Abstract
Gliomas are the most common and deadly type of primary brain tumor. In this study, we showed that cAMP response element-binding protein (CREB), a proto-oncogenic transcription factor that is overexpressed in gliomas, can promote gliomagenesis by modulating the expression of oncogenic microRNA-23a (mir-23a). First, we found that CREB is highly expressed in glioma tissues and cell lines. CREB is also essential for glioma cell growth and cell survival in vitro and is critical for gliomagenesis in vivo. Second, microRNA microarray, ChIP-chip, ChIP-quantitative PCR, and luciferase reporter assays showed that CREB directly binds to the regulatory sequences of mir-23a and enhance the expression of mir-23a. Moreover, mir-23a was confirmed as a functional downstream target of CREB in glioma cell growth and cell survival. Finally, using computational prediction followed by experimental confirmation, we identified PTEN, which is frequently silenced in gliomas, as a downstream target of mir-23a. Taken together, we propose that CREB promotes gliomagenesis and acts as a modulator of oncogenic mir-23a, which represses the tumor suppressor PTEN.
Gliomas are the most common brain tumors with high morbidity and mortality. In the past several decades, the prognosis for malignant gliomas has not significantly improved (1). Identifying the molecular mechanism underlying gliomagenesis is crucial for developing specific treatment strategies. In addition to the genomic mutation events that trigger the activation of oncogenes or the silencing of tumor suppressor genes (TSGs) (2–5), the dysregulation of microRNAs (miRNAs) is also a common epigenetic event in the development of gliomas (6).
miRNAs are single-stranded, small, noncoding RNAs that play important roles in many biological processes including tumorigenesis (7). During the initiation and progression of human cancers, miRNAs modulate cell proliferation, survival, and tumor angiogenesis, invasion, and metastasis (8–11). Dysregulation of miRNAs has been found in various types of human cancers including tumors occurring in breast, colon, lung, liver, and pancreas tumors, chronic lymphocytic leukemia, and malignant gliomas (6, 12–15). Many high-throughput screens have searched for dysregulated miRNAs in gliomas, and the function of such miRNAs has been primarily studied (16–22). However, the mechanism of the dysregulation of these miRNAs remains largely unknown.
cAMP response element-binding protein (CREB) is a proto-oncogenic transcription factor that promotes tumorigenesis in many cancers, including non–small-cell lung carcinoma (NSCLC), breast cancer, acute myeloid leukemia (AML), and hepatocellular carcinoma (HCC) (23–26). As a transcriptional activator, CREB generally enhances the expression of target genes (27). The targets of CREB are involved in various cell functions including metabolism, cell cycle, cell survival, and DNA repair (28). Researches on the targets of CREB used to be limited to protein-coding genes. Recently, some noncoding targets of CREB have also been identified during human neuronal differentiation and in liver cancer (29, 30); however, the noncoding targets of CREB deserve further investigation. In this study, CREB was up-regulated in gliomas and promoted gliomagenesis and acted as a coordinator of oncogenic miRNAs in glioma cells.
Results
CREB Is Up-Regulated in Gliomas and Is Essential for Gliomagenesis.
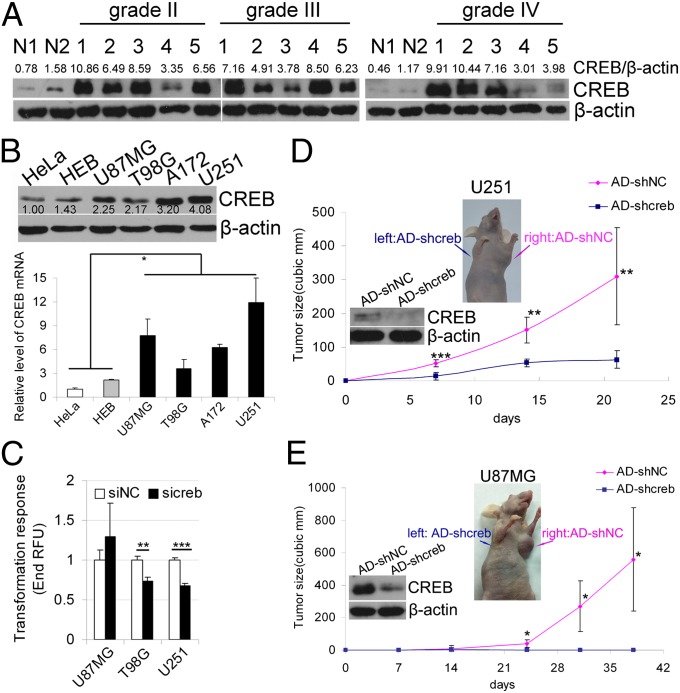
We first measured the CREB protein level in 15 glioma tissues and two normal brain tissues and found that CREB is highly expressed in glioma tissues from grade II to grade IV compared with normal brain tissues (Fig. 1A). The up-regulation of CREB protein was also found in glioma cell lines, U87MG, T98G, A172, and U251, compared with the human cervical carcinoma, HeLa and the human normal glial cell line, HEB (Fig. 1B). Coincident with the protein up-regulation, CREB mRNA was also up-regulated in glioma cell lines (Fig. 1B). We also found amplifications of the CREB gene copy number in U87MG cells and in a high grade (grade IV) glioma tissue (Fig. S1A). In addition, the result of prediction by TargetScan and PicTar algorithms suggests that CREB might be regulated by the miRNAs that are frequently down-regulated in gliomas, such as mir-181b (17, 31), mir-128 (17, 31–34), and mir-124 (21, 32–34) (Fig. S1B). The luciferase assay was used to test the interactions between these miRNAs and CREB 3′ UTR, and we found that mir-124 and mir-128 but not mir-181b repressed the activity of CREB 3′ UTR luciferase reporter (Fig. S1C). Moreover, the results of Western blotting and quantitative RT-PCR (QRT-PCR) showed that mir-124 and mir-128 significantly repressed the expression of CREB but had little effect on the mRNA level (Fig. S1D). Therefore, the down-regulation of these miRNAs might contribute to the high level of CREB in gliomas.
