. When active, they bind a number of specific effector proteins that mediate their diverse roles in transport vesicle formation, motility, docking, and fusion. A given cell may contain as many as 40 distinct Rab proteins (2), each localized to a distinct membrane microdomain and providing compartment identity to those membrane domains. Rab activity is regulated by GTPase activating proteins (GAPs) and guanine nucleotide exchange factors, which inactivate the Rab by catalyzing GTP hydrolysis and reactivate a Rab by facilitating release of bound GDP, respectively (3).
TBC1D16 is a member of the Tre2/Bub2/Cdc16 (TBC) domain-containing family of proteins. Members of this family have been associated with cancer and cell signaling events, and are important for intracellular receptor trafficking (4). TBC domains compose the catalytic core of several Rab GAPs (3, 5). More than 40 human TBC domain-containing proteins have been identified, and their substrate Rab specificities are slowly being assigned. TBC1D20 inactivates Rab1 and causes collapse of the Golgi complex (6, 7), TBC1D10 A-C acts on Rab35 (8, 9), TBC1D30 acts on Rab8A (10), and RUTBC1 is a GAP for Rab32 (11).
Metastatic melanoma is a leading cause of cancer deaths worldwide (12). TBC1D16 is up-regulated in melanoma and overexpressed in ∼60% of melanomas (www.genecards.org/cgi-bin/carddisp.pl?gene=TBC1D16). Micropthalmia-associated transcription factor, a master regulator of melanocyte development and melanoma progression, induces TBC1D16 expression (13). TBC1D16 is the second-highest scoring gene product identified using a computational framework integrating chromosomal copy number and gene expression data to detect aberrations that promote melanoma progression (14). Moreover, TBC1D16-overexpressing melanoma cells depend on TBC1D16 for growth (14). The function of TBC1D16 is unknown, however. We show here that TBC1D16 is a Rab4A GAP that regulates transferrin receptor recycling and EGF receptor (EGFR) signaling.
Results and Discussion
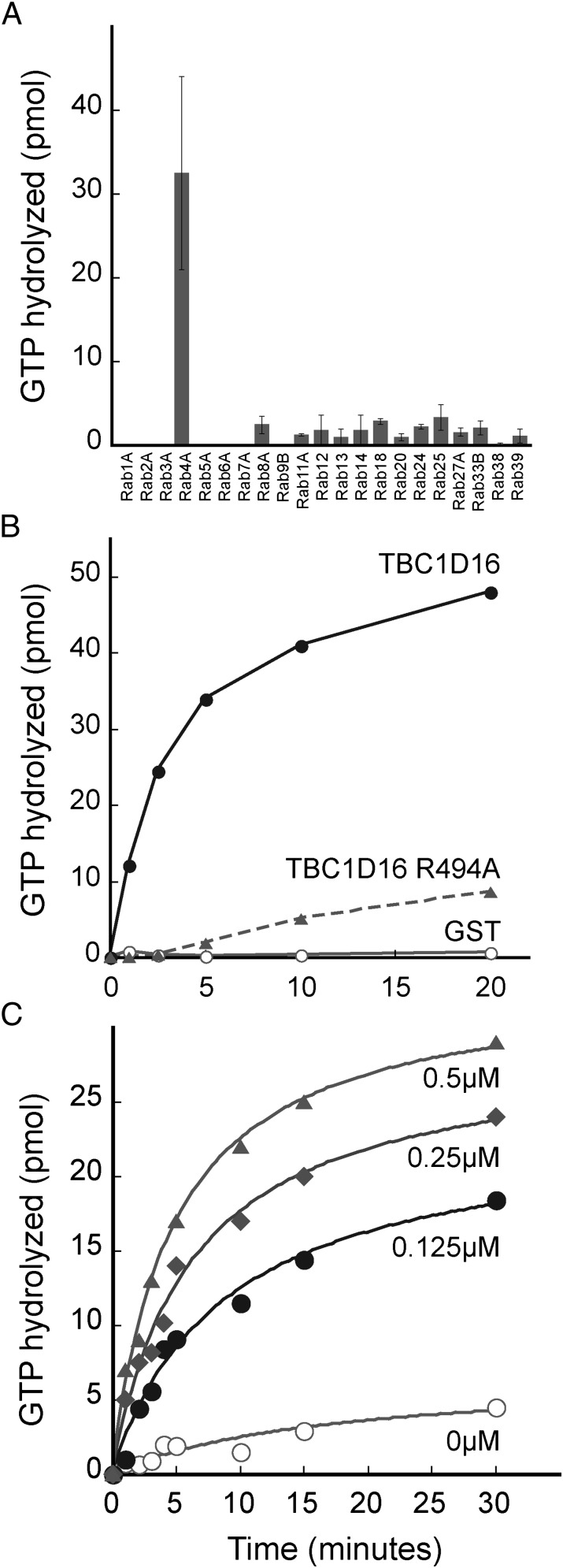
We screened 21 purified Rab GTPases as potential substrates for purified GST-TBC1D16, including Rab proteins that are enriched in melanoma cells (Rab27A and Rab38). We identified Rab4A as the preferred substrate for GST-TBC1D16 (Fig. 1A). Several Rab GAPs use an essential arginine residue to enhance the intrinsic GTPase activity of their corresponding Rab substrate(s) (5). We aligned the sequence of TBC1D16 with other Rab GAPs and identified arginine 494 as being potentially critical for TBC1D16 GAP function. Mutation of this arginine to alanine (TBC1D16 R494A) significantly impaired the ability of TBC1D16 to enhance Rab4A GTPase activity (Fig. 1B). Thus, TBC1D16 is a Rab4A GAP in vitro, and arginine 494 is indispensable for catalysis. We performed our initial experiments using equimolar amounts of Rab substrates and GAP enzyme, to ensure detection of activity. Reactions containing substoichiometric catalytic amounts of TBC1D16 also displayed potent enhancement of Rab4A GTPase activity in a concentration-dependent manner (Fig. 1C).
