Abstract
Aberrant activation of receptor tyrosine kinases (RTKs) is a common feature of many cancer cells. It was previously suggested that the mechanisms of kinase activation in cancer might be linked to transitions between active and inactive states. Here we estimate the effects of single and double cancer mutations on the stability of active and inactive states of the kinase domains from different RTKs. We show that singleton cancer mutations destabilize active and inactive states, however inactive states are destabilized more than the active ones leading to kinase activation. We show that there exists a relationship between the estimate of oncogenic potential of cancer mutation and kinase activation. Namely, more frequent mutations have a higher activating effect, which might allow us to predict the activating effect of the mutations from the mutation spectra. Independent evolutionary analysis of mutation spectra complements this observation and finds the same frequency threshold defining mutation hot spots. We analyze double mutations and report a positive epistasis and additional advantage of doublets with respect to cancer cell fitness. The activation mechanisms of double mutations differ from those of single mutations and double mutation spectrum is found to be dissimilar to the mutation spectrum of singletons.
Keywords: cancer mutation, receptor tyrosine kinase, protein structure, kinase activation, mutation spectra, double mutations
Introduction
Receptor tyrosine kinases (RTK) transduce signals from the extracellular matrix to the cytoplasm of a cell, they contain extracellular, trans-membrane and catalytic kinase domains and may include regulatory domains. RTK phosphorylation may lead to Ras activation and initiate the cascade of events which lead to regulation of gene expression implicated in cell division, cellular homeostasis and survival. It has been shown that kinases, especially receptor tyrosine kinases, are frequently mutated in cancer; a large fraction of all alterations in cancer represent point mutations (Wood, et al., 2007) and only a small fraction of mutations are inherited. Although somatic mutations contribute significantly to tumorigenesis, the large majority of them are considered to be neutral (so called “passenger” mutations) and only a few of them are under positive selection in cancer cells (so called “driver” mutations) (Greenman, et al., 2007; Wood, et al., 2007). Certain cancer mutation hot spots have been detected in several receptor tyrosine kinases, most of them located in the activation loop, P-loop and DFG loops. Moreover, driver mutations are more likely found to be associated with the functionally important regions in kinases than passenger mutations (Izarzugaza, et al., 2009). Although mutations in multiple genes have been observed in many cancers, interestingly it was shown that properties of cancer mutations are more similar to Mendelian disease mutations than to complex disease mutations pointing to the scenario when cancer progresses through a series of stepwise mutations each of which might provide some advantage to the tumor cells (Kaminker, et al., 2007; Vogelstein and Kinzler, 1993). Furthermore, according to the concept of “oncogene addiction”, cancer cells depend on the activity of a single or a few oncogenes for their proliferation and survival (Weinstein and Joe, 2008). Finding oncogenes is not a trivial task and one approach includes finding significant driver mutations. Various statistical methods have been applied in the attempt to find positively selected mutants and distinguish driver from passenger mutations but their predictive power remains limited and largely depends on the background mutation rate which is difficult to determine for each sample (Torkamani, et al., 2009).
The connection between cancer and kinase activation has been found fairly recently (Martin, 2004; Wan, et al., 2004). Such increased RTK activity may be caused by gene amplifications, enhanced transcription or translation and also by mutations. Discovery of cancer related mutations in several receptor tyrosine kinases (Paez, et al., 2004) and the analysis of their effect on kinase structure and activity revealed that some mutations may disturb autoinhibitory interactions and considerably accelerate catalysis (Dixit, et al., 2009b; Yun, et al., 2007). Moreover different mutations in receptor tyrosine kinases were shown to result in different binding selectivity to cancer drugs. Structural studies of kinases in complexes with small molecule drugs indeed revealed different structural perturbations in response to cancer mutations which might affect the equilibrium between active and inactive conformations (Engelman, et al., 2007; Eswaran and Knapp, 2010; Greulich, et al., 2005). This reflects the importance of the analysis of structure and dynamics of kinases in understanding the mechanisms of cancer mutations.
The interconversion between active and inactive states in kinases is highly regulated and kinases differ in their mechanisms of activation and inactivation, specifically those processes which lead to conversion of inactive to active forms. Interestingly, it has been shown that despite commonly conserved features, inactive conformations might be more structurally diverse than active conformations and could be targeted selectively by small molecule inhibitors, among them cancer drugs (Jura, et al., 2011; Schindler, et al., 2000). Activation mechanisms of kinases represent a spectrum with two extremes, with the CDK1 (MIM# 116940) and EGF (MIM# 131550) receptors on one side that are activated by allosteric effectors, namely by the formation of an asymmetric dimer in cases of EGFR (Zhang, et al., 2006) and by binding of cyclin in cases of CDK (De Bondt, et al., 1993; Jeffrey, et al., 1995). At the other extreme, SRC (MIM# 190090) kinases are normally inhibited by binding of SH2 and SH3 domains and activated spontaneously when this interaction is disrupted (Moarefi, et al., 1997). In general, ligand binding to the extracellular region controls dimerization of kinase domains and subsequent cross-phosphorylation of tyrosine in the activation loop. Phosphorylated tyrosine can form an electrostatic contact with the basic residues and stabilize the active state of kinase enabling phosphorylation of other tyrosine residues on the C-terminal tail which in turn mediate binding of SH2 and PTB domains of downstream signaling molecules (Hubbard and Miller, 2007). Moreover, it has been proposed that four hydrophobic residues forming the so-called “spine” contribute to the process of activation by coordinating the movements of the N- and C-lobes of the molecule (Kornev, et al., 2006).
