The recent discovery of bioorthogonal click chemistry has created a new field in chemical biology in which biomolecules that are not directly encoded in the genome can be monitored in living systems.[1] By hijacking a cell's biosynthetic machinery, a metabolic precursor functionalized with a bioorthogonal chemical tag is incorporated into target biomolecules, including glycans,[2] lipids,[3] proteins,[4] and nucleic acids.[5] Subsequently, a tailor-designed click reaction is employed to conjugate a complementary biophysical probe, which enables visualization[2] or enrichment of the target biomolecules for molecular identification.[6,7] Though both applications require exquisite selectivity to maximize signal to noise ratio, each has specific criteria to meet. For dynamic imaging studies, not only must the employed reaction proceed rapidly at physiological conditions to allow monitoring of events taking place on the minute time scale, it must also be nontoxic and non-interfering with the surrounding cellular milieu.[2,8] Contrarily, molecular identification applications, e.g. proteomics analysis, prioritize sensitivity over biocompatibility — in most cases only limited amounts of samples are available for analysis and the targets of interest may be low in abundance. Thus, reactions that enjoy fast kinetics at low substrate concentration (e.g. micromolar) are preferred. In view of these requirements, the choice of a tailored chemical tag and chemistry for a specific bioconjugation process is not trivial.
To date, azide is the most utilized bioorthogonal chemical tag for biomolecule-conjugate experiments due to its small size and inertness to most components in a biological environment.[9] Three bioorthogonal click reactions have been reported for labeling azide-tagged biomolecules. The Staudinger ligation, introduced by Bertozzi in 2000, covalently links the azide and an ester-functionalized triphenylphosphine via an amide bond.[10] Though highly specific for the azide, this reaction suffers from slow reaction kinetics and competing oxidation of the phosphine reagents.[11] By contrast, the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC), promoted by the Cu(I)-stabilizing ligand BTTES (Scheme 1),[12] and the strain-promoted azide-alkyne cycloaddition,[13] inherit the bio-benign characteristics of the Staudinger ligation but are further endowed with improved kinetics. With the discovery of BTTES, not only did we confer the canonical CuAAC[14,15] with biocompatibility, but also dramatically boost its reactivity. Similarly, by increasing the strain energy of cyclooctyne probes, Bertozzi and coworkers developed a biarylazacyclooctynone (BARAC) reagent, which showed significantly enhanced kinetics in live cell labeling experiments compared to the unfunctionalized cyclooctyne probes.[16] Though both reactions show great promise for molecular identification or imaging studies, no direct comparison of the bio-benign CuAAC and the BARAC-mediated cycloaddition, often referred to as copper-free click chemistry, has been performed to provide a framework for choosing the optimal reaction for these specific applications. Here, we compared these two bioorthogonal reactions in four applications: detecting purified recombinant glycoproteins and glycoproteins in crude cell lysates, and labeling of glycans on the surface of live cells and zebrafish embryos.
Scheme 1.