Fig. 1.
Highly expressed CREB is essential for gliomagenesis. (A) Western blotting analysis was used to detect CREB in normal brain tissues and glioma tissues. The results of Western blotting were quantified by densitometry and are shown as the ratios of CREB/β-actin protein levels (the values shown above the blots). (B Left) Western blotting and quantitative RT-PCR were used to detect CREB protein (Upper) and mRNA (Lower) level in the human cervical carcinoma cell line HeLa and human normal glial cell line HEB as well as in the four glioma cell lines (U87MG, T98G, A17,2 and U251). (B Right) The Western blotting results were quantified by densitometry and are shown as the ratios of CREB/β-actin protein levels. The mRNA levels of CREB are expressed as the mean ± SD, n = 3. (C) Knocking down CREB inhibits the anchor-independent growth in soft agarose of glioma cells. The cells were lysed, and the fluorescence was read (mean ± SD, n = 4). (D and E) Nude mice were injected s.c. with adenovirus-based CREB shRNA (AD-shcreb) and control shRNA (AD-shNC)-infected U251 cells (D) or U87MG cells (E) in the right and left flanks, respectively. The tumor volume was measured intermittently. Insets show the mice with tumors and the Western blotting results depicting the effect of CREB knockdown with AD-shcreb. Data are expressed as mean ± SD, n = 7 (U251) or n = 5 (U87MG). *P < 0.05; **P < 0.01; ***P < 0.001.
To investigate whether the high expression of CREB contributes to gliomagenesis, we knocked down endogenous CREB with specific siRNA (sicreb) or adenovirus-delivered shRNA (AD-shcreb) in three glioma cell lines (U87MG, T98G, and U251). We found that the anchor-independent growth of T98G and U251, but not U87MG, cells in soft agarose was inhibited by CREB knockdown (Fig. 1C). Furthermore, when transplanted s.c. into mice, U251 and U87MG cells infected with AD-shcreb formed much smaller tumors than cells infected with AD-shNC (Fig. 1 D and E).
Knocking Down CREB Suppresses Glioma Cell Growth and Survival.
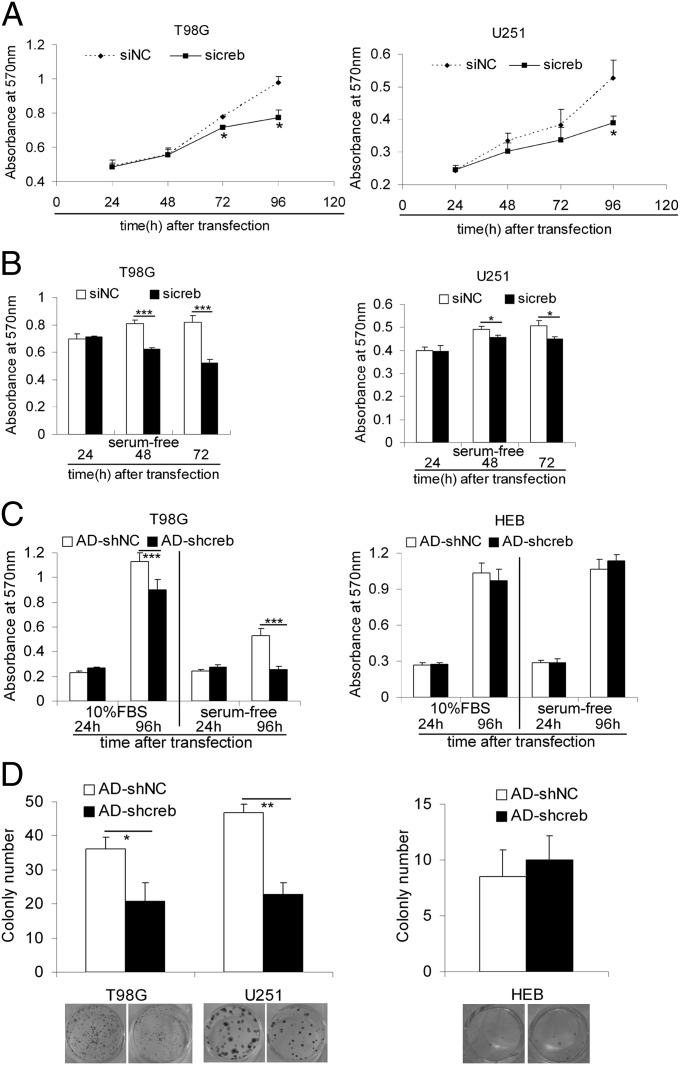
It has been reported that CREB promotes the proliferation, cell cycle progression, survival, and resistance to apoptosis in acute myeloid leukemia and lung cancer (23, 24, 26). Therefore, we investigate the effects of CREB knockdown on proliferation, survival, cell cycle progression, and apoptosis. CREB knockdown by sicreb or AD-shcreb was first verified by Western blotting (Fig. S2A). The 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay showed that the reduction of CREB slowed the growth of glioma cells and made them more sensitive to serum starvation than controls (Fig. 2 A and B and Fig. S2 B and C). A TUNEL assay was used to detect the apoptotic cells, and we found that knocking down CREB clearly induced apoptosis (Fig. S3A), especially in serum-free culture conditions (Fig. S3B). FACS assays showed that CREB knockdown resulted in slight cell cycle retardation (G0/G1 arrest in U87MG and T98G and G2/M arrest in U251) in glioma cells (Fig. S3C). Importantly, knocking down CREB with AD-shcreb inhibited the growth and survival of the T98G cells but not HeLa or HEB cells (Fig. 2C). Furthermore, the reduction of CREB decreased the colony formation ability of glioma cells (T98G and U251) but not HeLa or HEB cells (Fig. 2D and Fig. S2E). It is also worth mentioning that the sicreb and AD-shcreb targeted different sites on the CREB mRNA, two target sequences that are widely used for silencing CREB (24, 26, 35, 36), which should have reduced the possibility of an off-target effect. In addition, knocking down CREB with AD-shcreb#2 (targeting the same sequence as sicreb) produced similar effects on glioma cell growth, survival and colony formation (Fig. S4).