Although structural analyses showed significant differences in conformations between the active and inactive forms of kinase domains, only a few hotspot mutations have been analyzed by structural studies and dynamics simulations. The role of many rare as well as frequent mutations in the activation of kinases remains unclear. In this work we examined the energetic effect of cancer mutations on both active and inactive conformations of kinase domain in relation to their oncogenic potential to quantify the coupling between cancer mutations, kinase stability and activity. Oncogenic potential was measured as a number of samples where a given mutation was observed. Since cancer mutations can be observed not only as singletons, but also in doublets or triplets, we analyzed mutation patterns for single and double mutations in the kinase domain (the juxtamembrane, JM, region was also included if its structure was available). In accord with other studies we showed that cancer mutations had an activating effect on RTKs. Singleton cancer mutations overall destabilized both active and inactive states, however inactive states were destabilized more than the active ones so that active states were more populated. Interestingly, more frequently observed mutations had a higher activating effect for both single and double mutations which might allow one to predict the activating effect of the mutations from their mutation spectra. Independent evolutionary analysis of mutation spectra complemented this observation and found the same frequency threshold defining the mutation hot spots. Moreover, for many double mutations we found a positive epistasis or non-additive effect which pointed to the additional advantage of doublets for the tumor cell compared to singletons. The evolutionary analysis also demonstrated that non-synonymous single and multiple mutations in RTKs occured more often than expected by chance and led to selective advantages for tumor cells. In addition, the mutation spectrum of multiple mutations was found to be different from the spectra of singletons which hints at different underlying mechanisms of their origin.
Materials and Methods
Examining the frequency distribution of cancer mutations
We derived all mutations available for 58 different human RTKs from the COSMIC database v49 Release (Forbes, et al., 2008) which stores somatic mutations of cancer cells extracted from the primary literature. Amino acid sequences of the 58 RTKs and mutation numbering were obtained from the COSMIC database (Supp. Table S1). We divided the sequences into five regions: the extracellular region, transmembrane domain, juxtamembrane domain, kinase domain, and C-terminal tail. First, we determined the boundaries of the transmembrane domain. Second, to determine the boundaries of the kinase domain we generated a multiple sequence alignment of 58 sequences using the MAFFT program (Katoh and Toh, 2008) and mapped secondary structure elements onto the alignment using a crystal structure of the kinase domain of EGFR. We defined the first residue of the first beta-strand of the N-lobe as the first site of the kinase domain and the last residue of the last α-helix of the C-lobe as the terminal site of the domain. Finally, the extracellular region was defined between the N-terminal end and the transmembrane domain, the juxtamembrane region - between the transmembrane domain and the kinase domain, and the C-terminal tail - between the end of the kinase domain and the end of the protein. For each region, we counted the number of unique mutations and mutation sites, and calculated the number of unique mutation sites per residue in the protein. The evolutionary tree of 58 RTKs was constructed based on the multiple alignment of the kinase domain with the neighbor-joining method (Saitou and Nei, 1987) using MEGA5.
Assessing the effect of mutations on RTKs in active and inactive states
We obtained all crystal structures for RTKs available in the manually curated KEGG pathway database (Kanehisa, et al., 2004), and manually compiled a list of structures of the kinase domain in active and inactive states (altogether 18 structures). We then calculated the change in unfolding free energy (ΔΔG) of single amino acid substitutions on these structures. Perturbations in the unfolding free energy caused by mutations may result in changes of equilibrium constant and concentrations of RTKs indifferent conformations. ΔΔG was calculated using two modules of FoldX version 3.0 (Guerois, et al., 2002) which is among the top available methods to estimate the effect of mutations on protein stability; it reaches 0.64 sensitivity and 0.43 specificity (Khan and Vihinen, 2010) of prediction and reports a correlation coefficient between experimental and computed ΔΔG values in the range of 0.5–0.8 (Guerois, et al., 2002; Potapov, et al., 2009; Zhang, et al., 2012).
First we identified strained torsion angles and Van-der-Waals' clashes in the original structure and optimized side chains to provide a repaired structure (RepairPDB module). Then using the BuildModel module we optimized the configurations of the neighboring side chains of the mutation site and calculated the difference in stability (ΔΔG) between the repaired native structure and the mutant structure.
For each mutation and corresponding amino acid substitution, we calculated a ΔΔΔG value which represents the difference in effect of the mutation on the stability of the active and inactive states:
| (1) |
| (2) |
where ΔG(wt) and ΔG(mut) were the free energy of unfolding for wild type and structures with mutations respectively. The unfolded state of wild type and mutants were considered to be similar as described in previous studies (Zhang, et al., 2011; Zhang, et al., 2010). The ΔΔΔG value was calculated only when the mutation could be mapped on both structures in the active and inactive states. We also extracted combinations of two or more concurrent mutations from our dataset which we call thereafter “multiple” mutations. The ΔΔG values of the observed multiple mutations were calculated using the BuildModel module of FoldX. Negative and positive ΔΔG values corresponded to stabilizing and destabilizing effects of mutations respectively. In addition, we sampled the background ΔΔG distribution by selecting all possible single amino acid substitutions caused by single nucleotide substitutions for all sites in a protein (for analysis of single mutations) and by randomly selecting 1000 pairs of amino acid substitutions (for analysis of double mutations).
We also defined the positive epistatic (super-additive) effect of multiple mutations as follows:
| (3) |
Here ΔΔΔG(AB), ΔΔΔG(A) and ΔΔΔG(B) describe the effect of double and single mutations on activation respectively. Positive SA values correspond to the positive epistasis, namely, when the shift towards the active state upon introducing both mutations is considerably more than the sum of the effects of the single mutations. All data for single and double mutations produced in this study is available at ftp://ftp.ncbi.nih.gov/pub/panch/RTK/.
Predicting mutation hotspots
Mutation hotspot prediction in this study was based on a threshold (Sh) value for the number of mutations in a mutable site. The threshold and resulting hotspot sites were calculated for each mutation spectrum (mutation spectrum represents a distribution of a number of position with a given mutation frequency) separately using classification analysis described previously (Glazko, et al., 1998; Rogozin, et al., 2001). For this purpose the CLUSTERP program was used (ftp://ftp.bionet.nsc.ru/pub/biology/dbms/CLUSTERM.ZIP) which is based on the SEM subclass approach (Simulation, Expectation, Maximization). The algorithm tries to classify the mutation sites according to different mutation probabilities, and each site should belong to only one class. The mutation spectrum of each class is approximated by the Poisson distribution and an overall mutational spectrum is regarded as a mixture of Poisson distributions. Variations in mutation frequencies among sites of the same class are assumed to be due to random reasons (since mutation probability is the same for all sites in one class), but differences between mutation frequencies among sites from different classes are statistically significant. A class with the highest mutation frequency is called a hotspot class. The process which separates the mutation spectrum into classes is iterative and each iteration includes simulation, maximization and estimation procedures.
