Abstract
Dinoflagellates are an important component of the marine biota, but a large genome with high–copy number (up to 5,000) tandem gene arrays has made genomic sequencing problematic. More importantly, little is known about the expression and conservation of these unusual gene arrays. We assembled de novo a gene catalog of 74,655 contigs for the dinoflagellate Lingulodinium polyedrum from RNA-Seq (Illumina) reads. The catalog contains 93% of a Lingulodinium EST dataset deposited in GenBank and 94% of the enzymes in 16 primary metabolic KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, indicating it is a good representation of the transcriptome. Analysis of the catalog shows a marked underrepresentation of DNA-binding proteins and DNA-binding domains compared with other algae. Despite this, we found no evidence to support the proposal of polycistronic transcription, including a marked underrepresentation of sequences corresponding to the intergenic spacers of two tandem array genes. We also have used RNA-Seq to assess the degree of sequence conservation in tandem array genes and found their transcripts to be highly conserved. Interestingly, some of the sequences in the catalog have only bacterial homologs and are potential candidates for horizontal gene transfer. These presumably were transferred as single-copy genes, and because they are now all GC-rich, any derived from AT-rich contexts must have experienced extensive mutation. Our study not only has provided the most complete dinoflagellate gene catalog known to date, it has also exploited RNA-Seq to address fundamental issues in basic transcription mechanisms and sequence conservation in these algae.
Dinoflagellates are a group of freshwater and marine eukaryotes, and the marine photosynthetic species are important contributors to the ocean’s primary production (1). Dinoflagellates are notable for their symbioses with coral (2), the production of harmful algal blooms termed red tides (3), and their spectacular bioluminescence (4). Dinoflagellates also display several unusual cytological and biochemical features. For example, their chromosomes remain permanently condensed throughout the cell cycle (5) and electron microscopy of dinoflagellate nuclei shows a whorled structure termed a cholesteric liquid crystal (6). It seems likely that this unusual nuclear organization may impose restrictions on DNA replication and transcription, yet details of these processes are still unknown.
In part, study of dinoflagellate biochemistry is limited by a paucity of molecular tools, including the lack of a genome sequence and an inability to produce transgenic organisms (7). One of the difficulties encountered is a generally large DNA content, with ∼200 pg of DNA (roughly 60 times that of the haploid human cell) reported for Lingulodinium (8). Interestingly, two well-studied genes in this species are found in multiple copies arranged in tandem: peridinin-chlorophyll a-protein (PCP; ∼5,000 copies per nucleus) (9) and luciferase (146 copies) (10). The presence of multiple gene copies is expected to render genome sequence assembly difficult and unless the different copies are well conserved, will also result in a complex transcriptome profile. Interestingly, the spacer regions between the coding sequence of the tandem-arranged genes have no recognizable transcription factor–binding motifs, confounding attempts to understand how gene expression is regulated. This observation, in concert with the discovery of transsplicing in dinoflagellates, has led to the proposal that dinoflagellate transcripts are polycistronic (11). This proposal is largely derived from studies in kinetoplastids, in which transsplicing and polyadenylation are used to excise ORFs from polycistronic transcripts (12). Although attractive, this proposal has not yet been tested experimentally in dinoflagellates.
Recent developments in high-throughput sequencing technologies have opened up an opportunity to examine transcriptomes of organisms as potentially gene rich as the dinoflagellate Lingulodinium. We report here a transcriptome profile derived from Illumina sequencing, a technique commonly called RNA-Seq (13). In addition to providing a measure of gene expression, the different sequences can be assembled de novo to develop a transcript profile. We have used RNA-Seq to obtain the most comprehensive gene catalog for any dinoflagellate described to date and to address fundamental issues in gene conservation and expression.
Results
De Novo Assembly Is an Authentic Portrait of the Transcriptome.
RNA-Seq was used to generate 312 million sequence reads from a clonal Lingulodinium cell line. These reads were subsequently assembled to create a gene catalog containing 74,655 contigs longer than 300 bp, a cutoff determined by the fragment size range selected for sequencing. The size distribution of the sequences in the catalog shows an exponential decrease in sequence number as a function of length (Fig. S1A), and although this is different from the size distribution of the poly(A) RNA (Fig. S1B), it is not unusual for de novo RNA-Seq assemblies (14).
The gene catalog is GC-rich, as expected (8), with a predominance of GC at the third codon position. To gauge the extent to which the catalog is an authentic representation of the transcriptome, we compared it with a set of 2,111 GC-rich Lingulodinium ESTs. Our catalog contains 93% of these ESTs, with an average identity of 98% (Fig. S2). We also mapped the assembly onto 16 primary metabolic Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and found that the catalog contains 141 of the 150 enzymes (94%) expected (Table S1). Finally, we also tested the transcriptome for the presence of proteins involved in other basic metabolic processes, such as DNA replication, transcription, mRNA transport, translation, splicing, ribosome biogenesis, and mRNA surveillance (Table S2). For this analysis, we prepared a dataset containing authentic KEGG component sequences for the different processes from whole genomes of mammals, plants, apicomplexans, ciliates, and diatoms. This dataset was used to screen the Lingulodinium catalog (BLASTx E value < e−20). Overall, our transcriptome contains 64% of the proteins used by mammals, a value similar to what is found for the phylogenetically related apicomplexans (60% of mammalian sequences) and ciliates (63%). One notable exception is the absence of the transcription factor IID subunit TATA-binding protein (TBP), which is replaced in dinoflagellates by a TBP-like protein of different binding specificity (15). We conclude our catalog is a good representation of the transcriptome.