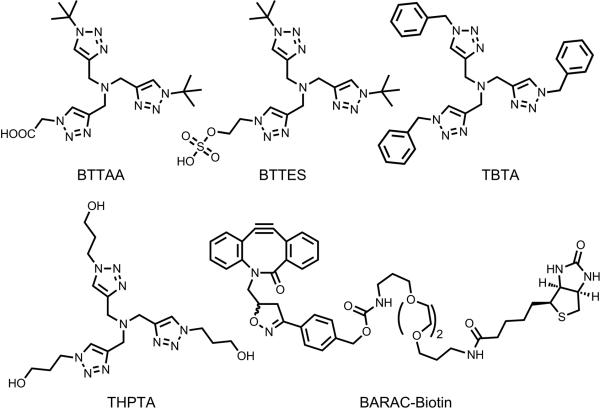
Structural formulae of tris(triazolylmethyl)amine-based ligands and BARAC-biotin. BTTAA=2-[4-({bis[(1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl]amino}methyl)-1H-1,2,3-triazol-1-yl]acetic acid, BTTES=2-[4-({bis[(1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl]amino}methyl)-1H-1,2,3-triazol-1-yl]ethyl hydrogen sulfate, TBTA=tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine, THPTA=tris[(1-hydroxypropyl-1H-1,2,3-triazol-4-yl)methyl]amine, BARAC=biarylazacyclooctynone
The canonical CuAAC protocol using TBTA and THPTA (Scheme 1) as Cu(I)-stabilizing ligands is associated with slow kinetics.[12,17] In our recent studies, we discovered that BTTES, a tris(triazolylmethyl)amine-based ligand for Cu(I), promoted the cycloaddition reaction rapidly in living systems.[12] BTTES contains two tert-butyl groups and a hydrogen sulfate, conferring the ligand an ideal balance between reactivity and solubility. When coordinating with the in situ generated Cu(I), the bulky tert-butyl groups are believed to prevent the polymerization of copper acetylides and thereby formation of unreactive species.[18] In the meantime, the hydrogen sulfate functionality that ionizes to a negatively charged sulfate at physiological pH secures the solubility of the BTTES-Cu(I) complex in aqueous mixtures and prevents cellular internalization of the coordinated copper. We found 50–75 μM of Cu(I) was sufficient to achieve robust labeling in mammalian cells and zebrafish embryos. However, minor developmental defects were observed when zebrafish embryos were treated under these conditions; ~10% embryos exhibited impaired posterior body development characterized by a shorter anterior-posterior axis. To improve labeling efficiency and biocompatibility of the CuAAC, a faster and less toxic catalyst is required.
We began our current study with ligand optimization by varying the ionizable substituent on the tris(triazolylmethyl)amine while keeping the two tert-butyl groups. We replaced the ethyl hydrogen sulfate of BTTES with an acetic acid group, producing a new ligand that we termed BTTAA (Scheme 1, see Supporting Information for synthetic details). At physiological pH, the acetic acid of BTTAA ionizes into acetate. The acetate functionality not only bears a negative charge, but it may also serve as an additional weak donor to coordinate with Cu(I), thus increasing the electron density of the metal center and facilitating the formation of the strained copper metallacycle and copper triazolide intermediate.[18b] Consequently, additional acceleration of the cycloaddition rate may be achieved.
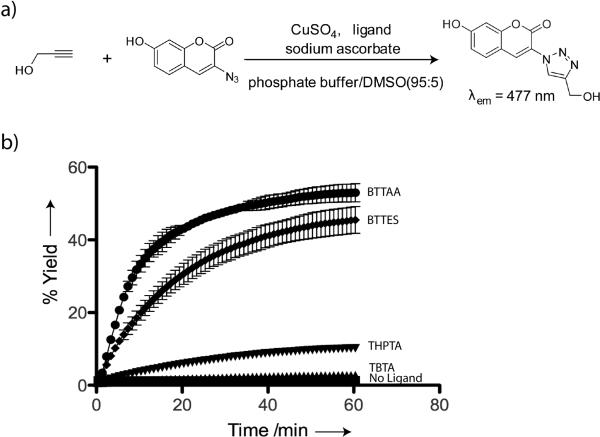
The relative reactivity of the canonical Cu(I) catalysts in the form of TBTA-Cu(I)[17] and THPTA-Cu(I)[19] and the new Cu(I) catalysts in the form of BTTES-Cu(I) and BTTAA-Cu(I) complexes was determined in a fluorogenic assay by reacting propargyl alcohol with 3-azido-7-hydroxycoumarin (Scheme 2a).[20] The azidocoumarin has very weak fluorescence that is quenched by the lone pair electrons from the internal nitrogen of the azide. The cycloaddition localizes the lone pair to the triazole ring, thus activating its fluorescence. Among the four ligands evaluated, BTTAA showed the highest activity in accelerating the CuAAC, followed by BTTES and THPTA, with TBTA showing the lowest activity. Greater than 45% cycloaddition product was formed using 50 μM Cu(I) within the first 30 min when the ligand-Cu(I) ratio was 6:1. By contrast, the THPTA- and TBTA-mediated reactions were significantly slower, yielding less than 15% cycloaddition products (Scheme 2b).