Analyzing doublet mutations
To study the properties of multiple mutation spectra, we used a sampling procedure repeated 1000 times. Each generated set of pseudo-multiple mutations had the size equal to the set of multiple mutations and mutations were randomly taken from the set of single mutations. The resulting set of generated pseudo-multiple mutations was compared to the observed set of multiple mutations. A Monte Carlo modification of the Pearson χ2 test of spectra homogeneity (Adams and Skopek, 1987) was used to compare distributions of multiple and pseudo-multiple mutations along the protein sequences. Calculations were done using the program COLLAPSE (Khromov-Borisov, et al., 1999). We used the above sampling procedure to estimate the expected number of doublets and the significance of their over- and under-representation. The number of doublets averaged over 1000 trials was used as an expected value. Statistical significance of over-represented doublets was estimated using p-value which referred to the probability to find the observed or larger number of a given doublet purely by chance (P(S≥O)). If p-value was less than 0.05, this pair was considered over-represented. The same logic was used for the analysis of under-represented doublets where the p-value P(O≥S) was calculated. This procedure was applied to pairs of mutations with non-zero expected values only. This severely limits the number of analyzed pairs, however increases the reliability of the analysis.
We also employed an analogy to the Hardy-Weinberg model (Crow, 1999; Hardy, 1908). The Hardy-Weinberg Principle is frequently used for allele and genotype frequencies: zygotic genotype frequencies are predictable from gamete frequencies, assuming random mating. In the case of double mutations we assume that frequencies of non-synonymous and synonymous substitutions are allele frequencies and those substitutions are randomly combined in doublets. We found that the observed number of “doublet heterozygosity” (doublets that contain one non-synonymous and one synonymous substitution) is significantly lower than expected under Hardy-Weinberg equilibrium (Supp. Table S2).
We used BioRuby (Goto, et al., 2010) and the Entrez Programming Utilities (Sayers, et al., 2011) to facilitate data manipulation and analyses throughout this study and Cytoscape (Shannon, et al., 2003) for the network visualization.
Results
The kinase and juxtamembrane domains have the largest density of cancer mutations
We obtained 9607 non-synonymous mutations observed in different cancer samples from the COSMIC database (Forbes, et al., 2008), including 1060 unique mutations from 841 unique mutation sites. Out of these 9607 mutations, zygosity information was available for 1534 mutations and 93% of them were heterozygous, namely contained the mutation in only one allele of the locus. The mutations were distributed over a wide range of human RTKs (Supp. Fig. S1) and EGFR and KIT (MIM# 164920), well known cancer causative genes, had more than 100 unique mutation sites. The majority of mutations and mutation sites (73%) were observed only in one cancer sample (patient), whereas about 5% of them were observed in ten or more samples (Supp. Fig. S2). The most frequently observed mutation in the dataset was the p.Leu858Arg mutation of EGFR, (observed in 2299 samples). These repeated mutations are more likely to be driver mutations in contrast to the mutations observed only in one sample, the large fraction of which might correspond to passenger mutations.
All mutations were classified depending on their oncogenic potential (which was estimated here as a frequency of samples where they were observed) into class “A” - for those observed in one sample, class “B” - for those observed in two to nine samples, and class “C” - for ten or more samples. This classification is consistent with our evolutionary analysis described below. We observed that the kinase domain and juxtamembrane region had the largest density of mutations out of all RTK regions, next came the extracellular domain, whereas mutations in the membrane domain and the C-terminal tail were relatively rare (Supp. Fig. S3). This result is consistent with the previous studies (Dixit, et al., 2009b; Izarzugaza, et al., 2009) although mutations were classified differently in the present study. Moreover, from Supp. Fig S3 one can see that the oncogenic potential varies greatly depending on the region of RTKs. For example, the percentage of mutation sites increased from 42% for class A to 57–59% for class B/C sites for the kinase domain and almost doubles (from 9% to 15–20%) in the case of the JM domain. In contrast, the number of mutation sites decreased with the oncogenic potential for the extracellular domain which points to many rare mutations in this region.
The effect of single mutations on stability and activity of kinase domain
Since the majority of mutations in class B and C are located in the kinase domain and their frequencies per residue are also high, we measured the effect of the mutations on the stability and activity of the kinase domain (in some cases the JM region was also included). We collected six pairs of structures of the same RTK kinase in the active and inactive states and structures from six additional RTK kinases available either in the active or inactive conformations (altogether seven crystal structures in the active state and eleven structures in the inactive state, Supp. Table S1). In total, 344 unique mutations were mapped onto active structures and 419 mutations were mapped onto inactive structures, of which 318 were mapped onto both of them.
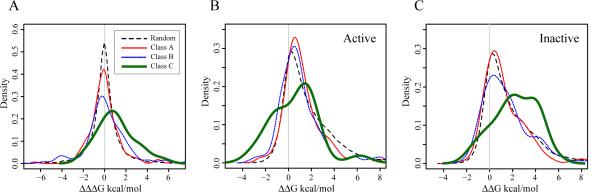
Figure 1A shows the distribution of ΔΔΔG values, which represents the difference in the unfolding free energies between the active and inactive states upon introducing the amino acid substitution corresponding to a cancer mutation (see Materials and Methods). Positive ΔΔΔG values correspond to the tendency for the activation by mutations when the equilibrium between active and inactive states is shifted towards active states. As can be seen from Figure 1A and Figure 2 the mean values of these distributions increase from −0.01 kcal/mol for class A, to +0.15 kcal/mol for class B, and +1.06 kcal/mol for class C cancer mutations. The distribution of class C is significantly shifted to the positive side (p = 0.001, one-sided Wilcoxon rank-sum test), compared to the background distribution (see Methods). Cancer mutations from the A and B classes do not show a significant effect on the stability of active or inactive states compared to random mutations (p = 0.71 and 0.25 respectively). This indicates that the frequently observed mutations have a different effect (the mean value of ΔΔΔG would be close to zero if the energetic effects of mutations on the active and inactive conformations were comparable) on the active and inactive states leading to the shift of equilibrium from inactive to active states.