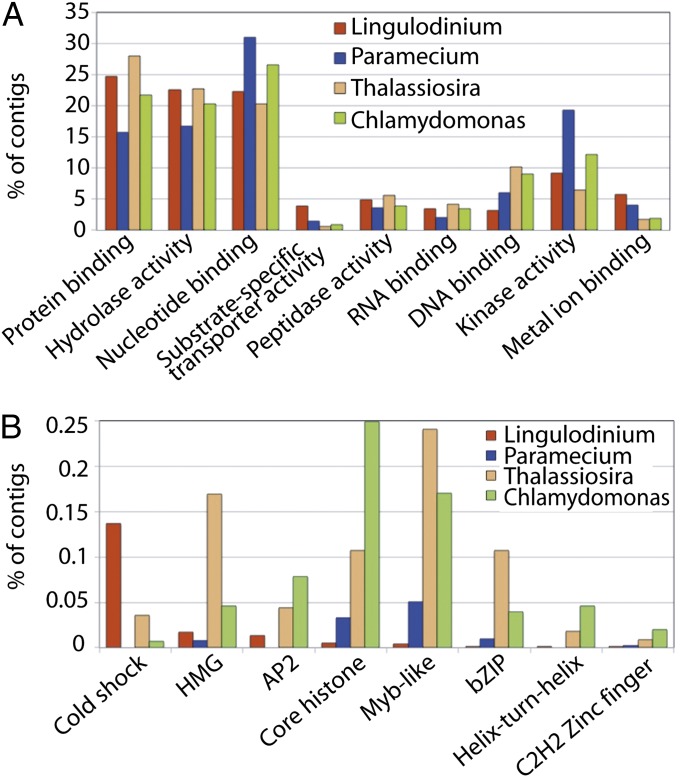
Using an E value cutoff of e−05, tBLASTn analysis of the catalog showed that 25% of the 74,655 sequences had an annotated match, 45% had a nonannotated match, and 30% lacked similarity to any known sequence in GenBank. The annotated sequences in the catalog, when classified into gene ontology (GO) categories, show an underrepresentation of proteins classified as DNA binding compared with the ciliate Paramecium, the diatom Thalassiosira, and the green alga Chlamydomonas (Fig. 1A). The gene catalog also was used to determine the number of matches to protein family (PFAM) DNA-binding domains (Fig. 1B). Lingulodinium has representatives for only half the known DNA binding domains found in ciliates and diatoms, and completely lack the heat-shock factor domains that comprise 0.8% of the diatom sequences. These domains present include the four core histone domains, a finding described elsewhere (16). Importantly, most (68%) of all the Lingulodinium DNA-binding domains fall into the class of cold-shock domains. However, because these domains also bind mRNA, and are better known for their role in posttranscriptional regulation in eukaryotes (17), we conclude that Lingulodinium shows a marked underrepresentation in the types of proteins and protein domains involved in regulating transcription.
Fig. 1.
Global analysis of the Lingulodinium assembly. (A) Gene ontology of annotated sequences in the transcriptome shows a decreased level of DNA-binding proteins and an increased level of substrate-specific membrane transporters compared with the ciliate Paramecium, the diatom Thalassiosira, and the green alga Chlamydomonas. (B) The number of protein family DNA-binding domains detected in Lingulodinium compared with those detected in Paramecium, Thalassiosira, and Chlamydomonas. DNA- binding domains not present in Lingulodinium (CCAAT, E2F, GATA, Helix-hairpin-helix, Helix–loop–helix, CBF, KN, HSF, Sigma-70, TAZ, CXC, and WRKY) are not included.
As a corollary to the underrepresentation of DNA-binding factors and domains, we reasoned that if dinoflagellates generally favor posttranscriptional regulation mechanisms, components involved in these processes might be enriched in sequences shared between different dinoflagellate species. We obtained ESTs for Karenia brevis and several Alexandrium species from GenBank and aligned the sequences to produce datasets containing 20,726 Karenia unigenes and 31,670 Alexandrium unigenes. We then searched these unigene datasets with our Lingulodinium transcriptome using tBLASTn (E value cutoff e−20). The 5,904 sequences shared among the three species were termed “core” dinoflagellate candidates (Fig. S3A). Compared with the Lingulodinium transcriptome, the core sequences are indeed depleted in DNA-binding proteins and are enriched in proteins with kinase activity (Fig. S3B). The ratio of core sequences to the total number in our catalog for different molecular functions shows a marked enrichment in translation factors, protein kinases, and protein phosphatases, whereas DNA-binding proteins are again underrepresented (Fig. S3C). This analysis supports the contention that dinoflagellates may preferentially regulate gene expression using posttranscriptional control mechanisms.
Tandem Gene Array Sequences Are Highly Conserved in the Transcriptome.
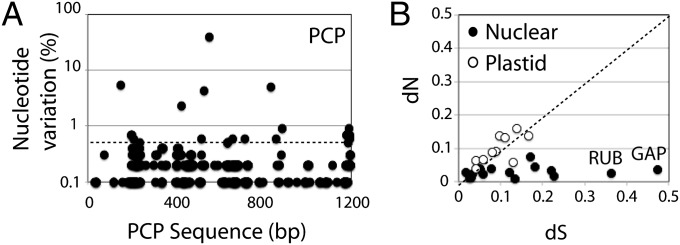
To assess the degree of sequence conservation in transcripts from the tandem array genes (TAGs), raw reads were assembled to a reference gene (mean coverage ∼10,000) and the number of variant nucleotides at each position was measured after trimming to remove low-quality bases. For PCP (∼5,000 copies), previous work has indicated that both the coding sequences and intergenic spacers are highly conserved (9), and this was confirmed in the transcript sequences at high coverage (Fig. 2A). When the entire PCP coding region is scanned nucleotide by nucleotide, few positions show a level of sequence variation greater than 0.5% (dotted line in Fig. 2A). To quantify the degree of sequence conservation, we counted the number of positions with different levels of variation (Fig. S4A). This spectrum of variation shows that more than half the positions have a level of variation corresponding to the Q30 (99.9% accuracy) of the sequencing reaction. This level of sequence conservation is similar to that observed for rRNA transcripts (Fig. S4B).