Fig. 1.
TBC1D16 is a GAP for Rab4A in vitro. (A) Biochemical screen of 21 Rabs. GAP activity was assayed using 150 pmol Rab loaded with γ-P32-GTP and 150 pmol GAP or GST control for 5 min at 25 °C. Depicted is the level of GAP-dependent, GTP hydrolysis observed. Error bars represent SD of duplicate determinations. (B) Reactions contained 150 pmol TBC1D16 (filled circles), TBC1D16 R494A (triangles), or GST (open circles) for the indicated times and 150 pmol Rab4A loaded with γ-P32-GTP. (C) Reactions were carried out as in B using TBC1D16 at the indicated concentrations with 1 μM Rab4A protein.
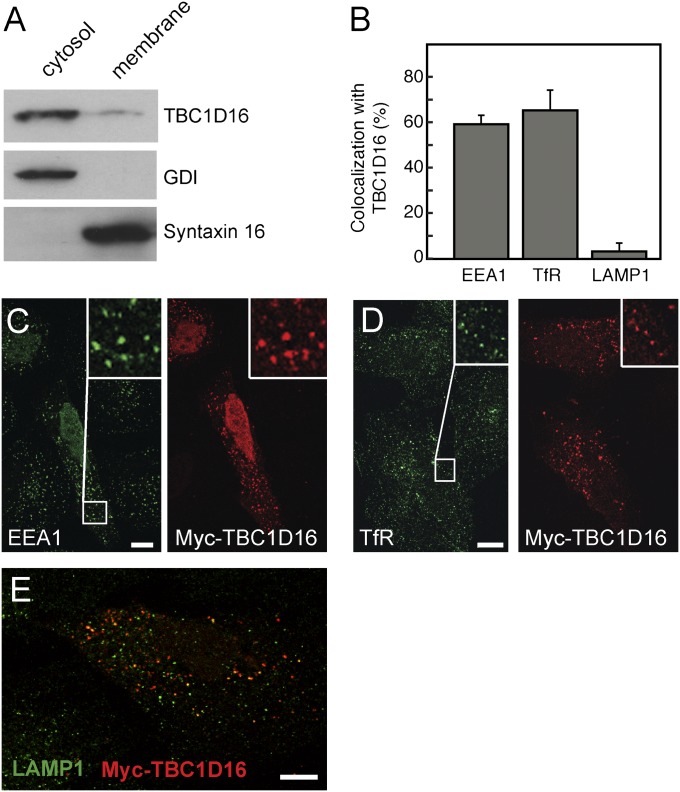
Rab4A is localized to early and recycling endosomes and is required for the rapid recycling of receptors from early endosomes to the cell surface (15). We expressed GFP-TBC1D16 in cells and evaluated its localization. Differential centrifugation (Fig. 2A) revealed that most GFP-TBC1D16 protein fractionated with cytosol, but a portion was membrane-associated. Cells expressing exogenous Myc-TBC1D16 were treated with liquid nitrogen to deplete cytosolic proteins and visualized by immunofluorescence microscopy (Fig. 2 B–E). TBC1D16 colocalized extensively with transferrin receptors and the early endosomal marker EEA1, but not with LAMP1, a marker for late endosomes and lysosomes (Fig. 2 B–E). This is consistent with TBC1D16's role as a Rab4A GAP as Rab4A localizes to early and recycling endosomes. Thus, membrane-associated TBC1D16 resides primarily on early endosomes, where it can act on Rab4A.
Fig. 2.