Scheme 2.
a) A fluorogenic assay for qualitative measurement of CuAAC kinetics. b) Conversion-time profiles of CuAAC in the presence of various ligands. Error bars represent the standard deviation of two replicate experiments.
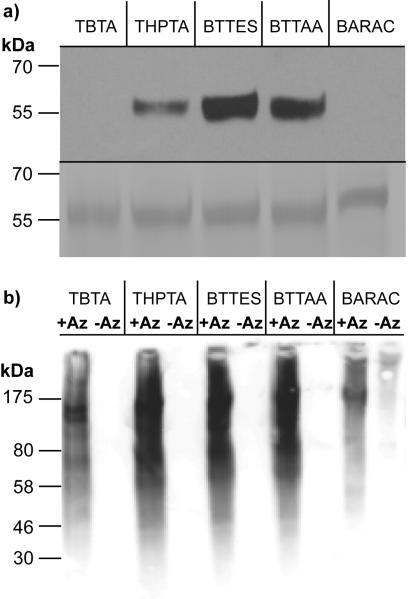
To evaluate the activity of the new ligand-Cu(I) complex and compare the click reactions in the context of biomolecular labeling experiments, the first system we chose to explore was a recombinant glycoprotein — Programmed Death 1-Immunoglobulin G Fc fusion (PD1-Fc). When expressed in mammalian cells, the Fc region of the recombinant protein is glycosylated and terminated with sialic acids. We cultured HEK-293 cells stably expressing PD1-Fc fusion protein in medium containing 50 μM sialic acid metabolic precursor — peracetylated N-azidoacetylmannosamine (Ac4ManNAz) for 4 days. The target protein was then isolated using protein G (a genetically engineered protein that contains Fc binding domains of protein G) agarose. To probe for the presence of the sialic acid-associated azide, the protein was reacted with biotin-alkyne via the CuAAC or BARAC-biotin via the copper-free click chemistry. For the CuAAC, 100 μM of biotin-alkyne was used as the coupling partner and the ratio of biotin-alkyne, ligand, CuSO4 and sodium ascorbate was held at 1:5:2.5:25, a labeling condition optimized in our lab. The copper-free click chemistry was performed with 100 μM of BARAC-biotin. The reactions were allowed to proceed for one hour and the labeled protein was analyzed by SDS-PAGE and Western blot. As quantified by ImageJ, reactions mediated by BTTES-Cu(I) and BTTAA-Cu(I) provided 2.6- and 2.1-fold stronger signal than the signal afforded by THPTA-Cu(I) for the fusion protein isolated from Ac4ManNAz-treated culture (Figure 1a). By contrast, no detectable signal was observed for the reactions mediated by TBTA-Cu(I) and BARAC.
Figure 1.
Comparison of the efficiency of the CuAAC and BARAC-mediated copper-free click chemistry in labeling recombinant proteins and crude cell lysates. a) Western blot analysis of PD1-Fc isolated from HEK cells treated with Ac4ManNAz (top panel). Total protein loading was confirmed by Coomassie staining (bottom panel). b) Western blot analysis of Ac4ManNAz-treated or untreated Jurkat cell lysates.
