Abstract
Posttranscriptional gene silencing (PTGS) mediated by siRNAs is an evolutionarily conserved antiviral defense mechanism in higher plants and invertebrates. In this mechanism, viral-derived siRNAs are incorporated into the RNA-induced silencing complex (RISC) to guide degradation of the corresponding viral RNAs. In Arabidopsis, a key component of RISC is ARGONAUTE1 (AGO1), which not only binds to siRNAs but also carries the RNA slicer activity. At present little is known about posttranslational mechanisms regulating AGO1 turnover. Here we report that the viral suppressor of RNA silencing protein P0 triggers AGO1 degradation by the autophagy pathway. Using a P0-inducible transgenic line, we observed that AGO1 degradation is blocked by inhibition of autophagy. The engineering of a functional AGO1 fluorescent reporter protein further indicated that AGO1 colocalizes with autophagy-related (ATG) protein 8a (ATG8a) positive bodies when degradation is impaired. Moreover, this pathway also degrades AGO1 in a nonviral context, especially when the production of miRNAs is impaired. Our results demonstrate that a selective process such as ubiquitylation can lead to the degradation of a key regulatory protein such as AGO1 by a degradation process generally believed to be unspecific. We anticipate that this mechanism will not only lead to degradation of AGO1 but also of its associated proteins and eventually small RNAs.
RNA silencing involves the processing of dsRNA by the enzyme Dicer into small RNAs, 21 to 25 nucleotides in length (1–3). One of the two RNA strands is then incorporated into a protein complex called RNA-induced silencing complex (RISC) that invariably contains a member of the highly conserved ARGONAUTE protein family (4, 5). The incorporated small RNA then guides the complex to partially or fully silence complementary RNA. RNA silencing is important for the regulation of development in animals and plants, but also plays an antiviral role in plants and invertebrates (including worms and flies). In this mechanism viral-derived small RNAs are incorporated into the RISC complex to guide degradation of the corresponding viral RNA (6). As a counter defense, viruses have evolved viral suppressors of RNA silencing (VSRs) that suppress the antiviral PTGS defense response (2, 7). VSRs counter host defense by different strategies, including binding to small interfering RNA (siRNA) or double-strand RNA (dsRNA) and inactivating components of the RNA silencing machinery.
Previous work has revealed that the VSR protein P0 from polerovirus encodes an F-box protein that hijacks the host S-phase kinase-associated protein1 (SKP1)-cullin 1 (CUL1)-F-box protein (SCF) ubiquitin-protein ligase (E3) to promote the degradation of AGO1, the key component of RISC (8–10). Although AGO1 ubiquitylation was not directly demonstrated in these studies, its degradation by the ubiquitin-proteasome system (UPS) was expected as it is well known that this system plays numerous and crucial roles in various pathogenic conditions, including interactions with pathogenic viruses (11, 12). However, the targeted degradation of AGO1 by P0 was discovered to be insensitive to inhibition of the proteasome (10) and is suspected to occur before RISC assembly by a still unknown process (13).
Results and Discussion
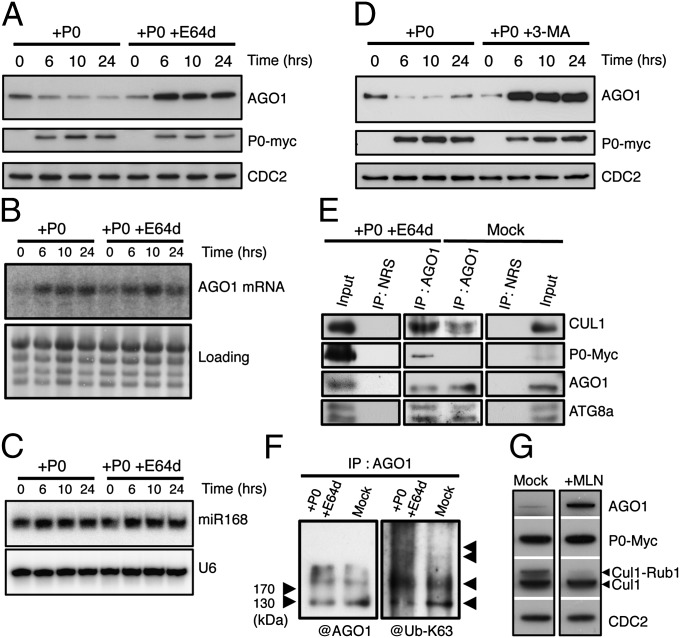
To investigate the mechanism of P0-mediated AGO1 degradation, we used stably transformed Arabidopsis XVE-P0BW transgenic lines in which P0 expression can be induced upon β-estradiol treatment (9). A kinetic analysis revealed a perfect correlation between P0 appearance and AGO1 protein turnover (Fig. S1). However, we also noticed that the process of AGO1 degradation requires several hours and does not lead to a total disappearance of the protein. Next we tested a panel of protease inhibitors. We found that the cysteine protease inhibitor E64d known to inhibit the degradation of autophagic cargo inside autolysosomes (14) led to AGO1 stabilization despite the presence of P0 (Fig. 1A). Overaccumulation of AGO1 protein in presence of E64d results from both a higher AGO1 transcript level in P0-induced plants (Fig. 1B) and impaired protein turnover.