Fig. 2.
CREB knockdown inhibits the growth and survival of glioma cells but not HEB cells. (A) MTT assays show the growth curve of T98G (Left) and U251 (Right) cell lines after transfection with the siRNA against CREB (sicreb) or the control siRNA (siNC). Data are expressed as mean ± SD, n = 5. *P < 0.05; **P < 0.01. (B) T98G (Left) and U251 (Right) cells transfected with sicreb or siNC were maintained in serum-free medium, and the MTT method was used to determine the cell survival. Data are expressed as mean ± SD, n = 5. *P < 0.05; ***P < 0.001. (C) T98G (Left) and HEB (Right) cells infected with AD-shcreb or AD-shNC were maintained in complete culture medium or serum-free medium, and MTT assays were performed to detect the cell growth and cell survival (mean ± SD, n = 5), ***P < 0.001. (D) The effects of knocking down CREB with AD-shcreb on the colony formation abilities of T98G, U251, and HEB cells. The cell colonies were counted and plotted. Data are expressed as mean ± SD, n = 5. *P < 0.05; **P < 0.01.
CREB Contributes to the High Expression of Oncogenic miRNAs.
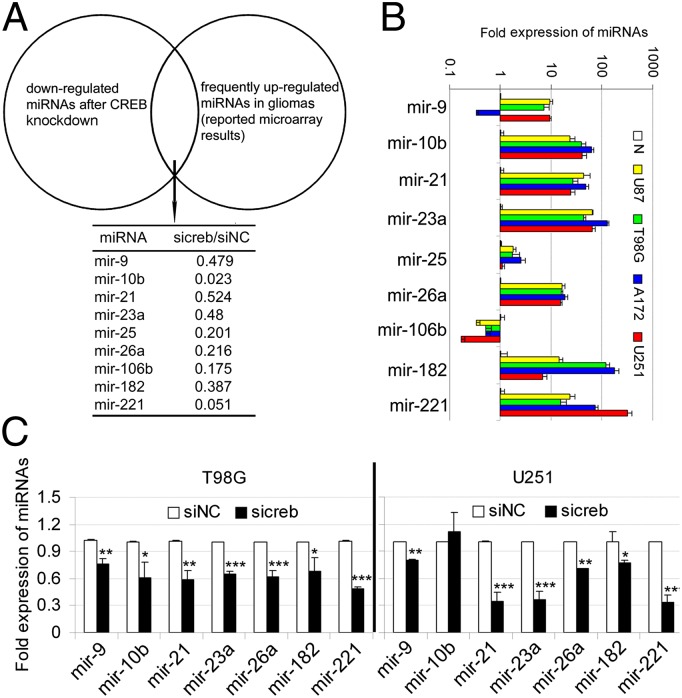
Some protein-coding genes are targets of CREB in human cancers (24, 26). We detected the mRNA levels of these target genes (CCNA1, CCND1, and BCL2) after knocking down CREB in U87MG, T98G, and U251 cells, except for that CCND1 mRNA decreased along with CREB mRNA in U87MG cells, no significant changes were found between the siNC and sicreb-transfected glioma cells (Fig. S5A). Therefore, additional targets needed to be identified, and our goal was to identify the noncoding targets, i.e., miRNAs. We performed genome-wide screening for miRNAs that were regulated by CREB in T98G cells. After transfecting T98G cells with sicreb or siNC, a microRNA microarray was performed to compare the expression of miRNAs between these two groups. Knocking down CREB led to a widespread decrease in the levels of miRNAs. To narrow our candidates, we focused on the known up-regulated miRNAs in gliomas that were supported by at least two studies (Table S1 and Fig. 3A). We validated the high expression of these miRNAs in glioma cell lines by QRT-PCR, and most of these miRNAs (mir-10b, mir-21, mir-23a, mir-26a, mir-182, and mir-221) were highly expressed in all four glioma cell lines. mir-9 was expressed highly in U87MG, T98G, and U251 cells but not in A172 cells. The level of mir-25 was only slightly higher in glioma cells, and mir-106b was decreased in glioma cells compared with the normal brain tissue (Fig. 3B). Using QRT-PCR on T98G and U251 cells, we confirmed the microarray result and found that all of the frequently up-regulated miRNAs in glioma cells, except mir-10b in U251 cells, decreased after CREB knockdown (Fig. 3C).