Figure 1. Effect of single mutations on stability of the kinase domain.
(A) The distribution of ΔΔΔG for unique mutations of class A is shown in red/thin line (187 mutations), class B in blue/stippled line (110 mutations), class C in green/bold line (21 mutations), and all possible random mutations caused by single nucleotide substitutions are shown in black dashed line. (B) The distribution of ΔΔG of active states. (C) The distribution of ΔΔG of inactive states. The probability density functions were smoothed using the Gaussian kernel.
Figure 2. The relationship between oncogenic and activation potentials.

Mean values and standard errors of ΔΔΔG plotted for each class of unique mutations.
We analyzed separately the energetic effect of cancer mutations on the active and inactive conformations. Figure 1B and 1C shows distributions of ΔΔG values for the active and inactive states respectively. All three distributions for active states are shifted to the positive values indicating that cancer mutations destabilize active structures however destabilize them significantly less than the random mutations (p = 0.033 for A, 0.0006 for B, and 0.017 for C classes respectively). As to the inactive structures, unlike cancer mutations from the A and B classes, only mutations from class C destabilize inactive structures significantly compared to random mutations (p = 0.025) (Fig. 1C). In short, mutations with high oncogenic potential have a tendency to destabilize inactive states to a greater degree than active states which may cause aberrant activations of RTKs in cancer cells (Figure 2). This effect is more pronounced for the KIT family of RTKs.
Assuming that homo or heterozygosity of RTK mutations should not affect the activity since mutant oncogene alleles are typically dominant, we indeed did not find any difference in ΔΔΔG distributions between heterozygous and homozygous mutations even though the most frequent mutations such as p.Leu858Arg in EGFR and p.Asp816Val or p.Leu576Pro in KIT were sometimes observed as homozygous.
Analysis of single mutation spectra
In this work, we analyzed two mutation spectra of the EGFR and KIT genes. The numbers of single mutations in EGFR and KIT were 2680 and 2034, respectively. Analysis of the mutation spectrum of the EGFR gene using CLUSTERP revealed four classes of sites (see Methods). The first class includes obvious “cold” sites with mutation frequencies (number of samples the same mutation was observed) less than or equal to 4, the second class includes sites with the mutation frequency less than 10, the third class - from 10 to 26, and the fourth class comprises obvious hotspot sites (mutation frequency > 49). The distribution of observed and expected mutation frequencies is shown in Supp. Fig. S4A. The second class does not contain hotspot sites since numerous sites with no mutations or just one mutation were also included in this class, while several obvious hotspots were present in the third class of sites. Thus ten mutations was chosen as the threshold value (Sh) for determining the mutation hotspot sites. Similar results were obtained for the KIT gene with 2034 mutations (Supp. Fig. S4B), analysis of four predicted classes of sites suggested the same threshold of ten mutations which is consistent with our class C derived in the previous section based on the analysis of oncogenic and activation potentials.
The great majority of EGFR and KIT mutations (>95%) occurred in non-synonymous codon sites and, accordingly, resulted in amino acid replacements in EGFR and KIT genes. The excess of non-synonymous over synonymous substitutions was statistically significant (P < 10−70 by the Fisher exact test) when compared to the random expectation under the neutral evolution model. Namely, using the modified Nei-Gojobori method, we estimated that ~73% of the substitutions were expected to occur in non-synonymous sites in the neutral regime. Preponderance of amino-acid replacement over silent substitutions is the signature of positive (directional) selection (Hurst, 2002). The large excess of non-synonymous substitutions seen in EGFR and KIT is similar to TP53 (MIM# 191170) and some other cancer-related genes (Glazko, et al., 2006) and is an unequivocal indication that, in tumors, the EGFR and KIT genes evolve under positive selection and preferentially accumulate mutations that lead to selective advantages for the tumor cells.
The effect of multiple mutations on stability and activity of kinase domain
We found 210 unique multiple mutations from 17 different RTKs, and about 73% of them were from the EGFR or KIT families (Supp. Fig. S5). The most frequent multiple mutation was the combination of p.Thr790Met and p.Leu858Arg in the EGFR protein, which was observed in 48 samples. We mapped 97 multiple mutations that consisted of 92 double mutations and 5 triple mutations on structures in active and inactive states (so-called “doublets”). Figure 3A shows distributions of ΔΔΔG values for multiple mutations and a background distribution of a randomly chosen 1000 pairs of mutations. The distribution corresponding to cancer mutations is significantly shifted to positive values compared to random doublet mutations (P << 0.01, one-sided Wilcoxon rank-sum test) pointing to a tendency for activation for doublets. Interestingly, the distribution of ΔΔG for active kinase states is significantly shifted to negative values (P << 0.01), compared to the background distribution implying the stabilization of the active state by double cancer mutations compared to random mutations (Fig. 3B) while the inactive state does not seem to be significantly destabilized by doublets (P = 0.33) (Fig. 3C).
Figure 3. Effect of multiple mutations on stability of the kinase domain.
(A) The distribution of ΔΔΔG for observed multiple mutations in red/solid line and random mutations in black dashed line. (B) The distribution of ΔΔG of active states. (C) The distribution of ΔΔG of inactive states. The probability density functions were smoothed using the Gaussian kernel.