Fig. 1.
P0-mediated degradation of AGO1 is blocked by autophagy inhibitors. AGO1 degradation kinetics were performed on 7-d-old XVE-P0BW-myc seedlings treated with β-estradiol (5 μM) for P0-myc induction. Autophagy was inhibited in its last steps using E64d (20 μM) (A) and AGO1, P0-myc, and CDC2 (loading control) protein accumulation levels were assayed by Western blot on a 24-h period. In a similar manner, AGO1 mRNA (B) and miR168 accumulation (C) was assayed by Northern blot analyses along P0-mediated degradation of AGO1 in presence or in absence of E64d (20 μM). Loading controls are methylene blue staining of the membrane for mRNA and U6 for small RNA blots. (D) Autophagy was inhibited in its first steps using the specific PI-3-kinase class III inhibitor 3-MA (5 mM) and AGO1, P0-myc, and CDC2 (loading control) protein accumulation levels were assayed by Western blot on a 24-h period. (E) Coimmunoprecipitation of AGO1 and SCFP0. XVE-P0BW-myc seedlings were treated with β-estradiol (5 μM) for P0-myc induction and E64d (20 μM) for at least 6 h before protein extraction. Plant extract were immunoprecipitated with an anti-AGO1 antibody and with normal rabbit serum (NRS). IP fractions were submitted to Western blot analysis using antibodies raised against the myc tag for P0 detection and against CUL1, AGO1, and ATG8a. (F) Ubiquitylation status of AGO1 was determined by Western blot analysis of IP fractions using an antibody specifically raised against K63-Ub. (G) Inhibition of SCF activity prevents P0-mediated degradation of AGO1. Seven-day-old XVE-P0BW-myc seedlings were pretreated with MLN-4924 (25 μM) for 3 h before P0-myc induction with β-estradiol (5 μM). The accumulation level of AGO1, P0-myc, CUL1, and CDC2 (loading control) was assayed by Western blot 24 h after P0 induction. Anti-CUL1 antibody detects two bands, the upper one corresponding to the NEDD8/RUB1-modified form of CUL1.
The higher AGO1 transcript level is likely mediated by the regulatory loop consisting of miR168-guided AGO1-catalyzed cleavage of AGO1 mRNA (15, 16), but it is noteworthy that the drug had no significant effect on miR168 accumulation (Fig. 1C). To further support the function of autophagy in the turnover of AGO1, we used 3-methyladenine (3-MA) that blocks autophagosome formation via the inhibition of type III phosphatidylinositol 3-kinases (PI-3K). Similar to the effect of E64d, 3-MA also led to a massive accumulation of AGO1 protein despite the presence of P0 (Fig. 1D).
The possibility that autophagy mediates P0-dependent AGO1 degradation is intriguing with respect to the proposed role of P0 in mediating ubiquitylation. Indeed previous work has shown that the viral F-box protein P0 interacts with the Arabidopsis SKP1-like1 and 2 (ASK1/2) both in vitro and in yeast cells, and that this interaction is required for the silencing suppressor activity of P0 (8). To further investigate the function of ubiquitylation in this process we immunoprecipitated AGO1 from extracts of plants treated with E64d in which P0 expression was induced or not (mock treated). These assays revealed that AGO1 efficiently coprecipitates with P0 as well as CUL1, suggesting that indeed P0 is a component of an E3 ligase targeting AGO1 in planta (Fig. 1E). It is noteworthy that in the absence of P0, AGO1 also precipitates CUL1, although less efficiently, suggesting that endogenous SCF-type ubiquitin E3 ligases may also regulate AGO1 as recently proposed (17). Furthermore, we observed an enrichment of polyubiquitin conjugates in AGO1 immunoprecipitates in the presence of P0. Specific ubiquitin antibodies revealed a significant enrichment in K63-linked chains (Fig. 1F). Whether AGO1 is directly ubiquitylated in a P0-dependent manner or whether other proteins that coimmunoprecipitate together with AGO1 are modified by ubiquitin remains unknown. Next we tested whether SCF-mediated ubiquitylation is required for P0-dependant AGO1 protein turnover. We took advantage of MLN-4924, a selective inhibitor of the neural precursor cell expressed, developmentally down-regulated 8 (NEDD8)/ubiquitin-related protein 1 (RUB1) conjugation pathway that controls the activity of cullin-really interesting new gene (RING) types of ubiquitin ligases in both mammals and plants (18, 19). P0 expression was induced in the Arabidopsis transgenic line in absence and in presence of 25 μM MLN-4924. In these conditions, the drug efficiently inhibited CUL1 neddylation and impaired AGO1 degradation (Fig. 1G). From these results we conclude that the process of AGO1 degradation requires ubiquitylation by an SCF-type E3 ligase.