TBC1D16 colocalizes with transferrin receptor and EEA1. (A) Immunoblot of GFP-TBC1D16 comparing membrane and cytosol fractions. Control proteins were GDI for cytosol and Syntaxin 16 for membranes. Equal proportions of cytosol and membrane fractions were compared. (B) Quantitation of colocalization of TBC1D16 with EEA1 (471 TBC1D16+ structures in 10 cells), transferrin receptor (TfR) (646 structures as for EEA1), and LAMP1 (582 structures as for EEA1). (C–E) Immunofluorescence microscopy of HeLa cells expressing Myc-TBC1D16 and costained for EEA1 (C), TfR (D), or LAMP1 (E). In D, HeLa cells were serum-starved and then allowed to endocytose human holotransferrin for 30 min before being processed. All cells were subjected to freeze-thaw in liquid nitrogen to deplete cells of cytosolic proteins. The insets at the upper right of C and D are magnified portions (boxes) of the images shown in C and D. A merged image is shown in E. (Scale bars: 10 μm.)
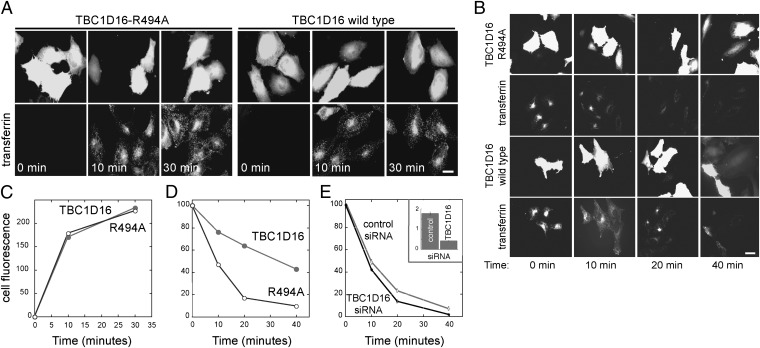
Rab GAPs can activate particular Rabs in vitro, but fail to act on those Rabs in cells (3). Thus, we examined the effect of TBC1D16 expression on Rab4A-dependent transferrin receptor recycling in cultured cells (16). We first tested transferrin endocytosis. Cells were pretreated with rhodamine-labeled transferrin on ice and then warmed for various times to monitor internalization. The amount of transferrin internalized was essentially identical in cells expressing GFP-TBC1D16 and cells expressing the catalytically inactive, mutant protein GFP-TBC1D16 R494A (Fig. 3 A and C). Thus, TBC1D16 expression did not influence Rab5-dependent endocytosis of transferrin.
Fig. 3.
TBC1D16 has no effect on transferrin internalization, but blocks transferrin recycling. (A) Cells transfected for 24 h with TBC1D16-R494A (Left) or WT TBC1D16 (Right) were incubated with rhodamine transferrin for the times indicated and then fixed for visualization. (B) Cells transfected as in A were pulsed with rhodamine transferrin for 30 min and then chased with unlabeled holotransferrin for the designated times to detect loss of internalized transferrin by recycling. (C) Quantitation of images obtained in an experiment carried out as in A. (D) Quantitation of images obtained in an experiment as in B. All experiments were performed three times; a representative example is shown. Each experiment involved counting 20 cells for each condition with <5% error observed at all time points. (Scale bars: 20 μm.) (E) UAC1273 cells at 48 h after transfection with control or fluorescent TBC1D16-siRNA were treated as in B. (Inset) Quantitative RT-PCR was used to determine relative TBC1D16 mRNA expression normalized to 18 S rRNA. The mean fluorescence per cell is shown. At least 80 cells were analyzed in each condition; only cells that incorporated fluorescent TBC1D16 siRNA were scored. Less than 5% error was observed at all time points.
To monitor receptor recycling, HeLa cells expressing GFP-TBC1D16 or GFP-TBC1D16 R494A were pulse-labeled with rhodamine-transferrin for 30 min at 37 °C and then chased with unlabeled holo-transferrin for various times and processed for immunofluorescence microscopy. Although cells expressing GFP-TBC1D16 R494A recycled most of the internalized transferrin by 20 min and had little remaining cell-associated fluorescence, cells expressing WT TBC1D16 retained significant amounts of transferrin after 20 min and as much as 40% of initial levels after 40 min (Fig. 3 B and D). In UAC1273 melanoma cells, siRNA depletion of TBC1D16 mRNA (monitored by quantitative PCR; Fig. 3E, Inset) slightly enhanced the rate of transferrin recycling (Fig. 3E). These experiments demonstrate that TBC1D16 interferes with the ability of cells to recycle transferrin, consistent with its role as a Rab4A GAP.