Fig. 3.
CREB contributes to the up-regulation of highly expressed miRNAs in glioma cells. (A) The T98G cells were transfected with sicreb or siNC, and a miRNA microarray was performed to compare the miRNA expression between the two groups. Knocking down CREB led to a widespread decrease of miRNAs in T98G cells, and only the miRNAs that were reported to be frequently up-regulated in gliomas by at least two studies are listed. The values of sicreb/siNC represent the fold changes of the miRNA levels determined by microRNA microarray. (B) The relative expression levels of miRNAs in a normal brain tissue (N) and four glioma cell lines (U87MG, T98G, A172, and U251) were measured by quantitative real-time PCR. (C) Quantitative RT-PCR was performed to confirm the microarray results (except mir-25 and mir-106b) in T98G and U251. Data are expressed as mean ± SD, n = 3. *P < 0.05; **P < 0.01; ***P < 0.001.
mir-23a Is a Direct Target of CREB.
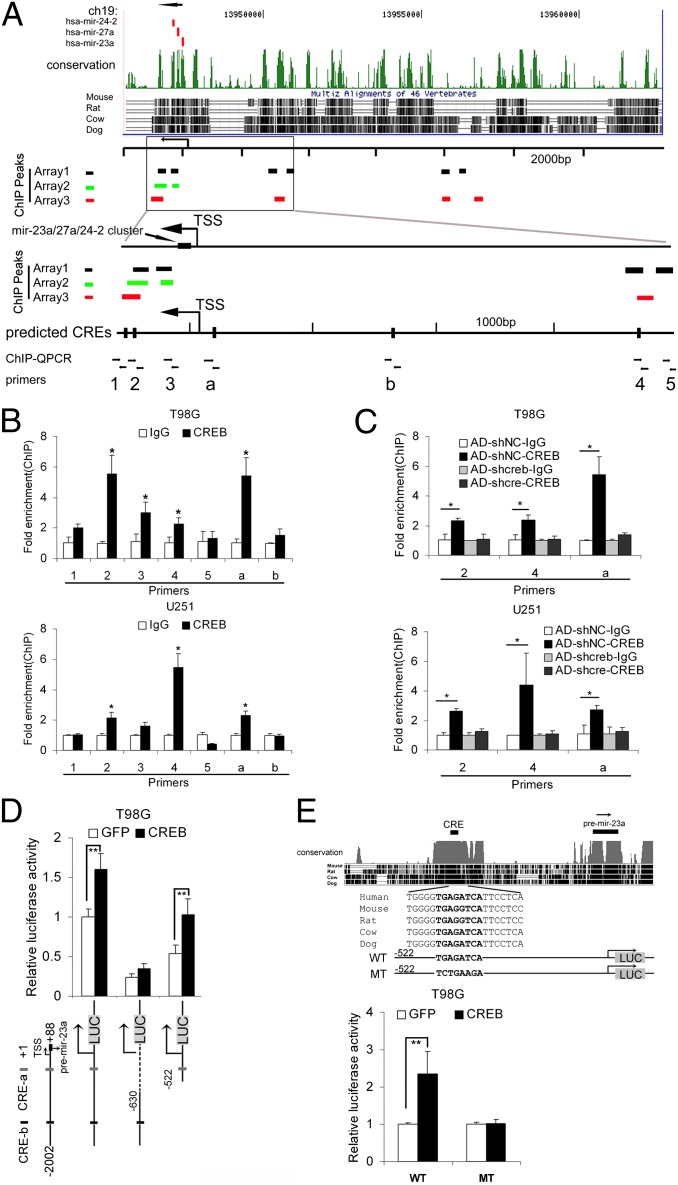
Most of the transcription start sites (TSSs) and promoter regions of these miRNA genes remain largely unknown; therefore, the ChIP-chip method was used to examine the direct interactions between CREB and its target miRNA genes in glioma cells. Three independent ChIP-chip assays were performed in T98G cells, and we found that the flanking sequences of several miRNA genes that were highly expressed in glioma (mir-130b, mir-148a, mir-188, mir-199b, mir-210, mir-23a, mir-23b, mir-362, mir-516a-1, mir-516a-2, mir-671, mir-9-1, mir-9-2, mir-9-3, and mir-106b-25 cluster) contained CREB binding peaks in at least two experiments. Interestingly and surprisingly, the upstream sequence of the mir-23a-27a-24-2 cluster was highly enriched in all three ChIP-chip assays. Eleven peaks were enriched in at least two assays, and seven of them were located within the −10 kb to 2 kb region (Table S2). To confirm the ChIP-chip results, ChIP-quantitative PCR (QPCR) was used to detect the occupancy of CREB on the flanking regions of the mir-23a-27a-24-2 gene cluster by using seven specific primers. Five primers were designed to amplify the five peaks that were enriched in the ChIP-chip assays and were located within the −4 kb to 2 kb region (primers 1–5), and two of them were designed to amplify the two predicted cAMP-response elements (CREs) in the −2 kb to 0 kb region (a and b) (Fig. 4A). The regions “2,” “4,” and “a” were significantly enriched by the CREB antibody in untreated and AD-shNC–infected T98G and U251 cells (Fig. 4 B and C), but not in AD-shcreb–treated cells with CREB knocked down (Fig. 4C).