Moreover, we found a positive epistasis for double mutations (Fig. 4). Indeed, the effect of multiple mutations on shifting the population of kinase conformations toward active ones is higher than a total of individual mutations (P << 0.01, one-sided Wilcoxon rank-sum test), suggesting that, overall, double mutations have a synergistic effect. This trend is especially pronounced for double mutations observed in more than one sample (Fischer exact test P = 0.02). In addition, we do not observe any correlation between ΔΔΔG values and the spatial distances between two mutations in the structures (Supp. Fig. S6), indicating that synergistic multiple mutations are not necessarily close to each other in space providing for their direct interactions.
Figure 4. Positive epistasis of double mutations.

Distribution of SA (difference between ΔΔΔG (multiple mutations) and ΔΔΔG (sum of corresponding mutations).
To understand the mechanism of multiple mutations we focused on doublets found specifically in EGFR and KIT. We constructed a mutation network, where nodes represented mutation sites and edges corresponded to double mutations. The network shows that L858 of EGFR is clearly the biggest hub, which connects to more than 20 nodes/mutation sites in doublets (Fig. 5) although this hub is even smaller than expected based on the single mutation frequencies (see next section). EGFR also has some hot spots that connect to more than five nodes, such as G719, S768 and L861, and all of them are in the kinase domain. On the other hand, KIT has many more concurrent mutations between residues in the kinase domain and the JM domain, showing many cliques and connections between multiple sites. This might reflect the difference between their regulation mechanisms. At the same time there are seven sites aligned in both KIT and EGFR that are involved in double mutations (shown in magenta in Figure 5).
Figure 5. The mutation network of EGFR and KIT for different RTK regions.
Each node represents one mutation site where at least one mutation can occur. The mutation sites of KIT are shown in blue/grey, EGFR in green/black, and aligned sites are shown in maroon/white. Residue numbers are given inside the nodes. The edge connects two sites that are concurrently mutated. Those edges with positive epistasis are shown in bold.
Analysis of doublet mutation spectra
The numbers of doublet mutations in EGFR and KIT were 207 and 76 pairs, respectively. Similar to the single mutations, the great majority of multiple mutations (>88%) occurred in non-synonymous codon sites implying that doublets are under positive selection. To compare the properties of multiple mutation spectra to single mutations, we used a sampling procedure (see Methods). The observed and simulated spectra of double mutations (generated from the spectra of single mutations) were significantly different (P << 0.01 for EGFR and KIT) which points to the different mechanisms underlying single and double mutation spectra. The major difference between observed and simulated mutation spectra for the EGFR gene came from the significantly smaller number of multiple mutations in the major hotspot position 858 (270 observed mutations versus 373 expected mutations, P << 0.01 by the Fisher exact test). Several positions with elevated frequency of multiple mutations, for example, the positions 709 (15 mutations), 719 (42 mutations) and 768 (22 mutations) in the EGFR gene were also observed. We listed doublets with a significant over- and under-representation in Table 1.
Table 1.
Significantly over- and under-represented double mutations in EGFR and KIT
| Protein | Mutation1 | Mutation2 | Observed | Expected | p-value |
|---|---|---|---|---|---|
| Over-represented pairs | |||||
|
| |||||
| EGFR | p.Leu833Val | p.Leu858Arg | 11 | 0.38 | 0.001 |
| EGFR | p.Gly719Cys | p.Ser768Ile | 6 | 0.17 | 0.005 |
| EGFR | p.Glu709Gly | p.Leu858Arg | 6 | 0.90 | 0.009 |
| EGFR | p.Thr790Met | p.Leu858Arg | 47 | 28.74 | 0.022 |
| KIT | p.Leu576Pro | p.Val654Ala | 4 | 0.18 | 0.031 |
|
| |||||
| Under-represented pairs | |||||
|
| |||||
| EGFR | p.Gly719Ser | p.Leu858Arg | 3 | 21.75 | 0.004 |
| EGFR | p.Leu858Arg | p.Leu861Val | 1 | 16.1 | 0.002 |
| KIT | p.Val560Asp | p.Asp816Val | 1 | 6.6 | 0.024 |
All mutation numbering was obtained from the COSMIC database. All mutations were mapped to a single version of each gene sequence, and are available in the Download section of the COSMIC database.
We also compared the frequency of double mutations in EGFR and KIT to the TP53 gene (Meng, et al., 1999) and found that the frequency of non-synonymous doublets was significantly higher in the EGFR and KIT genes compared to TP53 (Supp. Table S2). This result strongly suggests that multiple mutations in EGFR and KIT are driven by positive selection. Multiple mutations in TP53 are likely to be the result of transient mutagenesis (Drake, et al., 2005) and are unlikely to be under positive selection (Rodin and Rodin, 1998) and thus can be used as a null model (Drake, et al., 2005; Rodin and Rodin, 1998). Additional analysis using the Hardy-Weinberg Principle (see Methods) also suggests that strong positive selection is an important driving force for doublets. Under this model, we compared the observed level of “doublet heterozygosity” (doublets that contain one non-synonymous and one synonymous substitution) to what we expect under Hardy-Weinberg equilibrium. The observed “doublet heterozygosity” (2pq=51) is significantly lower than expected (2pq=82) (Supp. Table S2), this discrepancy is usually attributed to preferential fixation of double non-synonymous mutations due to positive selection for EGFR and KIT.
Interestingly, quite a few RTK mutations and mutation sites in our set are found only as a part of doublet mutations, namely 49% of mutations and 30% of mutation sites are observed only in doublets but not in singletons. For example, the p.Glu709Gly mutations in EGFR, observed in seven different patients, always appear with another mutation rather than as a single mutation. These mutations are likely to be secondary mutations.