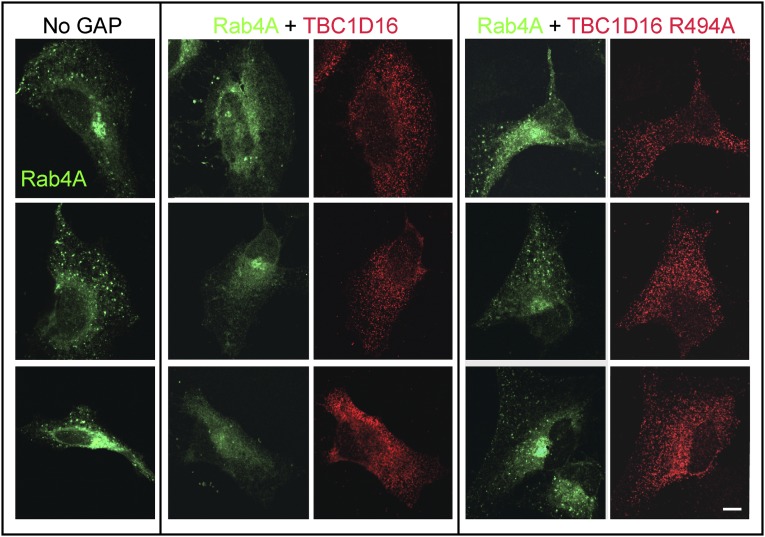
When Rabs are converted to their GDP-bound forms, they are targets for GDI-mediated membrane extraction (17). Indeed, GTP-Rabs are stabilized on membranes by binding of their specific effector proteins (18). If TBC1D16 were a Rab4A GAP in cells, it would be predicted to decrease the levels of Rab4A on endosomes. GFP-Rab4A decorated numerous punctate structures, and a perinuclear concentration specific to the exogenously expressed protein was also observed in some cells (Fig. 4, Left). In contrast, the Rab4A localization in cells expressing TBC1D16 (Fig. 4, Middle) was converted to a cytoplasmic haze and the punctate pattern was eliminated. Importantly, this change required the catalytic activity of TBC1D16 as cells expressing the inactive TBC1D16R494A showed the Rab4A puncta characteristic of untransfected cells (compare Fig. 4 Right and Left). These experiments strongly suggest that TBC1D16 acts on Rab4A in cells to convert membrane associated, GTP-Rab4A to a predominantly cytosolic, GDP-bearing form.
Fig. 4.
TBC1D16 expression alters the appearance of GFP-Rab4A in HeLa cells. HeLa cells were transfected with GFP-Rab4A alone (Left), GFP-Rab4A with WT Myc-TBC1D16 (Middle), or Myc-TBC1D16-R494A (Right). GAP proteins were detected using anti-Myc antibodies (red) after cytosolic depletion using liquid nitrogen. (Scale bar: 10 μm.)
Little colocalization was detected between GFP-Rab4A and exogenous, WT TBC1D16 or TBC1D16 R494A proteins (Fig. 4), despite the fact that TBC1D16 colocalized with EEA1 and transferrin receptors (Fig. 2B). It is likely that endogenous (and exogenous) WT TBC1D16 proteins act on GFP-Rab4A, causing its release from those membranes. The residual GFP-Rab4A puncta seen in cotransfected cells likely represent Rab4A-positive recycling intermediates that do not harbor TBC1D16. Colocalization of Rab4A and TBC1D16 is not expected, because GAPs have very low affinity for their Rab GTPase substrates (5) and in a few cases have been shown to localize to membranes by binding to a previous-acting Rab protein (3).
It is important to note that Rab17, Rab21, and Rab35 also participate in transferrin receptor trafficking (19–21). We cannot exclude the possibility that TBC1D16 also acts on these Rab proteins. However, the redistribution of Rab4A from membrane puncta to the cytosol (Fig. 4) demonstrates that TBC1D16 is a potent Rab4A GAP in cells.
The TBC1D16 protein has a predicted Mr of 86,372. A shorter isoform (predicted Mr, 47,124) also has been identified (14) largely comprising the TBC domain present within the C-terminal, 410 residues of TBC1D16 (357–767). Because the shorter form is often seen in melanoma, it was important to verify that this form has the same specificity for Rab4A. The purified, ∼45-kD form of TBC1D16 was similar to the full-length protein in its ability to stimulate Rab4A GTPase activity (Fig. S1A). Moreover, analogous to the full-length protein, expression of the shorter form had no effect on transferrin internalization (Fig. S1B), but did slow transferrin recycling (Fig. S1C).
The importance of TBC1D16 in melanoma (14) led us to explore its role in regulating the Rab4A-dependent trafficking of receptor tyrosine kinases. On addition of EGF, activated EGFRs are endocytosed by clathrin-coated vesicles and delivered to early endosomes; the receptors are then sequestered into the internal vesicles of multivesicular endosomes, before their degradation (22). Alternatively, they may recycle to the cell surface (22). Ligand release, intraendosomal sequestration, and degradation all turn off EGFR-mediated signaling events. We reasoned that the TBC1D16-regulated rate of receptor exit from early endosomes should influence EGFR trafficking and signaling.