To evaluate the efficacy of the CuAAC and BARAC-mediated copper-free cycloaddition in a more complex system, we sought to investigate bioconjugation of a biotin affinity probe to azide-tagged sialyl glycoproteins in crude cell lysates, the first step in enriching these proteins for glycoproteomic analysis. We introduced azides to the cell-surface sialyl glycoconjugates of Jurkat cells, a human T lymphocyte cell line, by culturing the cells with Ac4ManNAz. After 3 days, we lysed the cells in phosphate buffer with 1% NP40 to solubilize the membrane proteins. The cell lysates were then reacted with biotin-alkyne via the CuAAC or BARAC-biotin copper-free click chemistry using the same conditions specified for the labeling of the recombinant PD1-Fc.
Consistent with the observation by Cravatt and coworkers, we discovered that a significant portion of proteins precipitated out of the reaction buffer when the lysates were treated with TBTA-Cu(I) (Figure S1 in the Supporting Information).[21] By contrast, the reaction mixture remained homogeneous for the lysates treated with Cu(I) in complex with the other three ligands. As revealed by the anti-biotin Western blot (Figure 1b), the click reaction mediated by the three water soluble ligands provided significantly stronger glycoprotein labeling in lysates from cells treated with Ac4ManNAz than what was achieved using the TBTA-Cu(I) catalyst or BARAC-mediated cycloaddition. Importantly, the Cu(I)-mediated reactions exhibited remarkable selectivity with no background signals detectable for cell lysates harvested in the absence of the sugar. However, nonspecific biotinylation was observed for the BARAC-based copper-free click chemistry (Figure 1b), presumably generated by the reaction of the cyclooctyne with cysteine residues in the protein lysates.[22]
One advantage of CuAAC over BARAC-based cycloaddition is that the former reaction can also be used to detect biomolecules tagged with terminal alkyne residues. We compared the four Cu(I) catalysts for labeling peracetylated N-(4-pentynoyl)mannosamine (Ac4ManNAl)-treated cell lysates. Similarly, the strongest labeling signal was obtained using the BTTAA-Cu(I) catalyst whereas the weakest signal was observed with TBTA-Cu(I) (Figure S2).
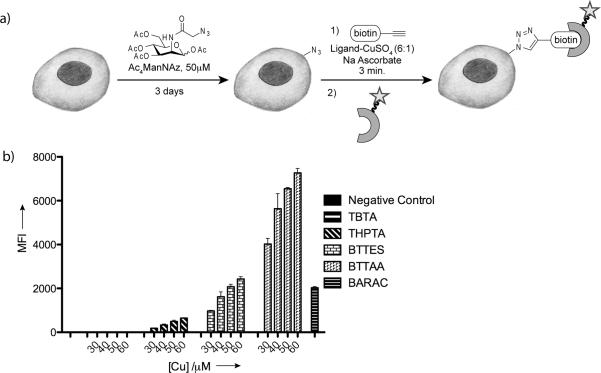
Next, we compared the efficacy of the CuAAC and copper-free click chemistry for labeling the azide-tagged glycoconjugates in live cells. We cultured Jurkat cells in medium supplemented with Ac4ManNAz, and then reacted the cells bearing the corresponding azido sialic acid (SiaNAz) with biotin-alkyne (45 μM) catalyzed by various concentrations of the four copper catalysts (catalyst formulation: [ligand]:[Cu]=6:1, 2.5 mM sodium ascorbate, optimized for live cell labeling)[12] or BARAC-biotin (45 μM) for 3 min at room temperature. The biotinylated cells were then analyzed by flow cytometry using Alexa Fluor 488-streptavidin. As shown in Figure 2, reactions mediated by the BTTAA-Cu(I) catalyst provided the strongest labeling of the SiaNAz-displaying cells, yielding cell-associated Alexa Fluor 488 signal 3 to 4-fold higher than the signal achieved by the BTTES-Cu(I) catalyst. As low as 30 μM of Cu(I) was sufficient to yield a significant signal. Both catalysts gave highly selective labeling with minimal background labeling observed in the absence of Cu(I). By contrast, six-fold weaker labeling was observed for cells treated with the THPTA-Cu(I) catalyst even at 2-fold higher concentration of Cu(I) (60 μM), and barely any labeling was detectable for cells treated with the TBTA-Cu(I). Noteworthy, BARAC-mediated cycloaddition showed a comparable level of efficiency for labeling cell-surface SiaNAz as the BTTES-promoted CuAAC ([Cu(I)]=50 μM).