Fig. 2.
Sequence variation among transcripts. (A) The nucleotide variation (% total reads with a nucleotide different from a Peridinin-Chlorophyll a-Protein (PCP) reference sequence at each position) is given along the PCP sequence. The dotted horizontal line at 0.5% variation is the threshold used for calculating dS and dN. (B) The ratio of nonsynonymous (dN) to synonymous (dS) changes is shown for NCBI reference sequences with greater than 1000-fold coverage. The dotted line (dN/dS = 1) represents neutral selection. Plastid-encoded (○) and nuclear-encoded (●) sequences are shown separately. The positions of RuBisCo (RUB) and glyceraldehyde-3-phosphate dehydrogenase (GAP) are indicated.
To verify whether there was a systematic bias toward synonymous mutations, raw reads were assembled onto several nuclear- and plastid-encoded reference gene scaffolds (average coverage >1,000). All positions with greater than 0.5% variant nucleotides were classified as synonymous or nonsynonymous and normalized to the total number of synonymous or nonsynonymous positions in the sequence (dS and dN) (Fig. 2B and Table S3). Curiously, although the dN/dS ratio is close to 1 for the plastid-encoded genes (Fig. 2B, open circles), nuclear-encoded genes (Fig. 2B, closed circles) show a clear tendency toward synonymous changes, indicating a purifying selection. It also is interesting that the two sequences with the largest number of synonymous mutations, the RuBisCo and glyceraldehyde-3-phosphate dehydrogenase sequences, are both thought to be derived from horizontal gene transfer (HGT) (18, 19).
Sequences of Potential Bacterial Origin Have the Same GC-Content as the Host.
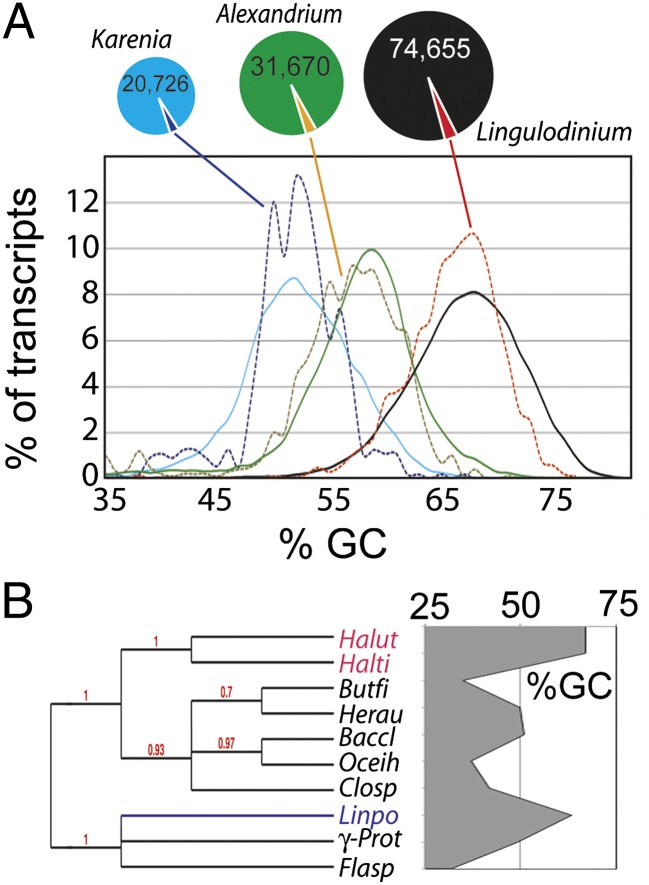
To pursue the possibility that sequences derived from HGT might act as a counterpoint to the high degree of sequence conservation observed for TAGs, we searched our catalog for suitable HGT candidates. We reasoned that sequences derived from HGT likely were transferred as single-copy genes and thus may not have been subject to the same sequence conservation mechanisms. A previous analysis of the dinoflagellate Karenia showed that 2.4% of the genes were of potential bacterial origin as determined by “best hit” BLAST searches, and 0.3% were uniquely found in bacteria and dinoflagellates (20). We also used these criteria to try to uncover examples of HGT in our catalog. First we compared the catalog to an in-house bacterial protein database using BLASTx, a search that returned 2,354 sequences (∼3%). Interestingly, similar searches using our Karenia and Alexandrium unigene datasets against the same bacterial protein databank yielded a similar fraction, and all the sequences recovered by the search have the same average GC-content as the ensemble of sequences in the species from which they are derived (Fig. 3A). We also note that of the 2,354 Lingulodinium sequences, most (∼80%) are targeted to the cytoplasm or to other cellular organelles (Fig. S5A), indicating only a few are likely to be mitochondrial or plastid components transferred to the nucleus from the endosymbiont (21). Most are enzymes, and the domain structure shows enrichment in nucleotide-binding or biosynthetic functions (Fig. S5 B and C).
Fig. 3.
Bacteria-like sequences in the transcriptomes of different dinoflagellates have GC-contents commensurate with the host. (A) The GC-content of the unigene catalogs, as well as for potential bacterial sequences, was compared for datasets from Lingulodinium and two other dinoflagellate species, Alexandrium spp. and Karenia brevis. A similar proportion of potential bacterial sequences (3%) is found in all three datasets. Solid lines represent the entire dataset, dotted lines the dataset of potential bacterial sequences. (B) The GC-content of an arabinofuranosidase (JO761275) in L. polyedrum (Linpo) is higher than the more closely related eubacterial sequences (Butyrivibrio fibrisolvens, Herpetosiphon aurantiacus, Bacillus clausii, Oceanobacillus iheyensis, Clostridium sp., an unidentified γ-proteobacterium, and Flavobacteriales sp.) and is more similar to the more distantly related archeal sequences (Halorhabdus utahensis and Halorhabdus tiamatea).