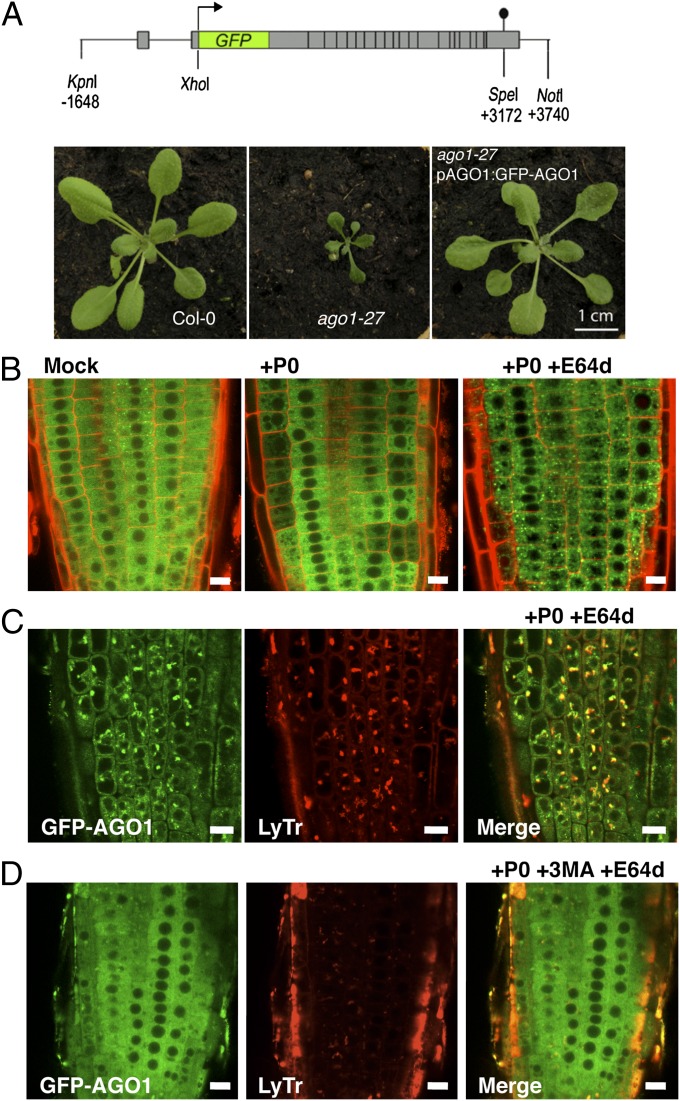
The subcellular localization of hmAGO2, the only human AGO protein that possesses endoribonuclease activity, was found localized in the cytosol and enriched at discrete cytoplasmic foci corresponding to P bodies (20–22). At present, very little is known about AGO1 subcellular localization in plants. To get insights on the AGO1 degradation process at the cellular level, we engineered a construct in which the GFP is fused to the N terminus of AGO1 and expressed the chimeric protein in an AGO1-defficient genomic context (Fig. 2A and Fig. S2). This construct was used to transform heterozygous weak (ago1-27) and strong (ago1-11) mutant alleles, respectively. Full suppression of both mutant phenotypes indicated that GFP-AGO1 protein is functional (Fig. 2A and Fig. S2). In both mutant backgrounds, the GFP signal was clearly visible in root tissues, where the GFP-AGO1 protein was detected in the cytosol, but excluded from the nucleus. It is noteworthy that the GFP-AGO1 signal was enriched in proximity of the nuclear envelope and this was especially visible in the ago1-11 complemented mutant line, in which endogenous AGO1 is entirely replaced by the GFP-AGO1 fusion protein (Fig. S2). It is interesting to note that the subcellular localization of GFP-AGO1 resembles HASTY, the Arabidopsis homolog of the mammalian transport receptors exportin 5 (23), suspected to be located at sites of nucleocytoplasmic mRNA export.
Fig. 2.