Discussion
It has been previously shown that cancer mutations may activate RTKs, sometimes in a ligand-independent way, and the mechanisms of kinase activation in cancer might be linked to transitions between the active and inactive states (Yun, et al., 2007; Zhang, et al., 2006). The crystal structures of the EGFR p.Leu858Arg mutant showed, for example, that this mutation prevents the activation loop from adopting the inactive, conformation (Yun, et al., 2007). It was suggested that the effect of the secondary EGFR p.Thr790Met mutation facilitates interconversion between the inactive and active conformations and enhances the stability of the active conformation relative to the inactive one (Yun, et al., 2008). More recently, the modeling of autoinhibited conformations and the effect of several frequent hotspot mutations revealed enhanced mobility near mutation sites which disrupted the local stabilizing interactions and in some cases allosterically altered the distribution of locally frustrated sites and destabilized the inactive form (Dixit, et al., 2009a; Dixit and Verkhivker, 2011; Dixit, et al., 2009b). It was shown that EGFR p.Thr790Met and EGFR p.Leu858Arg mutations also lead to the enhanced stability of the active state (Dixit and Verkhivker, 2009).
In this study we analyzed both the active and inactive states of the kinase domain from structures of different RTKs and estimated the effect of single and multiple cancer mutations on their stability. In accordance with the previous studies, using much larger dataset of mutations, we showed that, overall, single cancer mutations destabilized inactive states. We also showed that single cancer mutations destabilized active states of RTK, but to a lesser degree than the random mutations; moreover, to a lesser degree than the single cancer mutations destabilized inactive states. This led to kinase activation. The exception was the EGFR p.Thr790Met mutation which did not have a considerable overall effect (ΔΔΔG = −0.5 kcal/mol) alone and exhibited an activating effect as shown later as a double mutation together with EGFR p.Leu858Arg (see next section). Although destabilizing effect on both active and inactive states might potentially lead to misfolding and aggregation, the changes in unfolding free energy of about 1–3 kcal/mol might not compromise an overall fitness of a protein (Tokuriki and Tawfik, 2009). The homozygosity state of the mutation did not affect the obtained results, consistent with the premises that mutant oncogene alleles were typically dominant.
Moreover, we tried to answer the question if one can predict the activating effect of single or multiple mutations from their mutation spectra. Importantly we showed that there exists a relationship between the statistics-based estimate of oncogenic potential of mutation and its activation effect calculated based on thermodynamics principles. Namely, more frequent mutations have a somewhat higher activating effect. This effect is not linear, though, and for frequent mutations from more than ten samples the activity increases radically upon introducing mutations. From the Boltzmann distribution one can estimate the fraction of proteins with a given stability or stability effect of mutations (ΔΔG) which would follow a simple logistic function behavior. Indeed, previously the sigmoidal relationship was revealed between the destabilizing effects of mutations leading to monogenic diseases and the severity of the diseases (Yue, et al., 2005). In addition, recently an association was shown between the frequency of mutations and their potential functional impact calculated based on their evolutionary conservation (Reva, et al., 2011). Our analysis of thermodynamic properties is nicely complemented by the evolutionary analysis which uncovered four populations of mutations with different underlying mutation rates and we showed that mutation hotspots (potential driver mutations) can be reliably defined from the samples with frequencies higher than ten. It should be mentioned that although we analyzed local changes produced by cancer mutations in several RTK families differing by regulatory mechanisms we observed the common activating effect of cancer mutations in relation to its oncogenic potential. Thus our analysis complements previous observations. It implies that although increased RTK activity may be caused by gene amplifications, enhanced transcription or translation, both single and double point mutations might play a key role in kinase activation in cancer.
Moreover, a significant fraction of cancer-associated mutations comes in doublets or triplets. The origin of multiple mutations is still not very well understood, they can originate either through defects in DNA replication/repair systems or arise locally through transient mechanisms (Chen, et al., 2009; Chen, et al., 2011; Matsuda, et al., 2001; Pavlov, et al., 2006; Seidman, et al., 1987; Stone, et al., 2009). We found that 7.7% and 3.7% of all single cancer mutations of EGFR and KIT respectively represent double cancer mutations, compared to the previously reported 6% of double mutations in EGFR in lung cancers (Chen, et al., 2008). These numbers are substantially larger than the fraction of multiple somatic mutations in the lacI transgene in mouse somatic tissues (~1%) (Hill, et al., 2004). Furthermore, the fraction of multiple mutations in neighboring positions (tandem mutations) of the lacI gene was found to be higher compared to multiple mutations separated by one or more nucleotides (Hill, et al., 2004) whereas the opposite tendency was found in the EGFR gene where tandem mutations were rare (results not shown). In general, an excess of tandem mutations is a signature of various error-prone DNA polymerases and is expected to have a distinct DNA context specificity (Matsuda, et al., 2001; Pavlov, et al., 2006; Stone, et al., 2009). This is consistent with the distinct context properties of tandem lacI mutations in mouse somatic tissues and a lack of significant differences for spectra of single and non-tandem multiple lacI mutations (Buettner, et al., 2000; Hill, et al., 2004). However the mutation spectrum of multiple mutations in our study was found to be different from the spectrum of singletons which hints at different underlying mechanisms of their origin and suggests a role of clonal selection for multiple substitutions (Bazykin, et al., 2004). Although we did not find any evidence for prevalence of double substitutions to contact each other in three-dimensional protein structures of kinase monomers, such contacts may occur between different subunits of homodimers since dimerization is crucial for RTK functions in a cell. Further analysis is needed to study the effect of mutations on stability and function of protein complexes since many disease related mutations may disrupt protein interactions (Schuster-Bockler and Bateman, 2008; Teng, et al., 2009).
Similar to single mutations, double mutations were shown to activate the kinase domain. Moreover for many doublets a positive epistasis has been found which points to the additional advantage of doublets for the tumor cell compared to singletons. Some doublets have a drug resistance effect, one of the classical examples is the p.Thr790Met+p.Leu858Arg double mutant of EGFR showing strong resistance to gefitinib (Tam, et al., 2009). According to our study this double mutation is predicted to have a potential activating effect (ΔΔΔG = +1.9 kcal/mol) and quite high oncogenic potential (observed in 47 samples). Moreover, it exhibits a positive epistasis effect (SA = +1.14 kcal/mol). Some other mutations with high oncogenic potential and positive epistasis include p.Leu833Val + p.Leu858Arg (ΔΔΔG = +3.85 kcal/mol; SA = +1.4 kcal/mol) and p.Leu858Arg + p.Glu884Lys (ΔΔΔG = +4.57 kcal/mol and SA = +1.07 kcal/mol) of EGFR. The latter E884K mutation disrupts an ion pair between K884 and R958 and may change the downstream signaling significantly although its connection to the p.Leu858Arg mutation remains unclear (Tang, et al., 2009). In addition we observed many doublets which were not found as single mutations (about half of all doublets). One might hypothesize that such mutations are secondary mutations causing the previously mentioned differences between the single and double mutation spectra.