Among these 2,354 sequences, we next determined candidates for HGT following divergence of Lingulodinium by removing any sequences also found in the Karenia and Alexandrium datasets. This step reduced the number of potential HGT candidates to 1,422 sequences. Lastly, we tested whether any of these sequences were found uniquely in bacteria and dinoflagellates, using BLASTx to screen the GenBank nonredundant (nr) dataset. Because our goal was to find potential examples of HGT rather than to determine the full extent of HGT contribution to the Lingulodinium genome, only a 200-sequence subset of the 1,422 potential HGT sequences was tested; of these, 58 sequences returned only bacterial homologs in the top 100 BLAST hits.
To assess the possibility that the transcriptome might contain sequences resulting from bacterial contamination of our unialgal but not axenic cultures, we examined the initial 2,354 sequences for the presence of the characteristic 22-nucleotide transspliced leader (SL) sequence. The presence of a 5′ SL sequence constitutes an unambiguous marker for dinoflagellate nuclear transcripts. Bioinformatics searches found that 60 of the 2,354 putative bacterial sequences (2.5%) had a partial (10-nucleotide) match to the SL, whereas 1,420 sequences in the full catalog (2%) contained the same partial match. These percentages are low, presumably because read coverage is always low at the ends of our contigs and because many of the contigs are small and likely are only fragments of longer sequences. We also tested for the presence of the SL directly using a 5′ RACE reaction between an SL 5′ primer and a sequence-specific 3′ primer. Here we used 14 random sequences of the 58 found only in bacteria and Lingulodinium. Two of the 14 sequences—highly abundant in the transcriptome, based on read counts—and 2 of an additional 12 low-abundance transcripts were amplified successfully. By comparison, of 6 nonbacterial sequences, 4 highly abundant sequences were amplified whereas 2 low-abundance transcripts were not. We attribute the difficulty in amplifying low-abundance transcripts to the fact that the SL primer will bind to all transcripts, lowering its effective concentration and selectively disadvantaging amplification of low-abundance transcripts.
We then analyzed the phylogeny of the four Lingulodinium sequences that both contained an SL by 5′ RACE and had only bacterial homologs. As exemplified by the arabinofuranosidase gene (JO761275), three had AT-rich bacteria as their closest phylogenetic neighbors (Fig. 3B), whereas the fourth could not be assigned to a particular clade. Because any sequence derived from an AT-rich organism after divergence of Lingulodinium and Alexandrium has altered its GC-rich content, this indicates single-copy genes may not be subject to the same degree of sequence conservation as are TAGs.
Assessing the Potential for Polycistronic Transcripts.
Transcription in dinoflagellates is poorly understood, and the discovery of a transspliced leader at the 5′ end of all dinoflagellate transcripts (11), in conjunction with the unusual tandem arrangement of gene copies and the lack of recognizable promoter elements in the intergenic spacer (9), has led to the proposal that dinoflagellates may synthesize long polycistronic transcripts (22). In this model, mature transcripts from TAGs have a single origin of transcription and the individual ORFs are excised by transsplicing at their 5′ end and by cleavage followed by polyadenylation at the 3′ end. Interestingly, the model makes several predictions concerning the amount and type of RNA in the transcriptome that can be tested by deep sequencing.
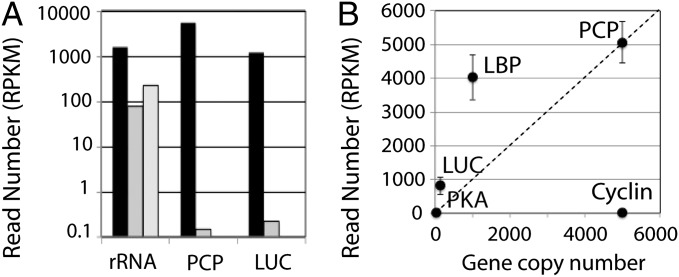
One prediction is that reads corresponding to the genomic sequence between coding sequences, termed intergenic spacer regions, should be present in the RNA. However, when reads are assembled to the genomic sequence of PCP, few reads are found corresponding to the spacer (Fig. S6 A and B); this is also found for the luciferase TAG (Fig. S6 C and D). Because there are no known introns in Lingulodinium, as a control we used the polycistronic rRNA precursor transcript containing two internal transcribed spacers (ITSs) that are excised during processing. The reads per kilobase of transcript per million reads (RPKM) values for mature rRNAs are 7–20 times greater than read counts for ITS1 and ITS2, respectively. In contrast, RPKM values for the coding sequences of luciferase and PCP are 5,000–36,000 times greater than for their respective spacer regions (Fig. 4A). We conclude that noncoding RNAs from TAGs do not accumulate to an appreciable extent in the transcriptome.
Fig. 4.
RNA-Seq does not support a polycistronic transcription mechanism. (A) Individual reads (as reads per kilobase per million [RPKM] (23)) aligning to the mature rRNA (dark bars) and each of the two internal transcribed spacers (gray bars) compared with both luciferase (LUC) and PCP coding sequences (dark bars) and intergenic spacers (gray bars). (B) Read counts (mean ± SD of four independent samples) plotted as a function of gene copy number for genes with a known copy number. LBP, luciferin-binding protein; PKA, protein kinase A.
A second prediction is that the number of mature transcripts should be roughly proportional to the number of gene copies. To test this, we counted the number of reads assembled to the reference sequences for the five genes for which gene copy numbers are known and counted RPKM (23). This analysis (Fig. 4B) shows not only that genes with different copy numbers may have similar transcript levels (compare protein kinase A with cyclin) but also that genes with similar copy numbers may have different abundance in the transcriptome (compare PCP with cyclin).