Subcellular localization of AGO1 along its degradation process. (A) The pAGO1:GFP-AGO1 construct complements ago1-27 allele phenotype. (B) Subcellular localization of functional GFP-AGO1 assayed by confocal microscopy. Seven-day-old seedlings were transferred from MS-agar plates to liquid MS medium supplemented with the indicated drugs and observed after overnight incubation (16–18 h). In the root tip of XVE-P0BW/GFP-AGO1 reporter lines GFP-AGO1 is localized exclusively in the cytoplasm of cells (Left). After P0 induction, the GFP-AGO1 signal decreases with a nonhomogenous pattern from cell to cell and is relocalized in vesicular-shaped structures (Middle). When P0 induction is combined with E64d (20 μM) treatment, GFP-AGO1 is stabilized and massively accumulates in these vesicular-shaped structures (Right). These speckles colocalize with acidic vesicles labeled with LysoTracker Red DND-99 (LyTr) (C) and their formation is significantly reduced if P0 induction and E64d treatment are combined with 3-MA (5 mM) (D). (Scale bars: 10 μm.)
We subsequently introduced the pAGO1:GFP-AGO1 construct in the LexA-VP16-ER (XVE)-P0BW line. Similarly to endogenous AGO1, the induction of P0 by β-estradiol triggered the degradation of the GFP-AGO1 fusion protein, although β-estradiol alone had no effect on GFP-AGO1 subcellular localization and/or stability (Fig. S3A). Note that the process of AGO1 degradation was not homogenous throughout the root, but occurred stepwise in some cells or group of cells (Fig. 2B), which most likely reflects a spatiotemporal variation in P0 induction by estradiol. Nevertheless, after long periods of P0 induction, we noticed that most (although not all) GFP-AGO1 protein disappeared from the root (Fig. S3A). P0 induction in the presence of E64d resulted in a massive appearance of GFP-AGO1 speckles in the cytosol (Fig. 2B and Fig. S3B). Confocal fluorescence studies showed that a number of these GFP-AGO1 bodies colocalized with LysoTracker Red (Fig. 2C and Fig. S3B), a red fluorescent dye that stains acidic compartments in live cells, including lysosomes, autolysosomes, and vacuoles. In agreement with 3-MA acting upstream of autophagic vesicle formation, we observed that this drug suppressed the formation of the GFP-AGO1 bodies in the presence of both P0 and E64d (Fig. 2D).
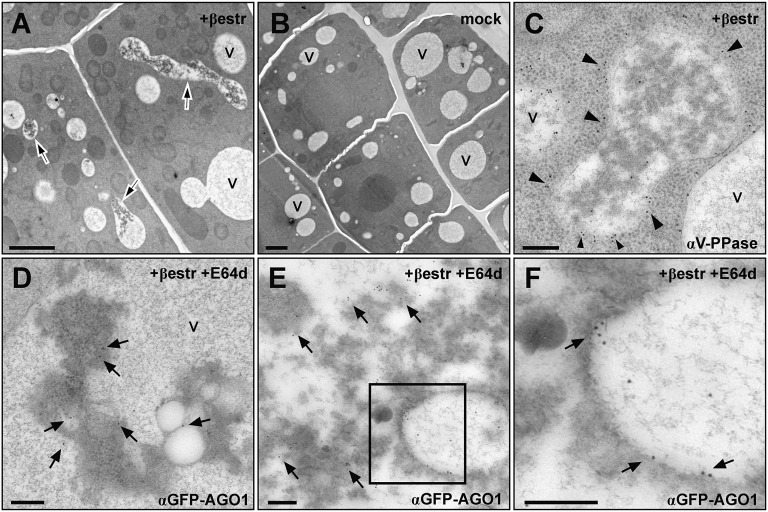
To further investigate the nature of these structures where AGO1 concentrate consecutively to P0 induction, we analyzed the XVE-P0BW/GFP-AGO1 reporter line treated with β-estradiol and E64d using transmission electron microscopy and immunogold labeling. This revealed that P0 triggered the accumulation of electron-dense, membrane-containing material inside vacuoles (Fig. 3). Moreover, GFP-AGO1 massively localized in this dense material often in close association with membranes (Fig. 3 D–F). These observations support the function of autophagy in P0-mediated AGO1 degradation, as proteins following this pathway terminate in the vacuole where they are degraded.
Fig. 3.
P0 expression leads to an accumulation of vacuolar inclusions containing GFP-AGO1. (A) Root-tip cells in which P0 expression was induced with β-estradiol (+βestr) for 12–16 h display electron-dense inclusions in several vacuole-like structures (arrows), and in mock treated cells (B) vacuoles do not contain this kind of inclusions. (A and B, scale bars: 2 μm). (C) The vacuolar identity of the compartments containing the inclusions was confirmed by immunodetection of the vacuolar pyrophosphatase (V-PPase); nanogold particles coupled to secondary antibodies uniformly label the limiting membrane (arrowheads). (D–F) Immunogold labeling of GFP-AGO1 after P0 induction and additional treatment with the protease inhibitor E64d (+βestr+E64d) revealed that AGO1 is present on membranous structures within the inclusions (arrows). V, vacuoles. (C–F, scale bars: 200 nm.)