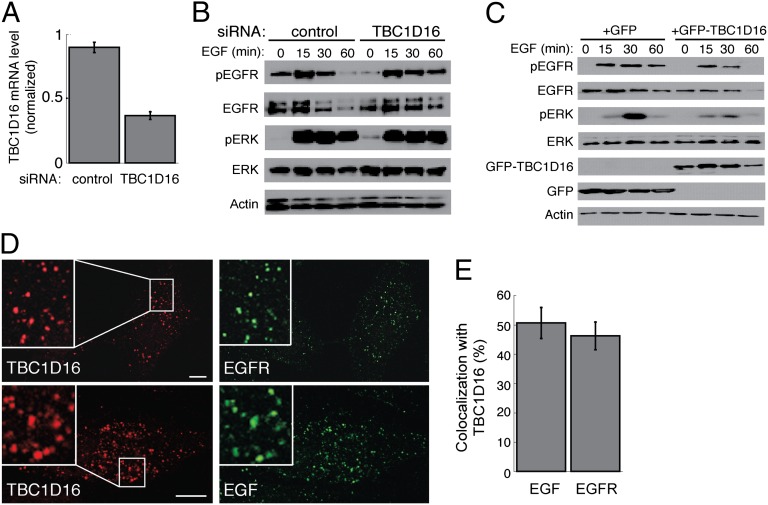
Serum-starved WM115 melanoma cells were stimulated with EGF for various times in the presence of either control or TBC1D16 siRNA (Fig. 5 A and B). Compared with control treated cells, cells depleted of TBC1D16 had higher levels of EGFR and phospho-EGFR at 60 min after EGF addition. Moreover, there was a slight increase in EGFR signaling, as determined by monitoring the levels of phosphorylated ERK protein, after 60 min of EGF treatment. WM115 cells, which express relatively high levels of EGFR, were unusual in that phospho-ERK levels remained high 60 min after addition of EGF, even in the control condition. Because we tested only a single siRNA, we cannot exclude off-target effects. However, in support of the conclusion that TBC1D16 influences EGFR trafficking, expression of GFP-TBC1D16, but not of GFP, facilitated EGFR degradation in HeLa cells (compare the 60-min time points in Fig. 5C). EGFR signaling was also reduced, with significantly less phospho-ERK and phospho-EGFR seen at 30 min and 60 min after EGF addition, respectively. These data support the conclusion that TBC1D16 interferes with Rab4A-mediated EGFR recycling, thereby promoting its degradation. In cells treated with EGF for 15 min, TBC1D16 colocalized with EGF and EGFR (Fig. 5 D and E), consistent with TBC1D16’s localization in early endosomes and its ability to regulate EGFR trafficking.
Fig. 5.
TBC1D16 regulates EGFR degradation, activation, and signaling. (A) Quantitative RT-PCR of TBC1D16 mRNA expression, normalized to 18S rRNA, in WM115 cells at 48 h after nucleofection with control or TBC1D16-siRNA. (B) Determination of phospho-EGFR (Y1068), EGFR, phospho-ERK (T183), ERK 1/2, and actin levels in WM115 cells (25 μg of total cell lysate) treated with 20 ng/mL EGF for indicated times at 48 h after nucleofection with control or TBC1D16 siRNA. Cells were serum-starved for 8 h and incubated with cycloheximide during the last 4 h before EGF stimulation. (C) HeLa cells (40 μg total lysate) at 24 h posttransfection with GFP or GFP-TBC1D16, analyzed as in B. (D) Colocalization of Myc-TBC1D16 with EGF and EGFR. At 24 h after transfection with Myc-TBC1D16, HeLa cells were were serum-starved for 30 min and then treated for 15 min with 100 ng/mL of Alexa Fluor 488 EGF for colocalization with EGF or 100 ng/mL of unlabeled human EGF for colocalization with EGFR. Insets show magnification of the images. (Scale bars: 10 μm.) (E) Quantitation of colocalization observed between EGF (618 TBC1D16+ structures) and EGFR (463 TBC1D16+ structures) with TBC1D16. The average colocalization for 10 cells is depicted.
In summary, this study demonstrates that TBC1D16 is a Rab4A GAP that interferes with Rab4A-dependent receptor recycling from endosomes back to the cell surface. Although TBC1D11/GAPCenA shows strong Rab4 GAP activity in vitro (8), TBC1D16 is the only protein known to function as a Rab4A GAP in cells. Rab4A is critical for the intracellular trafficking of potent signaling receptors, including EGFR, VEGFR-2, and PDGFRB (23–26). Thus, TBC1D16’s ability to regulate Rab4A controls EGF-induced EGFR degradation and signaling.
Our finding that TBC1D16 promotes EGFR degradation and decreases EGFR signaling does not explain how TBC1D16 facilitates melanomagenesis. TBC1D16 overexpression is known to enhance melanomagenesis, perhaps by altering the balance among multiple signaling pathways. Nevertheless, our finding that TBC1D16 regulates the ubiquitous Rab4A GTPase puts this enzyme in a key position to dictate receptor recycling from early endosomes, thereby regulating transferrin receptor recycling, EGFR trafficking, and EGFR signaling.
Materials and Methods
Plasmids.
TBC1D16 cDNA was obtained from American Type Culture Collection (MGC-25062). The long form (1–767) and short form (357–767) were generated by PCR amplification and ligated into pCS2+ vectors. The R494A construct was generated via Quick Change (Stratagene).