Analyses of mutation spectra and activating effects of different cancer mutations have important prognostic implications. Namely, it has been shown that some RTK activating mutants have been associated with a better survival prognosis and better response to RTK inhibitors (Jackman, et al., 2009). Indeed, cancer cells exhibiting mutant kinases become critically dependent on certain pathways (so called “oncogene addiction” (Weinstein, 2002)). For example, mutant EGFRs selectively activates Akt and signal transduction/activator of transcription (STAT) signaling pathways, which in turn promote cell survival. It explains the effectiveness of gefitinib inhibiting critical anti-apoptotic pathways in lung cancers with mutant EGFR genotypes (Sordella, et al., 2004). In our study we attempted to link the stability of RTKs with their oncogenic potential and differential activity which combined with other data on phosphorylation patterns for each mutant may provide insight into the mechanisms of activation of different pathways by cancer mutations and may help to design effective cancer drugs.
Supplementary Material
Acknowledgements
We thank Jeyanthy Eswaran, Joshua Cherry and Tom Madej for very helpful discussions. This work was supported by the Intramural Research Program of the National Library of Medicine at the U.S. National Institutes of Health. K.H. was supported by a JSPS Research Fellowship from the Japan Society for the Promotion of Science.
References
- Adams WT, Skopek TR. Statistical test for the comparison of samples from mutational spectra. J Mol Biol. 1987;194(3):391–6. doi: 10.1016/0022-2836(87)90669-3. [DOI] [PubMed] [Google Scholar]
- Bazykin GA, Kondrashov FA, Ogurtsov AY, Sunyaev S, Kondrashov AS. Positive selection at sites of multiple amino acid replacements since rat-mouse divergence. Nature. 2004;429(6991):558–62. doi: 10.1038/nature02601. [DOI] [PubMed] [Google Scholar]
- Buettner VL, Hill KA, Scaringe WA, Sommer SS. Evidence that proximal multiple mutations in Big Blue transgenic mice are dependent events. Mutat Res. 2000;452(2):219–29. doi: 10.1016/s0027-5107(00)00090-7. [DOI] [PubMed] [Google Scholar]
- Chen JM, Ferec C, Cooper DN. Closely spaced multiple mutations as potential signatures of transient hypermutability in human genes. Hum Mutat. 2009;30(10):1435–48. doi: 10.1002/humu.21088. [DOI] [PubMed] [Google Scholar]
- Chen JM, Ferec C, Cooper DN. Transient hypermutability, chromothripsis and replication-based mechanisms in the generation of concurrent clustered mutations. Mutat Res. 2011 doi: 10.1016/j.mrrev.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Chen Z, Feng J, Buzin CH, Sommer SS. Epidemiology of doublet/multiplet mutations in lung cancers: evidence that a subset arises by chronocoordinate events. PLoS One. 2008;3(11):e3714. doi: 10.1371/journal.pone.0003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF. Hardy, Weinberg and language impediments. Genetics. 1999;152(3):821–5. doi: 10.1093/genetics/152.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363(6430):595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- Dixit A, Torkamani A, Schork NJ, Verkhivker G. Computational modeling of structurally conserved cancer mutations in the RET and MET kinases: the impact on protein structure, dynamics, and stability. Biophys J. 2009a;96(3):858–74. doi: 10.1016/j.bpj.2008.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Verkhivker GM. Hierarchical modeling of activation mechanisms in the ABL and EGFR kinase domains: thermodynamic and mechanistic catalysts of kinase activation by cancer mutations. PLoS Comput Biol. 2009;5(8):e1000487. doi: 10.1371/journal.pcbi.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Verkhivker GM. The energy landscape analysis of cancer mutations in protein kinases. PLoS One. 2011;6(10):e26071. doi: 10.1371/journal.pone.0026071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Yi L, Gowthaman R, Torkamani A, Schork NJ, Verkhivker GM. Sequence and structure signatures of cancer mutation hotspots in protein kinases. PLoS One. 2009b;4(10):e7485. doi: 10.1371/journal.pone.0007485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Bebenek A, Kissling GE, Peddada S. Clusters of mutations from transient hypermutability. Proc Natl Acad Sci U S A. 2005;102(36):12849–54. doi: 10.1073/pnas.0503009102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, Zhao F, Vincent PW, Naumov GN, Bradner JE, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res. 2007;67(24):11924–32. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Knapp S. Insights into protein kinase regulation and inhibition by large scale structural comparison. Biochim Biophys Acta. 2010;1804(3):429–32. doi: 10.1016/j.bbapap.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;Chapter 10(Unit 10):11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko GB, Milanesi L, Rogozin IB. The subclass approach for mutational spectrum analysis: application of the SEM algorithm. J Theor Biol. 1998;192(4):475–87. doi: 10.1006/jtbi.1998.0668. [DOI] [PubMed] [Google Scholar]
- Glazko GV, Babenko VN, Koonin EV, Rogozin IB. Mutational hotspots in the TP53 gene and, possibly, other tumor suppressors evolve by positive selection. Biol Direct. 2006;1:4. doi: 10.1186/1745-6150-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto N, Prins P, Nakao M, Bonnal R, Aerts J, Katayama T. BioRuby: bioinformatics software for the Ruby programming language. Bioinformatics. 2010;26(20):2617–9. doi: 10.1093/bioinformatics/btq475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, Bulmer SE, Frank DA, Hahn WC, Sellers WR, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2(11):e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320(2):369–87. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Hardy GH. Mendelian Proportions in a Mixed Population. Science. 1908;28(706):49–50. doi: 10.1126/science.28.706.49. [DOI] [PubMed] [Google Scholar]