Figure 2.
Relative labeling efficiency of the bioorthogonal click reactions on live cells. a) Schematic representation of metabolic labeling and detection of cell-surface sialic acids using Ac4ManNAz and click chemistry. b) Flow cytometry analysis of cell surface labeling described in a) using Jurkat cells. Error bars represent the standard deviation of two replicate experiments.
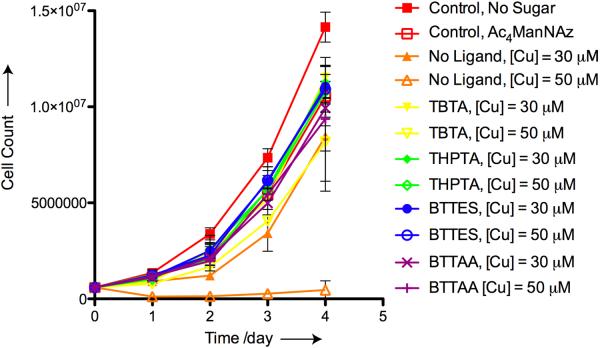
To evaluate if the copper catalysts cause any long-term perturbations to the treated cells, we labeled Jurkat cells bearing SiaNAz residues with biotin-alkyne in the presence of the four catalysts ([Cu] = 30 and 50 μM) for 3 minutes. The reactions were then quenched with bathocuproine sulphonate (BCS), a biocompatible copper chelator. For negative controls, unreacted cells cultured in the absence and presence of Ac4ManNAz were included. For a positive toxicity control we treated SiaNAz-bearing cells with Cu(I) in the absence of the ligands. All cells were cultured for 4 days post reaction. Viable cells, based on Trypan Blue assay, were counted each day. As shown in Figure 3, cells treated with the BTTAA-, BTTES- and THPTA-Cu(I) catalysts proliferated at rates similar to the untreated cells cultured with Ac4ManNAz. However, in the presence of 50 μM Cu(I), cells treated with the TBTA-Cu(I) catalyst showed a slower rate of proliferation than the cells treated with the other three catalysts, and greater than 50% cells underwent cell lysis in the presence of 75 μM of Cu(I) (data not shown). Noteworthy, incubation with 30 μM Cu(I) for 3 minutes in the absence of the ligands only induced minor toxicity to cells as revealed by the 20% slower proliferation rate of the treated cells on days 3 and 4. By contrast, when treated with 50 μM Cu(I) and no ligand greater than 90% of the cells underwent cell lysis within 24 hours and division of the remaining cells was significantly impaired. Combining this proliferation assay together with the live cell labeling results, we concluded that the BTTAA-Cu(I) catalyst is the optimal choice for cell surface azide-alkyne ligation.
Figure 3.
Cell proliferation assays indicating that tris(triazolylmethyl)amine-based ligands protect Jurkat cells from Cu(I)-induced long-term perturbation.