In Arabidopsis, it was recently shown that AGO1 is a peripheral membrane protein and that isoprenoid biosynthesis, important for membrane protein localization and trafficking, is required for miRNA function (24). Although it is still unknown to which membranes AGO1 associates in plant cells, we noticed its enrichment at proximity of the Golgi apparatus (Fig. S4). This is reminiscent of the situation in animal cells where both AGO and Dicer-like (DCL) localize and fractionate with membranes of the Golgi apparatus (25, 26). It is important to note that in none of our EM images did we observe a localization of AGO1 to multivesicular bodies (MVBs; as exemplified in Fig. S4), arguing against their involvement in routing AGO1 to the vacuole. This situation is different from receptor proteins of the plasma membrane such as Arabidopsis FLAGELLIN-SENSING 2 (FLS2), which upon flagellin perception becomes ubiquitylated and translocated into intracellular vesicles including MVBs to be eventually degraded in the vacuole (27–29).
Autophagic vesicles can be visualized in plant cells using GFP-ATG8a (30). ATG8 is covalently attached to the lipid phosphatidylethanolamine (PE) to produce ATG8-PE that is bound to autophagic membranes via its lipid moiety. To further address the identity of the GFP-AGO1 bodies, we coexpressed GFP-AGO1 and red fluorescence protein (RFP)-ATG8a fusion proteins in Nicotania benthamiana cells. Under these conditions, GFP-AGO1 signal was observed in the cytosol of transformed cells and only a few small structures of less than 1 μm in which both fluorescent proteins colocalized could be detected in transformed cells (Fig. S5). However, E64d induced the emergence of larger bodies (3 μm and above) containing both GFP-AGO1 and RFP-ATG8a proteins. Therefore, even in the absence of P0, inhibition of the autophagy pathway leads to AGO1 accumulation in autophagic vesicles (Fig. S5). These structures were also observed consistently when P0 was induced in these assays. In line with these results, we found that ATG8a coimmunoprecipitates with AGO1 in E64d-treated Arabidopsis plants whether P0 was induced or not (Fig. 1E).
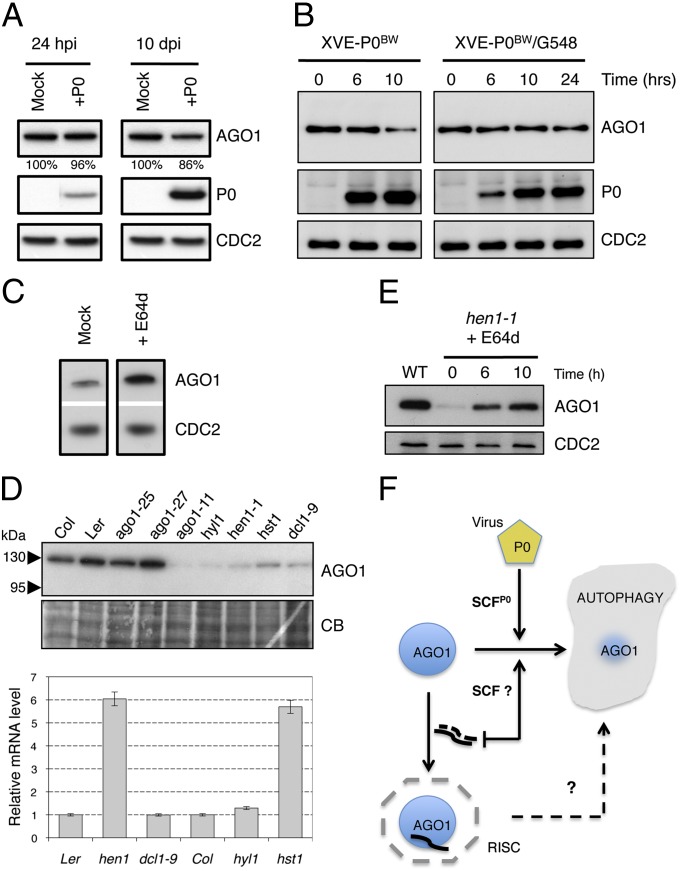
AMSH3, an Arabidopsis deubiquitinating enzyme processing both polyubiquitin K48- and K63-linked chains, was recently shown to be essential for vacuole biogenesis and its mutation leads to the accumulation of autophagosomes (31). AGO1 protein amount was at least fourfold enriched in this mutant, suggesting that its degradation in a nonviral context depends on AMSH3 function (Fig. S6A). When the pAGO1:GFP-AGO1 construct was expressed in homozygous amsh3-1 mutant, we consistently observed a strong accumulation of GFP-AGO1 in the root (Fig. S6B). Moreover, although GFP-AGO1 was difficult to detect in the upper parts of wild type Arabidopsis seedlings including cotyledon and primary leaves (Fig. S6C), the protein was stabilized in the cotyledons of the homozygous amsh3-1 mutant (Fig. S6D). A closer inspection of these cells revealed that GFP-AGO1 accumulated in vesicles that colocalized with the lipophilic dye FM4-64 (Fig. S6E), which were previously shown to accumulate in the amsh3-1 mutant. Next we investigated whether P0-dependent degradation of AGO1 was compromised in this mutant. Therefore, the XVE-P0BW-myc construct was introduced in the amsh3-1 mutant. Hence, P0 accumulation led only to a weak reduction in AGO1 protein amount even after 10 d of induction (Fig. 4A).