- Hill KA, Wang J, Farwell KD, Scaringe WA, Sommer SS. Spontaneous multiple mutations show both proximal spacing consistent with chronocoordinate events and alterations with p53-deficiency. Mutat Res. 2004;554(1-2):223–40. doi: 10.1016/j.mrfmmm.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Hubbard SR, Miller WT. Receptor tyrosine kinases: mechanisms of activation and signaling. Curr Opin Cell Biol. 2007;19(2):117–23. doi: 10.1016/j.ceb.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18(9):486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Izarzugaza JM, Redfern OC, Orengo CA, Valencia A. Cancer-associated mutations are preferentially distributed in protein kinase functional sites. Proteins. 2009;77(4):892–903. doi: 10.1002/prot.22512. [DOI] [PubMed] [Google Scholar]
- Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Janne PA, Riely GJ, Ruiz MG, Giaccone G, Sequist LV, Johnson BE. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15(16):5267–73. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376(6538):313–20. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42(1):9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Zhang Y, Waugh A, Haverty PM, Peters B, Sebisanovic D, Stinson J, Forrest WF, Bazan JF, Seshagiri S, et al. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 2007;67(2):465–73. doi: 10.1158/0008-5472.CAN-06-1736. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–98. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Khan S, Vihinen M. Performance of protein stability predictors. Hum Mutat. 2010;31(6):675–84. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- Khromov-Borisov NN, Rogozin IB, Pegas Henriques JA, de Serres FJ. Similarity pattern analysis in mutational distributions. Mutat Res. 1999;430(1):55–74. doi: 10.1016/s0027-5107(99)00148-7. [DOI] [PubMed] [Google Scholar]
- Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A. 2006;103(47):17783–8. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GS. The road to Src. Oncogene. 2004;23(48):7910–7. doi: 10.1038/sj.onc.1208077. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312(2):335–46. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- Meng L, Lin L, Zhang H, Nassiri M, Morales AR, Nadji M. Multiple mutations of the p53 gene in human mammary carcinoma. Mutat Res. 1999;435(3):263–9. doi: 10.1016/s0921-8777(99)00053-1. [DOI] [PubMed] [Google Scholar]
- Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385(6617):650–3. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV, Rogozin IB. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- Potapov V, Cohen M, Schreiber G. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Eng Des Sel. 2009;22(9):553–60. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin SN, Rodin AS. Strand asymmetry of CpG transitions as indicator of G1 phase-dependent origin of multiple tumorigenic p53 mutations in stem cells. Proc Natl Acad Sci U S A. 1998;95(20):11927–32. doi: 10.1073/pnas.95.20.11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin I, Kondrashov F, Glazko G. Use of mutation spectra analysis software. Hum Mutat. 2001;17(2):83–102. doi: 10.1002/1098-1004(200102)17:2<83::AID-HUMU1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39(Database issue):D38–51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289(5486):1938–42. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Schuster-Bockler B, Bateman A. Protein interactions in human genetic diseases. Genome Biol. 2008;9(1):R9. doi: 10.1186/gb-2008-9-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman MM, Bredberg A, Seetharam S, Kraemer KH. Multiple point mutations in a shuttle vector propagated in human cells: evidence for an error-prone DNA polymerase activity. Proc Natl Acad Sci U S A. 1987;84(14):4944–8. doi: 10.1073/pnas.84.14.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305(5687):1163–7. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- Stone JE, Kissling GE, Lujan SA, Rogozin IB, Stith CM, Burgers PM, Kunkel TA. Low-fidelity DNA synthesis by the L979F mutator derivative of Saccharomyces cerevisiae DNA polymerase zeta. Nucleic Acids Res. 2009;37(11):3774–87. doi: 10.1093/nar/gkp238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam IY, Leung EL, Tin VP, Chua DT, Sihoe AD, Cheng LC, Chung LP, Wong MP. Double EGFR mutants containing rare EGFR mutant types show reduced in vitro response to gefitinib compared with common activating missense mutations. Mol Cancer Ther. 2009;8(8):2142–51. doi: 10.1158/1535-7163.MCT-08-1219. [DOI] [PubMed] [Google Scholar]
- Tang Z, Jiang S, Du R, Petri ET, El-Telbany A, Chan PS, Kijima T, Dietrich S, Matsui K, Kobayashi M, et al. Disruption of the EGFR E884-R958 ion pair conserved in the human kinome differentially alters signaling and inhibitor sensitivity. Oncogene. 2009;28(4):518–33. doi: 10.1038/onc.2008.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Madej T, Panchenko A, Alexov E. Modeling effects of human single nucleotide polymorphisms on protein-protein interactions. Biophys J. 2009;96(6):2178–88. doi: 10.1016/j.bpj.2008.12.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19(5):596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Verkhivker G, Schork NJ. Cancer driver mutations in protein kinase genes. Cancer Lett. 2009;281(2):117–27. doi: 10.1016/j.canlet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9(4):138–41. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297(5578):63–4. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68(9):3077–80. doi: 10.1158/0008-5472.CAN-07-3293. discussion 3080. [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353(2):459–73. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11(3):217–27. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105(6):2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–49. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Norris J, Schwartz C, Alexov E. In silico and in vitro investigations of the mutability of disease-causing missense mutation sites in spermine synthase. PLoS One. 2011;6(5):e20373. doi: 10.1371/journal.pone.0020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Teng S, Wang L, Schwartz CE, Alexov E. Computational analysis of missense mutations causing Snyder-Robinson syndrome. Hum Mutat. 2010;31(9):1043–9. doi: 10.1002/humu.21310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang L, Gao Y, Zhang J, Zhenirovskyy M, Alexov E. Predicting folding free energy changes upon single point mutations. Bioinformatics. 2012;28(5):664–71. doi: 10.1093/bioinformatics/bts005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.