Fig. 4.
CREB directly regulates the expression mir-23a. (A) The results of three ChIP-chip assays were summarized. Eleven peaks were found in three ChIP-chip assays within the 10-kb region upstream of the mir-23a. The locations of the peaks are denoted as short bars (black, green, and red represent the three assays). To confirm the ChIP-chip results, we designed five primers to detect the occupancy of CREB within the 4-kb region upstream of the mir-23a (primers 1–5). We also designed two primers (a and b) to amplify the two predicted CREs within the 2-kb region upstream of the mir-23a, although no peaks were found in this region in the three ChIP-chip assays. (B) ChIP-QPCR analysis was used to confirm the ChIP-chip results in T98G and U251 cells with the specific primers mentioned above. (C) ChIP-QPCR analysis was performed with AD-shcreb/AD-shNC-infected T98G and U251 cells to determine the occupancy of CREB. Data are expressed as mean ± SD, n = 3. *P < 0.05, two-tailed unpaired Student’s t test, relative to IgG control. (D) Luciferase reporter assays were used to confirm the transcription-enhancing effect of CREB on the promoter region of the mir-23a-27a-24-2 gene cluster. The gray box (LUC) denotes the luciferase gene cassette. (E) The conservation of the promoter region of the mir-23a-27a-24-2 gene cluster is shown (Upper). The promoter regions containing the wild-type (WT) or mutated (MT) CRE were inserted upstream of the luciferase cassette, and luciferase reporter assays were performed. (E Lower) The normalized luciferase activity is expressed as the mean ± SD, n = 4. **P < 0.01.
Because the TSS of the gene cluster mir-23a-27a-24-2 is known (37), luciferase reporter assays were used to ensure that the binding of CREB to the promoter region contributes to the transcriptional activation. As shown in Fig. 4D, the transcription activity of the proximal promoter region of the mir-23a gene (−522 to +88), but not the distal region (−2002 to −630), was enhanced by CREB. Moreover, mutating the conserved CRE (CRE-a) totally abolished the enhancing effect of CREB overexpression on the luciferase reporter (Fig. 4E), indicating that CREB can directly regulate the transcription of mir-23a by binding to this CRE.
Although mir-23a is in a miRNA gene cluster (mir-23a-27a-24-2), the other two miRNA products (mir-27a and mir-24) in this cluster were not frequently up-regulated in gliomas (only one report mentions the high level of mir-27a in gliomas; ref. 22), possibly due to the posttranscriptional regulation of the processing of pri- and premiRNAs. We confirmed that the primary mir-23a transcript (pri-mir-23a) and the mature mir-23a were highly expressed in the four glioma cell lines compared with the HeLa and the HEB lines (Fig. S5B) without gene copy number changes (Fig. S5C).
miR-23a Is a Functional Downstream Target of CREB.
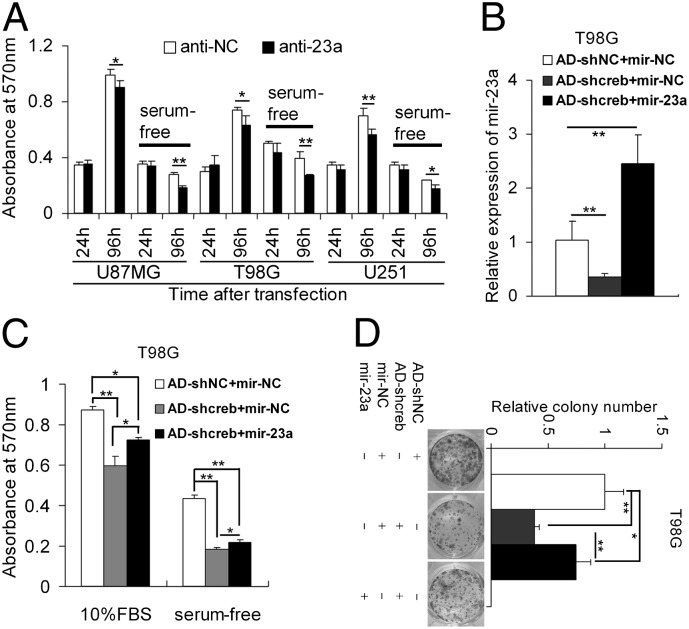
mir-23a has been identified as a highly expressed miRNA in glioma cell lines by our results and several previous studies (17–19, 21). Here, we found that mir-23a is a direct target of CREB. However, the function of mir-23a in gliomagenesis remains largely unknown. Therefore, we knocked down endogenous mir-23a with miRNA antagomirs (anti-23a) in glioma cells (Fig. S6A) and found that the reduction of mir-23a significantly inhibited the growth and survival of U87MG, T98G, and U251 cells in MTT assays (Fig. 5A). The T98G cells with knocked down mir-23a showed significantly lower colony formation ability than control T98G cells (Fig. S6B). To test whether mir-23a could mediate the function of CREB, rescue experiments were performed. Overexpression of mir-23a partly rescued the effect of CREB knockdown on the growth, survival, and colony formation ability of T98G and U251 cells (Fig. 5 B–D and Fig. S6 C–E). Moreover, AD-shcreb–infected U251 cells transfected with mir-23a mimics formed larger tumors than cells infected with AD-shcreb after transfection with mir-NC mimics in xenograft models, which suggests that mir-23a can partly rescue the inhibitory effect of CREB knockdown on the growth of U251 cells in vivo (Fig. S6F).