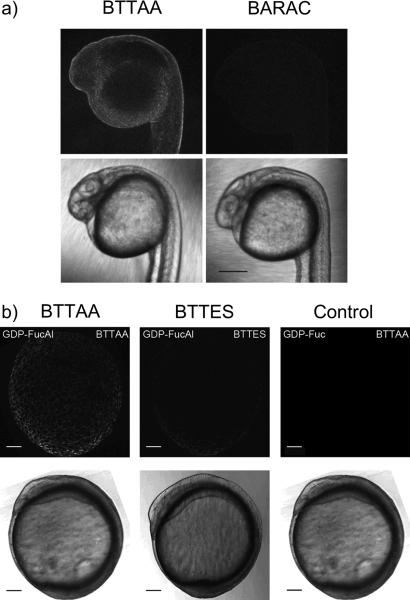
After establishing the relative reactivity of the copper catalysts and BARAC on cultured cells, we extended our comparison to the labeling of glycans in living systems. We chose zebrafish as a vertebrate model for this purpose. Zebrafish is transparent in the first 24 hours of its developmental program, which allows the labeled glycans to be detected via molecular imaging techniques. [23] Bertozzi and coworkers demonstrated that peracetylated N-azidoacetylgalactosamine (Ac4GalNAz) can be used to metabolically label O-linked glycans in zebrafish embryos by microinjecting or bathing the embryos in medium supplemented with this azido sugar.[24,25] Following their protocol, we microinjected embryos at the one-cell stage with Ac4GalNAz. Microinjection into the yolk sack at this stage allows the sugar to diffuse into all daughter cells during the developmental program. At 24 hours post fertilization (hpf), we reacted the Ac4GalNAz-treated embryos with biotin-alkyne (50 μM) catalyzed by BTTAA-Cu(I) ([Cu] = 45 μM) or BARAC-biotin (50 μM) for five minutes to detect the membrane-associated azides in cell surface O-linked glycans. Following the reactions, we incubated the biotinylated embryos with streptavidin-Alexa Fluor 488 and compared the efficacy of the BTTAA-mediated CuAAC and copper-free click chemistry using fluorescence confocal microscopy. As shown in Figure 4a, significant labeling was achieved with the BTTAA-Cu(I) catalyst whereas only weak labeling was detectable with BARAC-biotin.
Figure 4.
Relative efficiencies of CuAAC and copper-free click chemistry in labeling zebrafish glycans. a) One-cell embryos were microinjected with a single dose of Ac4GalNAz. At 24 hpf, the embryos were reacted with biotin-alkyne (50 μM) catalyzed by the BTTAA-Cu(I)-catalysts ([Cu] = 40 μM) or BARAC-biotin (50 μM) for 5 minutes, then probed with streptavidin-Alexa Fluor 488 and imaged using confocal microscopy. b) One-cell embryos were microinjected with a single dose of GDP-FucAl or GDP-Fuc (control) and allowed to develop to 9 hpf. The embryos were then reacted with Alexa Fluor 488-azide (50 μM) catalyzed by BTTAA-Cu(I) or BTTES-Cu(I) ([Cu] = 40 μM) for 3 min and imaged using confocal microscopy. Scale bar: a) 200 μm b) 100 μm. Top: 488 channel; bottom: brightfield.
Our previous work showed that an alkyne-bearing fucose analog, FucAl, can be successfully incorporated into the glycans of zebrafish embryos. Here, we also compared the efficiency of the BTTAA- and BTTES-Cu(I) catalysts for labeling these alkyne-tagged fucosides in live zebrafish embryos. To accomplish this task, we microinjected one-cell embryos with an alkyne-derivatized GDP-fucose analogue (GDP-FucAl). At 9 hpf, we reacted the embryos with Alexa Fluor 488-azide in the presence of BTTAA-Cu(I) or BTTES-Cu(I) for 3 min ([Cu] = 40 μM), and acquired fluorescent images using confocal microscopy. As quantified using ImageJ, BTTAA-Cu(I) provided 2.5-fold stronger signal than what was achieved using the BTTES-Cu(I) catalyst (Figure 4 b). After the labeling reactions, we followed the development of the BTTAA-treated embryos for 5 days and observed no developmental defects (n = 30, Figure S5), which confirmed the biocompatibility of the new Cu(I) catalyst formulation.