A third prediction is that a polycistronic transcript processed into its repeat units should produce equal numbers of each ORF. To test this, we examined the sequence variation in luciferase gene transcripts (Fig. S4C). This gene has 146 copies, and if transcripts from each gene accumulated to the same extent, any position where only one gene has a mutation would result in a sequence variation of 1/146 (∼0.7%). Thus, if many positions were mutated in only 1 of the 146 copies, our analysis would show a peak at 0.7% (simulated in Fig. S4D). Because a peak at 0.7% is not observed in our data, we conclude that transcripts from all 146 gene copies do not accumulate equally in the transcriptome.
Discussion
We used RNA-Seq to profile the transcriptome of the dinoflagellate Lingulodinium with the initial aim of providing a gene catalog to facilitate gene discovery. De novo assembly is difficult; therefore, to evaluate the quality and completeness of our transcriptome, we determined the coverage of Lingulodinium ESTs in GenBank and of 16 different primary metabolic KEGG pathways. This indicates that 93–94% of the transcriptome is represented in our catalog.
A global GO analysis of the catalog reveals some striking features in the types of genes present. Of particular interest are the DNA-binding proteins, which, unless they are very different in the dinoflagellates, are remarkably underrepresented. Little is known about the regulation of gene expression in dinoflagellates, but this observation suggests that transcriptional control may not be used as extensively as in other organisms. Interestingly, the most abundant DNA-binding domain, the cold-shock domain, is also associated with posttranscriptional regulation in eukaryotes (17) and thus may not function as a DNA-binding protein at all in Lingulodinium. The idea that dinoflagellates favor regulation of gene expression at a posttranscriptional level agrees with studies on circadian regulation of protein synthesis showing extensive translation control (24, 25). It also is interesting that although our transcriptome contains all four core histones, as well as a suite of histone-modifying enzymes, the histone RNA levels are low compared with higher plants, and the histone proteins are still below the level of detection using antibodies (16). Thus, dinoflagellates may not have extensive access to modified histones as a means of regulating transcription rates.
The possibility that very low levels of histones are present in the dinoflagellates is intriguing, as low levels of acetylated histone H3 are used to initiate polycistronic transcription in the kinetoplastids (26). Kinetoplastids transcribe a polycistronic RNA in both directions from a central point on the chromosome then excise the individual ORFs by addition of a transspliced leader (27). This similarity with the transsplicing of dinoflagellate transcripts (11) has led to the proposal that polycistronic transcription of TAGs might occur in dinoflagellates. Here, a TAG would be transcribed as a single transcript with multiple ORFs, and processed by transsplicing and polyadenylation to yield equal numbers of all individual ORFs. We have tested this model experimentally using our RNA-Seq data. First we examined the raw RNA read data for sequences that could be assembled to the intergenic spacer sequences, reasoning that polycistronic transcripts should produce intergenic regions and coding sequences in initially equal amounts. We anticipated these noncoding sequences, like introns, might be easily detectable, as intron sequences accumulate to roughly 1% of sequence reads in fission yeast (28) and are even more abundant in mammals (29). Because currently there are no known introns in Lingulodinium, we instead used read counts corresponding to the ITS, a spacer region excised during formation of mature rRNA, and found that the read ratio of ITS to mature rRNA is much higher than that of TAG spacer to mature ORF (Fig. 4A). One factor that may influence the spacer/coding sequence read ratio is a preferential loss of nonpolyadenylated spacer regions during poly(A) purification. However, poly(A) RNA is enriched only about 10-fold in our preparations, which clearly is insufficient to account for the differences observed. We also tested for a correlation between the number of genes in the tandem gene array and the amount of transcript, which would be expected if a TAG were in a single operon (or several coregulated operons) and different transcripts were not differentially degraded. However, we found that transcript abundance does not directly correlate with copy number (Fig. 4B). Lastly, we also devised a test to determine whether all the genes in the 146-gene luciferase tandem array are expressed equally. We predicted that because a mutation in one of the copies in the array would have a sequence variation (0.7%), if a sizable number of positions were different in one of the 146 gene copies, we would see a peak of nucleotide variation at 0.7% (Fig. S4D). However, this predicted peak of variation was not seen in our data (Fig. S4C). Considering all these findings, we conclude there is no support for the existence of polycistronic transcripts in Lingulodinium.
Interestingly, TAG transcripts are remarkably well conserved. This sequence conservation is seen at the nucleotide level, as the variation at each position of the gene sequence is low. This may suggest that, as for ribosomal genes, a tandem gene arrangement may be conducive to the conservation of sequence by gene conversion. Indeed, the levels of sequence variation found in the transcriptome for PCP (∼5,000 copies) are similar to those observed for rRNA. It is important to note that this variation is contained within the raw reads; thus a new assembly of reads to a reference sequence appears to be the only means of recovering all the variant nucleotides in their correct proportions. We also observed extensive sequence conservation at the deduced protein level, as variations in nuclear-encoded gene sequences appear to be biased toward synonymous mutations. It is an intriguing question how mutations that lead to deleterious changes in the amino acid sequence of the protein might be traced back to the gene that encodes them in the context of a large gene family. It is possible that purifying selection might operate against a deleterious mutation only if it becomes fixed in the gene array by gene conversion.
By itself, a TAG arrangement is not sufficient to confer a high degree of sequence conservation, as considerable sequence diversity has been observed for TAGs in other dinoflagellates (30–35). This may involve the number of gene copies, as the ∼36 PCP copies in Symbiodinium have multiple nonsynonymous mutations in the coding sequence (30) in contrast to the ∼5,000 almost identical PCP copies in Lingulodinium polyedrum (Fig. 2). Furthermore, the proximity of the elements in the TAG also seems important, as actin copies in Amphidinium are found in two separate genomic clusters with different nucleotide sequences, intron lengths, and intergenic spacer sizes (32). This suggests concerted evolution is allowed within TAG clusters but not between two different clusters of the same gene.