Fig. 4.
P0-mediated degradation of AGO1 is compromised in amsh3-1 mutant and in TOR-overexpressing plants and the endogenous pathway for AGO1 degradation also relies on autophagy. (A) P0-dependant degradation of AGO1 in XVE-P0BW-myc/amsh3-1 line. (Left) AGO1 protein accumulation level 24 h after P0 induction (10 μM β-estradiol) on 11-d-old seedlings. (Right) AGO1 accumulation level in 10-d-old seedlings that have been germinated and grown on MS-agar dish containing 10 μM β-estradiol. (B) AGO1 degradation kinetics performed on 7-d old XVE-P0BW-myc and XVE-P0BW-myc/G548 seedlings treated with β-estradiol (5 μM) for P0-myc induction. Because P0 induction is delayed in the XVE-P0BW-myc/G548 line, we extended this kinetic to 24 h. AGO1, P0-myc, and CDC2 (loading control) protein contents were assayed by Western blot. (C) E64d treatment (20 μM for 24 h) on wild-type Col-0 seedling leads to a higher accumulation of AGO1 protein. (D) Mutants affected in miRNA maturation and production pathways show reduced level of AGO1. (Upper) AGO1 protein accumulation in mutants and wild-type controls assayed by Western blot using the anti-AGO1 antibody. Coomassie blue staining is given as loading control. (Lower) AGO1 mRNA accumulation in each mutant assayed by quantitative RT-PCR. (E) The hen1-1 seedlings treated with E64d (20 μM) show AGO1 protein reaccumulation. AGO1 and CDC2 (loading control) protein contents were assayed by Western blot at the indicated time point. (F) Model for AGO1 turnover in a viral and nonviral context.
In environmental favorable conditions, the target of rapamycin (TOR) pathway promotes plant growth and restrains catabolic processes such as mRNA degradation and autophagy in all eukaryotes (32). Thus, we introduced XVE-P0BW-myc construct in a TOR-overexpressing mutant line [G548 (33)] showing reduced autophagy activity. Although P0 induction was slightly delayed in the P0-myc/G548 line, its expression was unable to promote AGO1 degradation (Fig. 4B). Overall, our data support a role of autophagy in P0-mediated degradation of AGO1.
Next, we asked whether AGO1 degradation by the autophagy pathway is restricted to P0 function or whether this pathway in a nonviral context may also degrade endogenous AGO1, as suggested by a higher accumulation of the protein in the amsh3-1 mutant (Fig. S6) and its colocalization with ATG8a in E64d-induced vesicles in the absence of P0 (Fig. S5). In line with this assumption, we noticed a higher accumulation of AGO1 protein level in wild-type seedlings when autophagy was chemically inhibited (Fig. 4C).