In summary, the parallel comparison of the premium bioorthogonal click reactions, namely the strain-promoted copper-free click chemistry and the ligand-accelerated CuAAC, verifies the great potential of the latter as a highly effective ligation tool for broad biological applications. With the discovery of a new accelerating ligand for CuAAC, not only is faster kinetics over the known catalysts achieved, but more importantly, it allows for effective bioconjugation with suppressed cell cytotoxicity by further lowering Cu(I) loading in the catalyst formulation. The reaction conditions optimized here are shown to be the most effective in four biological settings, i.e. labeling of recombinant glycoproteins, glycoproteins in crude cell lysates, on live cell surfaces, and in the enveloping layer of zebrafish embryos. An additional advantage of the bio-benign CuAAC is that it liberates the bioconjugation from the limitation where ligations could only be accomplished with azide-tagged biomolecules. Now terminal alkyne residues can also be incorporated into biomolecules and detected in vivo. Overall, the reported ligand-accelerated CuAAC represents a powerful and highly adaptive bioconjugation tool for biologists, which holds great promise for further improvement with the discovery of more versatile catalyst systems.
Supplementary Material
Footnotes
This work was supported by the National Institutes of Health grants GM080585 (P.W.). C.B. is supported by the NIH training grant T32 GM007491. Part of the ligand synthesis was performed as a User Project at the Molecular Foundry, Lawrence Berkeley National Laboratory, which was supported by the Office of Science, Office of Basic Energy Sciences, U.S. Department of Energy, under contract DE-AC02-05 CH11231.
References
- [1].Baskin JM, Bertozzi CR. Qsar Comb. Sci. 2007;26:1211–1219. [Google Scholar]
- [2].Laughlin ST, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Jao CY, Roth M, Welti R, Salic A. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15332–15337. doi: 10.1073/pnas.0907864106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, Lopez CB, Hang HC. Nat. Chem. Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Nat. Protc. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]; b) Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, Beatty KE, Connor RE, Yoo TH, Dieterich DC, Schuman EM, Tirrell DA. Nat Chem Biol. 2009;5:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Salic A, Mitchison TJ. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jao CY, Salic A. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hanson SR, Hsu TL, Weerapana E, Kishikawa K, Simon GM, Cravatt BF, Wong CH. J. Am. Chem. Soc. 2007;129:7266–7267. doi: 10.1021/ja0724083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang YY, Ascano JM, Hang HC. J. Am. Chem. Soc. 2010;132:3640–3641. doi: 10.1021/ja908871t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- [9].Sletten EM, Bertozzi CR. Angew. Chem. 2009;121:7108–7133. doi: 10.1002/anie.200900942. Angew. Chem. Int. Ed. 2009, 48, 6974-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- [11].a) Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Poloukhtine AA, Mbua NE, Wolfert MA, Boons GJ, Popik VV. J. Am. Chem. Soc. 2009;131:15769–15776. doi: 10.1021/ja9054096. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ning X X, Guo J, Wolfert MA, Boons GJ. Angew. Chem. 2008;120:2285–2287. doi: 10.1002/anie.200705456. Angew. Chem. Int. Ed. Engl. 2008, 47, 2253-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. J. Am. Chem. Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. ACS Chem. Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- [14].Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. 2002;114:2708–2711. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. Angew. Chem. Int. Ed. 2002, 41, 2596-2599. [DOI] [PubMed] [Google Scholar]
- [15].Tornoe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- [16].Jewett JC, Sletten EM, Bertozzi CR. J. Am. Chem. Soc. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org. Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- [18].a) Wu P, Fokin VV. Aldrich. Acta. 2007;40:7–17. [Google Scholar]; b) Hein JE, Fokin VV. Chem. Soc. Rev. 2010;39:1302–1315. doi: 10.1039/b904091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hong V, Presolski SI, Ma C, Finn MG. Angew. Chem. 2009;121:10063–10067. doi: 10.1002/anie.200905087. Angew. Chem. Int. Ed. 2009, 48, 9879-9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sivakumar X, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. Org. Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- [21].Weerapana E, Speers AE, Cravatt BF. Nat. Protoc. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- [22].Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1821–1826. doi: 10.1073/pnas.0911116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- [24].Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR, C. R. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10360–10365. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.