Fig. 5.
mir-23a is a functional downstream target of CREB. (A) Knocking down mir-23a with specific inhibitors (antagomirs, anti-23a) inhibited the growth and survival of glioma cells. Data are expressed as mean ± SD, n = 4. **P < 0.01; ***P < 0.001. (B–D) Overexpression of mir-23a partly rescued the growth-inhibiting effect of CREB knockdown. T98G cells were transfected with synthetic mir-23a mimics (mir-23a) or control mimics (mir-NC) after infection with AD-shcreb or AD-shNC. The relative expression of mir-23a was determined by quantitative RT-PCR (mean ± SD, n = 3) (B). The transfected/infected cells were subjected to MTT assays (C) and colony formation assays (D). Data are expressed as mean ± SD, n = 4. *P < 0.05; **P < 0.01.
PTEN Is a Downstream Target of mir-23a.
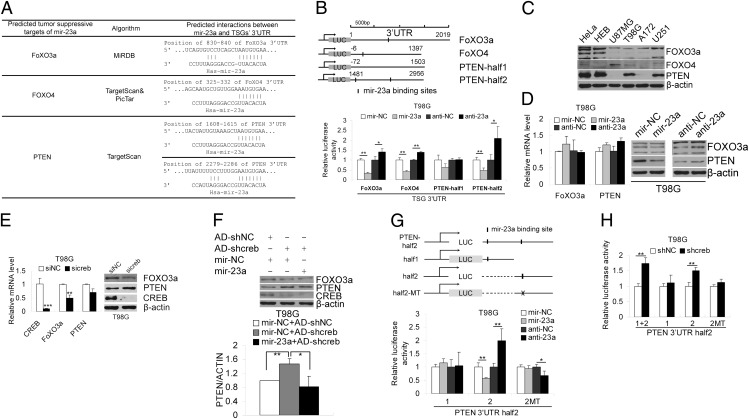
Usually miRNAs function by targeting protein-coding genes. Therefore, we investigated the direct targets of mir-23a. Target predictions were primarily performed by using three algorithms (TargetScan, PicTar, and MiRDB). Many protein-coding genes were predicted to be the targets of mir-23a. Among them, several TSGs (i.e., FOXO3a, FOXO4, and PTEN) that are involved in gliomagenesis attracted our attention (38) (Fig. 6A). Luciferase assays were used to test the interactions between the miRNAs and the 3′ UTRs of these TSGs in T98G cells. Overexpressing and knocking down mir-23a significantly decreased and increased the FOXO3a, FOXO4, and PTEN 3′ UTR (half2) luciferase reporter activities, respectively (Fig. 6B). CREB and mir-23a are highly expressed in glioma cells and, accordingly, the targets of mir-23a are expected to be down-regulated in glioma cells. Through Western blotting, we found that FOXO3a and PTEN, but not FOXO4, were down-regulated in glioma cells (Fig. 6C). Ectopic expression of mir-23a and knockdown of mir-23a, respectively, repressed and increased the protein levels, but not the mRNA levels, of FOXO3a and PTEN in T98G cells (Fig. 6D), suggesting that FOXO3a and PTEN were potential downstream targets of mir-23a. Because knockdown of CREB can significantly down-regulate the expression of mir-23a in glioma cells, the targets of mir-23a were expected to be up-regulated after knocking down CREB. PTEN protein increased, accompanied by a slightly decrease in its mRNA level, after CREB knockdown. Surprisingly, knocking down CREB led to decreases in the mRNA and protein levels of FOXO3a (Fig. 6E). Importantly, overexpression of mir-23a in CREB-knockdown T98G cells decreased the induction of PTEN protein expression (Fig. 6F), suggesting that CREB affected PTEN protein expression by, at least partly, regulating the expression of mir-23a. Because there are two predicted binding sites for mir-23a in the PTEN 3′ UTR, luciferase reporter assays were used to test which site is functional. The results showed that the second site (position 2279–2286) interacted with mir-23a, and mutating this binding site abolished the repression of PTEN 3′ UTR-luciferase expression by mir-23a (Fig. 6G). Furthermore, knocking down CREB increased the activity of the luciferase reporter containing the PTEN 3′ UTR-half2 and -half2-half2 but not the PTEN 3′ UTR-half2-half1 or -half2-half2-MT, suggesting that CREB modulates repressional activity at the PTEN 3′ UTR by regulating the expression of mir-23a.