Protein Purification.
Rabs were purified from Rosetta2 (DE3) cells (Novagen) grown at 37 °C to A600 = 0.4–0.6. Cells were induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside for 3 h at 30 °C. His-tagged Rabs were purified using nickel-nitrilotriacetic acid (Qiagen) and desalted using PD-10 columns (GE Healthcare) in 250 mM NaCl, 25 mm Hepes (pH 7.4), 10 mM β-mercaptoethanol, and 10% (vol/vol) glycerol. GST-Rabs were purified with glutathione-Sepharose 4 FF (GE Healthcare), concentrated using an Amicon Ultra spin concentrator (Millipore), and stored in 10% (vol/vol) glycerol at −80 °C. GST-TBC1D16 proteins were produced using Arctic Express RIL (Agilent Technologies) induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside 24 h at 11 °C.
Cell Culture and Transfection.
HeLa cells were transfected with FuGENE 6 (Promega). WM115 cells were nucleofected with 300 nM control siRNA (GUUCAAUAGGCUUACUAAUUU) or 5′ fluorescently labeled TBC1D16 siRNA (Dy547-GCCAGAGGAUGAGGAGAAUU) using program X-001, Solution V, and a nucleofector 2 apparatus (Lonza). UAC1273 cells were transfected with siRNA using Dharmafect (Reagent 1; Thermo Scientific). Cells were maintained in DMEM supplemented with penicillin/streptomycin, glutamine, and 7.5% (vol/vol) FCS.
Antibodies and Reagents.
Rabbit anti-GFP, rabbit anti-actin, Alexa Fluor 488 goat anti-mouse, Alexa Fluor 488 goat anti-rabbit and mouse anti-transferrin receptor antibodies, Alexa Fluor 488 transferrin, Alexa Fluor 488 EGF, and tetramethylrhodamine transferrin were obtained from Invitrogen. Human holotransferrin was obtained from Sigma-Aldrich. Rabbit anti-EGFR conjugated to Alexa Fluor 488 and phospho-EGFR (Tyr1068) rabbit mAbs were obtained from Cell Signaling. Rabbit anti-Syntaxin 16 and mouse anti-EEA1 were obtained from BD Biosciences, chicken anti c-Myc was obtained from Bethyl Laboratories, and rabbit anti-GDI was raised in rabbits. Goat anti-rabbit HRP was obtained from Bio-Rad. Human EGF, rabbit anti-ERK 1/2, and rabbit anti–phospho-MAPK (pT183) were obtained from Promega. Mouse anti-LAMP1 mAb ascites was obtained from the Developmental Studies Hybridoma Bank at the University of Iowa.
Cell Lines.
UAC1273 is a low-passage human melanoma cell line derived from tumor samples collected between 1990 and 1997 at the University of Arizona Medical Center. Samples were completely deidentified, and excess tissue was removed at the time of therapeutic resection. Each patient signed a general surgical consent form allowing use of tissues for research. Tumor samples were procured in compliance with the University of Arizona Institutional Review Board’s regulations, with cells obtained from M.B.P. WM115 cells were obtained from the Wistar Institute Melanoma Repository.
GAP Assays and Cell Fractionation.
Purified Rab GTPases were loaded with [γ-32P]GTP (18), and GAP assays were performed as described previously (11). For membrane/cytosol fractionation, at 24 h after transfection, HeLa cells were washed twice in PBS, then collected in cold lysis buffer [10 mM Hepes (pH 7.4), 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM ATP, and 1× protease inhibitor mixture (Roche)]. Lysates were passed 20 times through a 25-gauge needle and rotated end over end for 15 min at 4 °C. Postnuclear supernatant was obtained by centrifugation at 835 × g for 7 min at 4 °C. The supernatant was then centrifuged at 213,000 × g for 15 min at 4 °C in a Beckman Coulter TLA 100.2 rotor. The membrane fraction was resuspended in ice-cold lysis buffer plus 1% (vol/vol) Triton.
Transferrin Receptor Uptake and Recycling.