Because P0 was proposed to promote AGO1 degradation more efficiently before it is incorporated in the RISC complex (13), we speculated that disturbing normal RISC assembly might lead to AGO1 degradation. Efficient RISC assembly requires the incorporation of small RNAs in both animal and plant cells (34–36). Thus, we selected mutations that are known to affect miRNA biogenesis and accumulation, including mutations in the double-stranded RNA-binding protein DRB1 (also known as HYL1) and Dicer homolog DCL1 mediating processing of most miRNA precursors (37, 38), the RNA methyltransferase HEN1 (39) critical for miRNA stability and HASTY (HST), the Arabidopsis ortholog of Exp5 required for the nuclear export and/or stability of miRNAs (40). RNA and protein samples were extracted from wild type and the respective mutant lines and subjected to mRNA and protein analyses. It is striking to note that AGO1 protein abundance was strongly reduced in all mutants that affect miRNA biogenesis and accumulation (Fig. 4D). This reduction of AGO1 protein accumulation was not the consequence of decreased AGO1 transcript levels, as the AGO1 mRNA level was similar to wild type in dcl1-9 and hyl1 and was even significantly increased in hen1 and hst. To determine whether the decrease in AGO1 protein accumulation was the result of its active turnover by the autophagy pathway, we treated hen1-1 seedlings with E64d. Indeed blocking autophagy by this drug at least partially reestablished AGO1 protein accumulation in this mutant background (Fig. 4E). From these results we conclude that AGO1 is also degraded by the autophagy pathway in a nonviral context, at least when miRNA production or stability is compromised. Altogether our results support a model (Fig. 4F) in which a viral SCFP0 E3 ligase promotes the degradation by autophagy of a specific protein such as AGO1. We also provide evidence that in the absence of P0 endogenous SCF(s) trigger(s) AGO1 degradation by the same pathway, particularly under conditions in which RISC assembly is compromised. Further experiments will reveal whether AGO1 and eventually other ARGONAUTE proteins are also targeted to the same pathway during cellular stress, a situation where miRNA/siRNA populations quickly change and RISC reprogramming is expected.
Materials and Methods
Chemical Treatments.
Constructs and quantitative PCR are indicated in SI Experimental Procedures and the corresponding list of primers in Table S1. For chemical treatments, plants were germinated on Murashige and Skoog (MS)-agar plates. Seven-day-old seedlings were then transferred onto liquid MS medium-containing drugs. E64d (Sigma) was used at the final concentration of 20 μM; 3-MA (Sigma) was prepared freshly for each experiment following this procedure: powder was solubilized in deionized water under gentle heating (45 °C) to a concentration of 100 mM and immediately diluted in liquid MS medium to a concentration of 5 mM. MLN-4924 (Active BioChem) was used at the final concentration of 25 μM. β-Estradiol was used at final concentration of 5 or 10 μM.
Protein Immunoprecipitation.
Plant samples were homogenized in the extraction buffer [50 mM Tris at pH 7.6, 150 mM NaCl, 0.1% Nonidet P-40, GM-132 proteasome inhibitor (Sigma), complete protease inhibitors mixture (Roche)] and insoluble material was removed by centrifugation (30 min, 12,000 g, 4 °C). Lysate was precleaned by incubation with protein A-agarose beads (Roche) at 4 °C for 30 min. The supernatant was then incubated with either normal rabbit serum (RS; Sigma) or anti-AGO1 (Agrisera) serum prebound to protein A-agarose beads overnight at 4 °C. Immunoprecipitates were washed three times with the extraction buffer, eluted from the beads with sample buffer and analyzed by Western blot.
Protein Analysis and Western Blotting.
Total proteins were extracted from 7 d-old seedlings or from plant leaves using denaturing buffer as described in Büche et al. (2000) (41); 10 μg of total protein extracts were separated on SDS–PAGE [15% (wt/vol) acrylamide] gels and blotted onto Immobilon-P membrane (Millipore). For ATG8a detection, total protein extracts were separated on 15% acrylamide gels containing 8 M urea or on Novex NuPAGE Bis-Tris 4–12% gradient gels (Invitrogen). AGO1 protein was detected using the anti-AGO1 antibody (Agrisera) diluted 1:40,000 (v:v). P0-3xMyc protein was detected using anti-myc antibody (Roche) diluted 1:10,000 (v:v). CDC2 protein was detected using anti-PSTAIR antibody (Santa Cruz Biotechnology) diluted 1:5,000 (v:v). ATG8a protein was detected using anti-ATG8A antibody (Abcam) diluted 1:1,000. Cullin-1 protein was detected using anti-CUL1 antibody (42) diluted 1:10,000. K63-ubiquitilation was detected using anti-Ub-K63 antibody (eBioscience) diluted 1:500 (v:v).
Microscopy.
Confocal plan images were acquired using a Zeiss LSM700 confocal laser microscope (Carl Zeiss). propidium iodide (Sigma) was used for cell-wall staining at a concentration of 50 μg/mL For FM4-64 (Invitrogen) staining, seedlings were incubated in MS medium containing 1 μM FM4-64 for 10 min in the dark and then transfer into fresh MS medium without FM4-64 for at least 30 min before observation. In case of Lysotracker Red DND-99 (Invitrogen) staining, the molecule was added directly to the liquid MS medium (100 nM final) in which plantlets were incubated 10 min before observation.