Fig. 6.
mir-23a directly target PTEN. (A) The tumor suppressor genes FOXO3a, FoXO4, and PTEN were predicted to be potential targets of mir-23a. (B) The 3′ UTRs of the three target genes containing mir-23a binding sites were cloned downstream of luciferase reporter gene. The first half of the PTEN 3′ UTR (PTEN-half1) containing no mir-23a binding sites was used as a negative control. Luciferase reporters were cotransfected with synthetic miRNA mimics or antagomirs into T98G cells. Forty-eight hours later, the normalized luciferase activity was determined (mean ± SD, n = 4). *P < 0.05; **P < 0.01; ***P < 0.001. (C) Western blotting was used to detect FOXO3a, FOXO4, and PTEN in HeLa, HEB and the four glioma cell lines (U87MG, T98G, A172, and U251). (D) T98G cells were transfected with synthetic mir-23a mimics (mir-23a), inhibitors (anti-23a), or negative controls (mir-NC or anti-NC). The protein and mRNA of FOXO3a and PTEN were detected by Western blotting and quantitative RT-PCR, respectively. (E) The mRNA and protein of FOXO3a and PTEN were detected after knocking down CREB with sicreb in T98G cells. (F) T98G cells were transfected with synthetic mir-23a mimics or control mimics followed by infection with AD-shcreb or AD-shNC, and FOXO3a, PTEN, and CREB were detected by Western blotting and quantified by densitometry and plotted (mean ± SD, n = 3). *P < 0.05; **P < 0.01. (G) The 3′ UTR (half2) of PTEN, harboring two potential mir-23a binding sites, was itself divided into two halves (1, 2), each containing one of the two mir-23a binding sites. These partial 3′ UTRs of PTEN with either the wild-type or a mutated mir-23a binding site were cloned downstream of the luciferase reporter gene, and luciferase assays were performed to test the interactions between mir-23a and the 3′ UTRs parts. (H) The luciferase reporters containing the PTEN 3′ UTR-half2 (1+2), PTEN 3′ UTR-half2-1 (1) or PTEN 3′UTR-half2-2 (2) or PTEN 3′ UTR-half2-2-MT (2MT) were cotransfected with shcreb or shNC. The normalized luciferase activity is expressed as the mean ± SD, n = 4. *P < 0.05; **P < 0.01.
Discussion
The mechanism of the overexpression of CREB in tumorigenesis is poorly understood. We found amplifications of the CREB gene copy number in one case grade IV tissue and in the U87MG cell line. However, these findings appear in contrast with a previous study that detected no gene copy number changes in a cohort of glioblastoma multiforme (GBM) samples of TGCA compared with normal brain controls (39), suggesting that the gene copy number amplification is not a universal mechanism of the up-regulation of CREB in gliomas. Reduced levels of miRNAs, the master regulators of protein-coding genes, usually lead to up-regulation of their target genes (40–43). The frequently down-regulated miRNAs (mir-124 and mir-128) in gliomas were predicted and confirmed to target the 3′ UTR of CREB, implying that the down-regulation of these miRNAs may lead to a high CREB level in gliomas. Although CREB mRNA was only slightly up-regulated in the aforementioned cohort of GBM samples (49% with log2 tumor/normal ratio ≥ 0.5) (39), its regulation by miRNAs might be one cause of the CREB high level in the absence of significant genomic or transcriptional aberrations.
Even though CREB is highly expressed in some types of tumors and is associated with malignancy, we present evidence of the crucial role of CREB in the growth of glioma cells. Although knocking down CREB only had a moderate growth-inhibitory effect on glioma cells in vitro, it largely inhibited tumor formation by glioma cells in a xenograft model. Importantly, the growth-inhibitory effect of knocking down CREB seemed to be limited to glioma cell lines; the growth and colony formation ability of HeLa and HEB lines were almost unaffected, suggesting that CREB knockdown is selectively lethal to glioma cells.
The oncogenic roles of miRNAs have been characterized in various types of tumors, including gliomas (44), but little is known about the mechanism of their dysregulation. Uncovering the reason for their dysregulation might make it possible to return the expression levels of the oncogenic miRNAs to normal. Similar to protein-coding genes, miRNA genes themselves are subject to sophisticated control. The factors determining the expression of miRNAs include genomic amplification, transcriptional regulation, processing, editing, and decay (45). It seems that dysregulation at the transcriptional level is one of the major factors and, indeed, some reports have characterized the transactivators or suppressors of miRNAs, for example, c-MYC and p53 (46–48). We found that CREB is a direct transcriptional regulator of the oncogenic mir-23a, and we also showed that knocking down CREB has a widespread silencing effect (direct or indirect) on the expression of oncogenic miRNAs (including the bona fide oncogenic miRNAs mir-21, mir-26a, and mir-221) in glioma cells, so we postulate that CREB might act as a coordinator modulating the expression of oncogenic miRNAs. If we can silence a group of oncogenic miRNAs together, the anticancer effect could be more efficient. From this point of view, CREB could be a potentially valuable molecular target for glioma therapy.
mir-23a is highly expressed in glioma tissues and cell lines. To our knowledge, only one report has mentioned the oncogenic role of mir-23a, whose knockdown resulted in a reduction in the glioma cells’ colony formation ability in soft agar (18). We identified mir-23a as a growth enhancer of glioma cells and a regulator of the tumor suppressor PTEN. Considering the pivotal role of PTEN, we believe that the aberrant expression of mir-23a contributes significantly to gliomagenesis. In addition to PTEN, some other glioma suppressors such as LRRC4, SPRY2, and CAMTA1 are predicted to be targets of mir-23a (prediction results from miRecords: http://mirecords.umn.edu/miRecords). Because of the widespread regulatory function of miRNAs, CREB could affect the expression of many more genes, including critical glioma suppressors.
In summary, our results show that the highly expressed CREB transcription factor can promote gliomagenesis by modulating the expression of the oncogenic mir-23a, which can directly target the tumor suppressor PTEN. These findings also suggest that the proto-oncogene CREB can repress the tumor suppressors by modulating the expression of TSG-targeting miRNAs.