At 24 h after transfection of HeLa cells with GFP-tagged constructs or 48 h after transfection of UAC1273 cells with 20 nM siRNA, cells (on 22 × 22-mm coverslips) were washed in serum-free MEM. HeLa cells were incubated in serum-free uptake medium [SFUM; MEM plus 20 mM Hepes (pH 7.4) and 0.1% BSA] and UAC1273 cells in DMEM containing 1% (vol/vol) FCS for 30 min at 37 °C to deplete transferrin. Cells were chilled at 4 °C for 10 min, then incubated for 20 min at 4 °C in SFUM or DMEM/1% FCS plus 25 μg/mL of tetramethyrhodamine transferrin (Molecular Probes). Cells were rapidly warmed to 37 °C, and washed once in 20 mM acetic acid and 500 mM NaCl (pH 3.0) at various times. For transferrin uptake experiments, cells were subsequently washed twice in PBS and fixed for 20 min in 3.7% paraformaldehyde containing 20 mM Hepes (pH 7.4). After quenching in DMEM plus 10 mM Hepes (pH 7.4), cells were washed twice in PBS, rinsed in water, and mounted using Mowiol. For transferrin recycling, HeLa and UAC1273 cells were loaded with transferrin as described above at 37 °C for 30 min, then washed twice in SFUM and further incubated with 1 mg/mL of unlabeled holotransferrin at 37 °C for 0, 10, 20, or 40 min in SFUM (HeLa cells) or DMEM/1% FCS (UAC1273 cells). HeLa cells were imaged at room temperature using a Zeiss Axiophot 2 microscope fitted with a 1.3-NA 100× Plan-Neofluar oil immersion objective; images were captured with a Hamamatsu Orca R2 camera and AxioVision software version 7.1 (Zeiss). The smaller UAC1273 cells were imaged using a Leica TCS SP2 SE confocal scanner in conjunction with a Leica DM6000 B upright scope (with attached Leica HCX PL apochromatic 63×/NA 1.4 objective) and a Leica CTR 6000 confocal control box, which was controlled by Leica Control Software. Total cellular fluorescence was quantified using ImageJ software. A line was drawn around each cell, and the total integrated density corresponding to cellular fluorescence was determined. Integrated density values were divided by total cell area to obtain fluorescence values normalized for cell size. For cells expressing GFP-tagged TBC1D16 proteins, the 20 cells exhibiting the highest normalized integrated density value for GFP fluorescence were analyzed. Transfected UAC1273 cells were analyzed by counting 80 or more cells corresponding to each condition and time point (Fig. 3E). Only cells that incorporated the fluorescently labeled TBC1D16 siRNA were included in these calculations.
Colocalization/Immunofluorescence Studies.
HeLa cells grown on 22 × 22-mm coverslips were transfected with Myc-TBC1D16 for 24 h, then washed with SFUM and serum-starved in SFUM at 37 °C for 30 min. The cells were then incubated with SFUM containing 25 μg/mL of human holo-transferrin for 30 min, 100 ng/mL Alexa Fluor 488-EGF for 15 min, or 100 ng/mL human EGF for 15 min for colocalization with transferrin receptor, EGF, and EGFR, respectively. Cells were permeabilized and cytosolic proteins depleted following an adapted procedure of Seaman (27) designed to permit antibody access to the cytoplasm for EM. Coverslips were washed once with cold PBS and twice with cold glutamate buffer [25 mM Hepes (pH 7.4), 25 mM KCl, 2.5 mM Mg(OAC)2, 5 mM EGTA, and 150 mM glutamate]. Coverslips were then dried on a Kimwipe, and cells were permeabilized by immersion in liquid nitrogen for 5 s. The coverslips were thawed for 1 min on a Kimwipe, washed twice in cold glutamate buffer, and fixed in 3.7% paraformaldehyde for 15 min. They were then washed twice in DMEM plus 10 mM Hepes, then incubated at room temperature for 15 min. All subsequent steps were performed at room temperature. After three washes in PBS, cells were placed in 0.3% Triton X-100, 5% BSA, and PBS for 1 h, and then incubated with antibodies diluted 1:500 in 0.1% Triton X-100/1% BSA/PBS for 1 h. Cells were washed three times with PBS, incubated with the appropriate secondary antibodies diluted 1:1,000 in the aforementioned solution for 1 h, and again washed three times with PBS and mounted on slides using Mowiol. Adobe Photoshop was used to process images. For quantitation of colocalization, TBC1D16-positive vesicles were individually counted in 10 cells and assessed for costaining.
EGFR Degradation and Signaling Assays.
At 24 h after HeLa cells were transfected with GFP constructs, or 48 h after WM115 cells were transfected with siRNA, the cells were serum-starved in SFUM (HeLa cells) or DMEM containing 1% FCS (WM115 cells) for 8 h at 37 °C. During the last 4 h, 25 μg/mL of cycloheximide was added to the media to prevent protein synthesis. Cells were placed on ice for 30 min, 20 ng/mL of human EGF was added, and cells were kept on ice for an additional 30 min. Subsequently, cells in 10-cm tissue culture dishes were moved to a 37 °C incubator for the times indicated. Then, the cells were moved to a cold room and washed three times with cold PBS, then lysed in ice-cold RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1 mM EDTA, 0.5% sodium deoxycholate) plus 2 mM sodium orthovanadate and protease inhibitors (PMSF, leupeptin, aprotinin, and pepstatin A). Lysates were passed 10 times through a 20-gauge needle, then incubated end over end at 4 °C for 15 min. Supernatants were obtained after spinning at 18,188 × g for 10 min at 4 °C. Protein concentrations were measured with a bicinchoninic acid assay.
Quantitative RT-PCR.