As a contrast to the sequence conservation observed for TAGs, we sought genes that at one time may have been low or single copy and thus may have been allowed to mutate. To this end, we searched the transcriptome for sequences potentially derived from bacteria, because many marine bacteria are AT-rich and because HGT would have placed these sequences in a GC-rich environment. We reasoned that if any of these genes were originally AT-rich but are now GC-rich, Lingulodinium must be able to extensively modify the sequence of single-copy genes. We found several examples of sequences with AT-rich phylogenetic relatives and an unambiguous dinoflagellate origin based on the presence of the SL. Interestingly, because roughly a third of the sequences with similarity to bacterial proteins are found with matches to only bacterial sequences in the top 100 BLAST hits, up to ∼0.6% of Lingulodinium sequences may have a potential bacterial origin. Although this percentage is substantially lower than the 7.5% of sequences with a bacterial origin reported for the diatom Phaeodactylum tricornutum (36), this clearly is an interesting avenue to pursue in future work.
Our RNA-Seq–derived gene catalog contains 74,655 unique sequences that would agree well with gene content estimates based on extrapolations of genome sizes (37) if all genes were present in a single copy. However, the smallest gene family known so far, protein kinase A, has 30 copies (38) and other genes have even higher copy numbers. To accommodate 75,000 genes of 1 kbp with an average gene copy number of 30 within the 200-pg nuclear DNA (∼2 × 1011 bp) we would have to assume a gene density of 1%. So far, the only report of a large genomic DNA fragment sequence (230 kb) is in Heterocapsa, and this indicates a gene density of only 0.2% (39). Furthermore, the size distribution of our sequences, biased toward short sequences (Fig. S2), also suggests the total number of genes will be less than ∼75,000.
We report here the most extensive transcriptome profile yet presented for a dinoflagellate, and our analysis of the gene catalog suggests dinoflagellates may favor posttranscriptional regulation of gene expression. We also have used the read data to explore the nature of TAGs and the mechanisms used for their expression. In particular, we have found no evidence for the polycistronic transcripts found in kinetoplastids, another organism with rampant transsplicing. It appears that unraveling the mechanism of transcription in dinoflagellates will require extensive mining of databanks such as our Lingulodinium transcriptome as well as biochemical analyses to provide functional tests for DNA-binding activities.
Methods
Cell Culture.
Unialgal but not axenic L. polyedrum (CCMP 1936, previously Gonyaulax polyedra) was obtained from the National Center for Marine Algae. Clonal cell cultures derived from a single cell were grown in f/2 medium prepared from Instant Ocean under 12 h light (40 μmol photons m−2 s−1 cool white fluorescent light) and 12 h darkness at a temperature of 18 ± 1 °C as described (40).
RNA Purification and Sequencing.
Lingulodinium cultures were harvested at midday (LD 6) and midnight (LD 18) under a light–dark cycle, the corresponding times under constant light (LL 6 and LL 18), and from a culture taken at LD 18 but grown without added nitrate for 4 d. Cells were concentrated by centrifugation (500 × g for 1 min), washed with fresh seawater, and recentrifuged to reduce bacterial contamination. Total RNA was isolated by extracting cell pellets with TRIzol (Invitrogen) and enriched for poly(A) RNA using the PolyATract mRNA isolation system (Promega). RNA samples were subjected to quality control assessment using a Bioanalyzer (Agilent), and sequencing used an mRNA-Seq sample preparation kit (Illumina). Each sample was sequenced on a single lane of a Genome Analyzer IIX platform at the McGill University and Génome Québec Innovation Centre. In total, 312 × 106 76-base paired-end reads (24-Gb total sequence) were obtained. RNA samples used for sequencing also were used for 5′ and 3′ RACE using the SMARTer RACE cDNA amplification kit (Clontech) using the manufacturer’s protocol, except that for 5′ RACE, a version of splice-leader sequence modified to accommodate multiple splicing events (41) (5′-TGGCTCAAGCCATTTTGGCTCAAG-3′) replaced the forward primer supplied in the kit.
Sequence Assembly and Analysis.
After the sequences were filtered to exclude low-quality bases, reads were assembled with Velvet and Oases. Hash lengths of 21–61 were tested, and k-mers of 41 were retained for assembly. The original Velvet/Oases assembly resulted in 200,045 contigs, 88,655 of which were >300 bp (the fragment size selected for sequencing). Some contigs contained more than one mRNA, possibly because of alternative splicing. To facilitate further analyses, only one transcript (best BLAST hit and/or longest sequence) was retained per contig to yield 74,337 contigs. In parallel, small-scale assemblies (∼3 × 106 reads) were prepared from each sample using Geneious (42) at the default settings and screened against the Velvet/Oases assembly by BLASTn. These small-scale Geneious assemblies contained 318 generally high-coverage transcripts absent from the Velvet/Oases assembly, and these were added back to produce a final dataset of 74,655 contigs >300 bp. All assembled sequences have been deposited in GenBank (accession nos. JO692619–JO767447).