Materials and Methods
Samples and Cell lines.
All human normal brain tissue and glioma samples were obtained from the Department of Neurosurgery, Beijing Tiantan Hospital. All human materials were used in accordance with the policies of the institutional review board at Beijing Tiantan Hospital. The four human glioma cell lines used here (U87MG, T98G, A172, and U251) were purchased from American Type Culture Collection. The human cervical carcinoma cell line HeLa was obtained from the China Center for Type Culture Collection (Wuhan, China). HEB was kindly provided by Guangmei Yan (Department of Pharmacology, Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou, China). See SI Materials and Methods for details.
See more material and methods in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. William Lennarz for helpful discussion. Support was received from “973” project Grants 2011CBA01104, 2007CB946902, and 2009CB825403; National Sciences Foundation of China Grants 30825023 and 31071203; Program for New Century Excellent Talents Grant NCET-07-0505; and a “111” project grant.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207787109/-/DCSupplemental.
References
- 1.Saika K, Katanoda K. Comparison of time trends in brain and central nervous system cancer mortality (1990-2006) between countries based on the WHO mortality database. Jpn J Clin Oncol. 2011;41:304–305. doi: 10.1093/jjco/hyr004. [DOI] [PubMed] [Google Scholar]
- 2.James CD, Carlbom E, Nordenskjold M, Collins VP, Cavenee WK. Mitotic recombination of chromosome 17 in astrocytomas. Proc Natl Acad Sci USA. 1989;86:2858–2862. doi: 10.1073/pnas.86.8.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Libermann TA, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stokoe D. Pten. Curr Biol. 2001;11:R502. doi: 10.1016/s0960-9822(01)00303-7. [DOI] [PubMed] [Google Scholar]
- 6.Pang JC, Kwok WK, Chen Z, Ng HK. Oncogenic role of microRNAs in brain tumors. Acta Neuropathol. 2009;117:599–611. doi: 10.1007/s00401-009-0525-0. [DOI] [PubMed] [Google Scholar]
- 7.Yue J, Tigyi G. MicroRNA trafficking and human cancer. Cancer Biol Ther. 2006;5:573–578. doi: 10.4161/cbt.5.6.2872. [DOI] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 10.Nicoloso MS, Calin GA. MicroRNA involvement in brain tumors: From bench to bedside. Brain Pathol. 2008;18:122–129. doi: 10.1111/j.1750-3639.2007.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 12.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varnholt H, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 14.Yu SL, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Fulci V, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 16.Zhou X, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–155. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 17.Ciafrè SA, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 18.Rao SA, Santosh V, Somasundaram K. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol. 2010;23:1404–1417. doi: 10.1038/modpathol.2010.135. [DOI] [PubMed] [Google Scholar]
- 19.Malzkorn B, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125:1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 21.Jiang L, et al. miR-182 as a prognostic marker for glioma progression and patient survival. Am J Pathol. 2010;177:29–38. doi: 10.2353/ajpath.2010.090812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huse JT, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aggarwal S, Kim SW, Ryu SH, Chung WC, Koo JS. Growth suppression of lung cancer cells by targeting cyclic AMP response element-binding protein. Cancer Res. 2008;68:981–988. doi: 10.1158/0008-5472.CAN-06-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng JC, et al. CREB is a critical regulator of normal hematopoiesis and leukemogenesis. Blood. 2008;111:1182–1192. doi: 10.1182/blood-2007-04-083600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cougot D, et al. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J Biol Chem. 2007;282:4277–4287. doi: 10.1074/jbc.M606774200. [DOI] [PubMed] [Google Scholar]
- 26.Shankar DB, et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell. 2005;7:351–362. doi: 10.1016/j.ccr.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laneve P, et al. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 2010;38:6895–6905. doi: 10.1093/nar/gkq604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slaby O, et al. MicroRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma. 2010;57:264–269. doi: 10.4149/neo_2010_03_264. [DOI] [PubMed] [Google Scholar]
- 32.Silber J, James CD, Hodgson JG. microRNAs in gliomas: Small regulators of a big problem. Neuromolecular Med. 2009;11:208–222. doi: 10.1007/s12017-009-8087-9. [DOI] [PubMed] [Google Scholar]
- 33.Godlewski J, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 34.Silber J, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobroff AS, et al. Silencing cAMP-response element-binding protein (CREB) identifies CYR61 as a tumor suppressor gene in melanoma. J Biol Chem. 2009;284:26194–26206. doi: 10.1074/jbc.M109.019836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellegrini M, et al. Expression profile of CREB knockdown in myeloid leukemia cells. BMC Cancer. 2008;8:264. doi: 10.1186/1471-2407-8-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Meir EG, et al. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia H, et al. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med (Berl) 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 42.Chen L, et al. MicroRNA-181b targets cAMP responsive element binding protein 1 in gastric adenocarcinomas. IUBMB Life. 2012;64:628–635. doi: 10.1002/iub.1030. [DOI] [PubMed] [Google Scholar]
- 43.Kong WQ, et al. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J. 2012;279:1252–1260. doi: 10.1111/j.1742-4658.2012.08519.x. [DOI] [PubMed] [Google Scholar]
- 44.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 46.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hassan MQ, et al. A network connecting Runx2, SATB2, and the miR-23a∼27a∼24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci USA. 2010;107:19879–19884. doi: 10.1073/pnas.1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.