For transmission electron microscopy, 4- to 5-d-old Arabidopsis root tips were cut from the seedlings and submerged in freezing media (200 mM sucrose, 10 mM trehalose, 10 mM Tris buffer, pH 6.6), transferred into planchettes (Wohlwend) and frozen in a high-pressure freezer (HPM010; Bal-Tec). Freeze substitution was performed in a Leica EM AFS2 freeze substitution unit (Leica Microsystems) in dry acetone supplemented with 0.4% uranyl acetate at −85 °C for 16 h before gradually warming up to −60 °C over a 5-h period. After washing with 100% ethanol for 60 min, the roots were infiltrated and embedded in Lowicryl HM20 (intermediate steps of 30, 50, and 70% HM20 in ethanol, 1 h each). The resin was polymerized with UV light in the freeze substitution apparatus (−60 °C 24 h, from −60 °C to 0 °C 24 h, 0 °C 24 h). Ultrathin sections were cut on a Leica Ultracut S (Leica) and incubated with antibodies against GFP (Agrisera) or the V-PPase (CosmoBio) at a dilution of 1:2,000 or 1:4,000, respectively, followed by incubation with 10-nm gold-coupled secondary antibodies (BBInternational) at a dilution of 1:50 in PBS supplemented with 1% BSA. Sections were examined in a JEM1400 transmission electron microscope (JEOL) operating at 80 kV. Micrographs were recorded with a FastScan F214 digital camera (TVIPS).
Supplementary Material
Acknowledgments
We thank Christophe Robaglia for supplying TOR-overexpressing lines. Funding was provided by Centre National de la Recerche Scientifique, European Network of Excellence (NoE) Rubicon LSHG-CT-2005-018683 and Laboratoires d'Excellence (LABEX) NetRNA Grant ANR-10-LABX-36.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209487109/-/DCSupplemental.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 3.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 4.Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 5.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Ding S-W. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 7.Burgyán J, Havelda Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011;16:265–272. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Pazhouhandeh M, et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci USA. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol. 2007;17:1615–1621. doi: 10.1016/j.cub.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 10.Baumberger N, Tsai C-H, Lie M, Havecker E, Baulcombe DC. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol. 2007;17:1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 11.Levy A, Dafny-Yelin M, Tzfira T. Attacking the defenders: Plant viruses fight back. Trends Microbiol. 2008;16:194–197. doi: 10.1016/j.tim.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Gustin JK, Moses AV, Früh K, Douglas JL. Viral takeover of the host ubiquitin system. Front Microbiol. 2011;2:161. doi: 10.3389/fmicb.2011.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Csorba T, Lózsa R, Hutvágner G, Burgyán J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 2010;62:463–472. doi: 10.1111/j.1365-313X.2010.04163.x. [DOI] [PubMed] [Google Scholar]
- 14.Asanuma K, et al. MAP-LC3, a promising autophagosomal marker, is processed during the differentiation and recovery of podocytes from PAN nephrosis. FASEB J. 2003;17:1165–1167. doi: 10.1096/fj.02-0580fje. [DOI] [PubMed] [Google Scholar]
- 15.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22:3879–3889. doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earley K, Smith M, Weber R, Gregory B, Poethig R. An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence. 2010;1:15. doi: 10.1186/1758-907X-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 19.Hakenjos JP, et al. MLN4924 is an efficient inhibitor of NEDD8 conjugation in plants. Plant Physiol. 2011;156:527–536. doi: 10.1104/pp.111.176677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister G, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 22.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 23.Bollman KM, et al. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development. 2003;130:1493–1504. doi: 10.1242/dev.00362. [DOI] [PubMed] [Google Scholar]
- 24.Brodersen P, et al. Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:1778–1783. doi: 10.1073/pnas.1112500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cikaluk DE, et al. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell. 1999;10:3357–3372. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tahbaz N, et al. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otegui MS, Spitzer C. Endosomal functions in plants. Traffic. 2008;9:1589–1598. doi: 10.1111/j.1600-0854.2008.00787.x. [DOI] [PubMed] [Google Scholar]
- 29.Lu D, et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332:1439–1442. doi: 10.1126/science.1204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isono E, et al. The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell. 2010;22:1826–1837. doi: 10.1105/tpc.110.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobrenel T, et al. Regulation of plant growth and metabolism by the TOR kinase. Biochem Soc Trans. 2011;39:477–481. doi: 10.1042/BST0390477. [DOI] [PubMed] [Google Scholar]
- 33.Deprost D, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 35.Haley B, Tang G, Zamore PD. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 36.Iki T, et al. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- 38.Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park M-Y, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Büche C, Poppe C, Schäfer E, Kretsch T. eid1: A new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell. 2000;12:547–558. [PMC free article] [PubMed] [Google Scholar]
- 42.Shen WH, et al. Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell. 2002;13:1916–1928. doi: 10.1091/mbc.E02-02-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.