Comparison of the catalog with known L. polyedra ESTs used a 65% GC-rich unigene dataset (BP742156-4266) (43), 56% of which had matches to other dinoflagellate ESTs (tBLASTn, e−10). A second dataset also annotated as L. polyedra (CD809360-810879) was not used because it was 54% GC-rich and only 42% had matches to other dinoflagellate ESTs. Sequence annotations and mapping to GO (44) and the KEGG pathways (45) were performed using Blast2Go (46). Interspecies comparison of GO and PFAM domains (47) was made by similarly annotating Paramecium tetraurelia (48), Thalassiosira pseudonana (49), and Chlamydomonas reinhardtii (50) predicted gene models. Phylogenetic analysis was performed using the top 100 or 250 hits from BLAST (51) searches of the GenBank nonredundant database. Protein phylogenies were reconstructed using an online pipeline at Phylogeny.fr (52) that aligns the sequences using MUSCLE, curates them using GBlocks, performs phylogenetic analysis using PhyML, then renders a tree using TreeDyn.
To detect genes of potential bacterial origin, a three-step protocol was followed. First, candidates were selected by comparing the transcriptome with an in-house bacterial protein database (prepared by downloading all available protein sequences from a National Center for Biotechnology Information database of whole genomes for three α-proteobacteria [Rhodopseudomonas palustris, Sinorhizobium sp., Azorhizobium sp.], one β-proteobacterium [Burkholderia dolosa], two γ-proteobacteria [Pseudoalteromonas tunicata, Alteromonas macleodii], one Bacteroidetes [Flavobacteriales bacterium], and two cyanobacteria [Prochlorococcus marinus, Synechococcus elongatus]) using BLASTx with E values <e−30. Next, any sequences with a match (tBLASTx, E values <e−30) to the Karenia or Alexandrium datasets were removed. In the last step, candidates were compared with the GenBank nr database using BLASTx to assess how many had matches only to bacteria in the top 100 sequences.
To assess sequence variation in the contigs, we obtained full-length Lingulodinium sequences from the nucleotide database at GenBank to use as reference sequences. These were used as scaffolds to assemble raw reads using the Geneious reference assembly function at the default settings (42) and using a cutoff of 0.5% for SNP determinations. The number of synonymous and nonsynonymous mutations was calculated as dS and dN by dividing by the total number of synonymous and nonsynonymous positions in each sequence (53).
To find sequences common to several dinoflagellate species, EST datasets for Karenia brevis (66,657 sequences) and Alexandrium spp. (50,302 sequences) were downloaded from GenBank, and each was assembled separately using Geneious at default settings to produce unigene datasets containing 20,726 and 31,670 sequences for Karenia and Alexandrium, respectively. The Lingulodinium dataset was compared with these datasets using BLASTx with E values <e−30.
Supplementary Material
Acknowledgments
The authors thank S. Kholmogorova and S. Benribague for helpful discussions and Dr. Tao Jin (Beijing Genomics Institute) for review of the manuscript. Financial support from National Science and Engineering Research Council (NSERC) of Canada Grants 171382-03 and 194560 (to D.M. and B.F.L.) is acknowledged. S.D.-B. and M.B. are studentship recipients from Fonds pour la Formation de Chercheurs et l’Aide à la Recerche and NSERC, respectively.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JO692619–JO767447).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206683109/-/DCSupplemental.
References
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 2.Gordon BR, Leggat W. Symbiodinium-invertebrate symbioses and the role of metabolomics. Mar Drugs. 2010;8:2546–2568. doi: 10.3390/md8102546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glibert PM, Anderson DM, Gentien P, Granéli E, Sellner K. The global, complex phenomena of harmful algal blooms. Oceanography (Wash DC) 2005;18:132–141. [Google Scholar]
- 4.Wilson T, Hastings JW. Bioluminescence. Annu Rev Cell Dev Biol. 1998;14:197–230. doi: 10.1146/annurev.cellbio.14.1.197. [DOI] [PubMed] [Google Scholar]
- 5.Costas E, Goyanes V. Architecture and evolution of dinoflagellate chromosomes: An enigmatic origin. Cytogenet Genome Res. 2005;109:268–275. doi: 10.1159/000082409. [DOI] [PubMed] [Google Scholar]
- 6.Chow MH, Yan KT, Bennett MJ, Wong JT. Birefringence and DNA condensation of liquid crystalline chromosomes. Eukaryot Cell. 2010;9:1577–1587. doi: 10.1128/EC.00026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S. Genomic understanding of dinoflagellates. Res Microbiol. 2011;162:551–569. doi: 10.1016/j.resmic.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 8. Spector D, ed (1984) Dinoflagellates (Academic, New York), p 545.