Total RNA from UAC1273 or WM115 cells was isolated at 48 h after cells were transfected with control or TBC1D16 siRNA using an RNAeasy Mini Kit (Qiagen). Total RNA (1 μg) was reverse-transcribed with an iScript cDNA Synthesis Kit (Bio-Rad). Approximately 0.5% of each reaction was added to quantitative RT-PCR reactions in a total volume of 10 μL containing 5 μL of 2× SYBR Green Master Mix (Life Technologies) and 0.2 μL each of forward (5′CGGACAGCAA-CGGCCTCCTG-3′) and reverse (5′-CCCAGGTCCACGCGGAACAC-3′) primers to a final concentration of 10 μM. Detection and data analysis were performed with an ABI PRISM 7900 sequence detection system (Life Technologies), using 18S rRNA as an internal control. Expression levels were calculated using the relative standard curve method to determine RNA quantity, normalized to 18S rRNA.
Supplementary Material
Acknowledgments
We thank Dr. Dana Pe’er for sharing results before publication, Dr. Ryan Nottingham for guidance with GAP analysis, and Carmel Schindelhaim for help with microscopy. This work was supported by National Institutes of Health Grants DK37332 (to S.R.P.) and T32 HL007970 (to B.S.G.); and Stanford Cancer Institute Developmental Cancer Research Award 1149282-105-DHALS (to B.S.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.J.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204540109/-/DCSupplemental.
References
- 1.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen UT, et al. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat Chem Biol. 2009;5:227–235. doi: 10.1038/nchembio.149. [DOI] [PubMed] [Google Scholar]
- 3.Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuda M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci Rep. 2011;31:159–168. doi: 10.1042/BSR20100112. [DOI] [PubMed] [Google Scholar]
- 5.Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- 6.Sklan EH, et al. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem. 2007;282:36354–36361. doi: 10.1074/jbc.M705221200. [DOI] [PubMed] [Google Scholar]
- 7.Haas AK, et al. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs E, et al. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J Cell Biol. 2007;177:1133–1143. doi: 10.1083/jcb.200612068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu C, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nottingham RM, Ganley IG, Barr FA, Lambright DG, Pfeffer SR. RUTBC1 protein, a Rab9A effector that activates GTP hydrolysis by Rab32 and Rab33B proteins. J Biol Chem. 2011;286:33213–33222. doi: 10.1074/jbc.M111.261115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko JM, Fisher DE. A new era: melanoma genetics and therapeutics. J Pathol. 2011;223:241–250. doi: 10.1002/path.2804. [DOI] [PubMed] [Google Scholar]
- 13.Hoek KS, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 14.Akavia UD, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Sluijs P, et al. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell. 1992;70:729–740. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- 16.Yamashiro DJ, Tycko B, Fluss SR, Maxfield FR. Segregation of transferrin to a mildly acidic (pH 6.5) para-Golgi compartment in the recycling pathway. Cell. 1984;37:789–800. doi: 10.1016/0092-8674(84)90414-8. [DOI] [PubMed] [Google Scholar]
- 17.Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 18.Aivazian D, Serrano RL, Pfeffer SR. TIP47 is a key effector for Rab9 localization. J Cell Biol. 2006;173:917–926. doi: 10.1083/jcb.200510010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zacchi P, et al. Rab17 regulates membrane trafficking through apical recycling endosomes in polarized epithelial cells. J Cell Biol. 1998;140:1039–1053. doi: 10.1083/jcb.140.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson JC, et al. A role for the small GTPase Rab21 in the early endocytic pathway. J Cell Sci. 2004;117:6297–6311. doi: 10.1242/jcs.01560. [DOI] [PubMed] [Google Scholar]
- 21.Kouranti I, Sachse M, Arouche N, Goud B, Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 2006;16:1719–1725. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 22.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 23.Posner BI, Laporte SA. Cellular signalling: Peptide hormones and growth factors. Prog Brain Res. 2010;181:1–16. doi: 10.1016/S0079-6123(08)81001-1. [DOI] [PubMed] [Google Scholar]
- 24.Gampel A, et al. VEGF regulates the mobilization of VEGFR2/KDR from an intracellular endothelial storage compartment. Blood. 2006;108:2624–2631. doi: 10.1182/blood-2005-12-007484. [DOI] [PubMed] [Google Scholar]
- 25.Bruns AF, Bao L, Walker JH, Ponnambalam S. VEGF-A-stimulated signalling in endothelial cells via a dual receptor tyrosine kinase system is dependent on co-ordinated trafficking and proteolysis. Biochem Soc Trans. 2009;37:1193–1197. doi: 10.1042/BST0371193. [DOI] [PubMed] [Google Scholar]
- 26.Hellberg C, Schmees C, Karlsson S, Ahgren A, Heldin CH. Activation of protein kinase C alpha is necessary for sorting the PDGF beta-receptor to Rab4a-dependent recycling. Mol Biol Cell. 2009;20:2856–2863. doi: 10.1091/mbc.E08-12-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.