- 9.Le QH, Markovic P, Hastings JW, Jovine RV, Morse D. Structure and organization of the peridinin-chlorophyll a-binding protein gene in Gonyaulax polyedra. Mol Gen Genet. 1997;255:595–604. doi: 10.1007/s004380050533. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Hastings JW. The structure and organization of the luciferase gene in the photosynthetic dinoflagellate Gonyaulax polyedra. Plant Mol Biol. 1998;36:275–284. doi: 10.1023/a:1005941421474. [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, et al. Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci USA. 2007;104:4618–4623. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imboden MA, Laird PW, Affolter M, Seebeck T. Transcription of the intergenic regions of the tubulin gene cluster of Trypanosoma brucei: Evidence for a polycistronic transcription unit in a eukaryote. Nucleic Acids Res. 1987;15:7357–7368. doi: 10.1093/nar/15.18.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldmeyer B, Wheat CW, Krezdorn N, Rotter B, Pfenninger M. Short read Illumina data for the de novo assembly of a non-model snail species transcriptome (Radix balthica, Basommatophora, Pulmonata), and a comparison of assembler performance. BMC Genomics. 2011;12:317. doi: 10.1186/1471-2164-12-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillebault D, et al. A new class of transcription initiation factor, intermediate between TBPs and TLFs is present in the marine unicellular organism: The dinoflagellate Crypthecodinium cohnii. J Biol Chem. 2002;277:40881–40886. doi: 10.1074/jbc.M205624200. [DOI] [PubMed] [Google Scholar]
- 16.Roy S, Morse D. A full suite of histone and histone modifying genes are transcribed in the dinoflagellate Lingulodinium. PLoS ONE. 2012;7:e34340. doi: 10.1371/journal.pone.0034340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mihailovich M, Militti C, Gabaldón T, Gebauer F. Eukaryotic cold shock domain proteins: Highly versatile regulators of gene expression. Bioessays. 2010;32:109–118. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]
- 18.Morse D, Salois P, Markovic P, Hastings JW. A nuclear-encoded form II RuBisCO in dinoflagellates. Science. 1995;268:1622–1624. doi: 10.1126/science.7777861. [DOI] [PubMed] [Google Scholar]
- 19.Fagan T, Woodland Hastings J, Morse D. The phylogeny of glyceraldehyde-3-phosphate dehydrogenase indicates lateral gene transfer from cryptomonads to dinoflagellates. J Mol Evol. 1998;47:633–639. doi: 10.1007/pl00006420. [DOI] [PubMed] [Google Scholar]
- 20.Nosenko T, Bhattacharya D. Horizontal gene transfer in chromalveolates. BMC Evol Biol. 2007;7:173. doi: 10.1186/1471-2148-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackett JD, et al. Migration of the plastid genome to the nucleus in a peridinin dinoflagellate. Curr Biol. 2004;14:213–218. doi: 10.1016/j.cub.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 22.Lukes J, Leander BS, Keeling PJ. Cascades of convergent evolution: the corresponding evolutionary histories of euglenozoans and dinoflagellates. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9963–9970. doi: 10.1073/pnas.0901004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 24.Morse D, Milos PM, Roux E, Hastings JW. Circadian regulation of bioluminescence in Gonyaulax involves translational control. Proc Natl Acad Sci USA. 1989;86:172–176. doi: 10.1073/pnas.86.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fagan T, Morse D, Hastings JW. Circadian synthesis of a nuclear-encoded chloroplast glyceraldehyde-3-phosphate dehydrogenase in the dinoflagellate Gonyaulax polyedra is translationally controlled. Biochemistry. 1999;38:7689–7695. doi: 10.1021/bi9826005. [DOI] [PubMed] [Google Scholar]
- 26.Thomas S, Green A, Sturm NR, Campbell DA, Myler PJ. Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics. 2009;10:152. doi: 10.1186/1471-2164-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons M, Nelson RG, Watkins KP, Agabian N. Trypanosome mRNAs share a common 5′ spliced leader sequence. Cell. 1984;38:309–316. doi: 10.1016/0092-8674(84)90552-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 29.Ameur A, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 30.Reichman JR, Wilcox TP, Vize PD. PCP gene family in Symbiodinium from Hippopus hippopus: low levels of concerted evolution, isoform diversity, and spectral tuning of chromophores. Mol Biol Evol. 2003;20:2143–2154. doi: 10.1093/molbev/msg233. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Hou Y, Lin S. Isolation and characterization of proliferating cell nuclear antigen from the dinoflagellate Pfiesteria piscicida. J Eukaryot Microbiol. 2006;53:142–150. doi: 10.1111/j.1550-7408.2005.00085.x. [DOI] [PubMed] [Google Scholar]
- 32.Bachvaroff TR, Place AR. From stop to start: Tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS ONE. 2008;3:e2929. doi: 10.1371/journal.pone.0002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe CD, et al. The transcriptome of the novel dinoflagellate Oxyrrhis marina (Alveolata: Dinophyceae): Response to salinity examined by 454 sequencing. BMC Genomics. 2011;12:519. doi: 10.1186/1471-2164-12-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachvaroff TR, Concepcion GT, Rogers CR, Herman EM, Delwiche CF. Dinoflagellate expressed sequence tag data indicate massive transfer of chloroplast genes to the nuclear genome. Protist. 2004;155:65–78. doi: 10.1078/1434461000165. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Dungan CF, Lin S. Introns, alternative splicing, spliced leader trans-splicing and differential expression of pcna and cyclin in Perkinsus marinus. Protist. 2011;162:154–167. doi: 10.1016/j.protis.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 37.Hou Y, Lin S. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: Gene content estimation for dinoflagellate genomes. PLoS ONE. 2009;4:e6978. doi: 10.1371/journal.pone.0006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salois P, Morse D. Characterization and molecular phylogeny of a protein kinase cDNA from the dinoflagellate Gonyaulax. J Phycol. 1997;33:1063–1072. [Google Scholar]
- 39.McEwan M, Humayun R, Slamovits CH, Keeling PJ. Nuclear genome sequence survey of the Dinoflagellate Heterocapsa triquetra. J Eukaryot Microbiol. 2008;55:530–535. doi: 10.1111/j.1550-7408.2008.00357.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Jensen L, Højrup P, Morse D. Synthesis and degradation of dinoflagellate plastid-encoded psbA proteins are light-regulated, not circadian-regulated. Proc Natl Acad Sci USA. 2005;102:2844–2849. doi: 10.1073/pnas.0406522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slamovits CH, Keeling PJ. Widespread recycling of processed cDNAs in dinoflagellates. Curr Biol. 2008;18:R550–R552. doi: 10.1016/j.cub.2008.04.054. [DOI] [PubMed] [Google Scholar]
- 42.Drummond A, et al. 2011. Geneious, Version 5.4. Available at http://www.geneious.com.
- 43.Tanikawa N, Akimoto H, Ogoh K, Chun W, Ohmiya Y. Expressed sequence tag analysis of the dinoflagellate Lingulodinium polyedrum during dark phase. Photochem Photobiol. 2004;80:31–35. doi: 10.1562/2004-03-12-RA-110.1. [DOI] [PubMed] [Google Scholar]
- 44.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okuda S, et al. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36(Web Server issue):W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 47.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 49.Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 50.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.