Abstract
HIV-1 reverse transcriptase (RT) has been an attractive target for the development of antiretroviral agents. Although this enzyme is bi-functional, having both DNA polymerase and ribonuclease H (RNH) activities, there is no clinically approved inhibitor of the RNH activity. Here, we characterize the structural basis and molecular interaction of an allosteric site inhibitor, BHMP07, with the wild type (WT) RNH fragment. Solution NMR experiments for inhibitor titration on WT RNH showed relatively wide chemical shift perturbations, suggesting a long-range conformational effect on the inhibitor interaction. Comparisons of the inhibitor-induced NMR chemical-shift changes of RNH with those of RNH dimer, in the presence and absence of Mg2+, were performed to determine and verify the interaction site. The NMR results, with assistance of molecular docking, indicate that BHMP07 preferentially binds to a site that is located between the RNH active site and the region encompassing helices B and D (the “substrate-handle region”). The interaction site is consistent with the previous proposed site, identified using a chimeric RNH (p15-EC) [Gong, el (2011) Chem. Biol. Drug Des. 77, 39-47], but with slight differences that reflect the characteristics of the amino acid sequences in p15-EC compared to the WT RNH.
Keywords: Reverse transcriptase, ribonuclease H, HIV, acylhydrazone, inhibitor, enzyme, protein, NMR, dimer, molecular docking
INTRODUCTION
HIV-1 reverse transcriptase (RT) is a multifunctional enzyme with both DNA polymerase and ribonuclease H (RNH) activities. Approximately half of the current, clinically approved drugs for the treatment of HIV-1 infection target RT, specifically, the DNA polymerase activity of the enzyme (3-8). Like the DNA polymerase activity, RNH function is essential for viral replication and, thus, plays a crucial role in HIV-1 pathogenesis (9-12). Although effort has been devoted to developing RT-RNH inhibitors (13-21), currently, no clinically approved anti-AIDS drugs based on RNH inhibition are available. In part, this significant absence may be due to the fact that the structural basis of the RNH-inhibitor interaction is not well understood.
Recently, crystal structures of RNH active-site inhibitors in complex with RT or an RNH fragment (22-24) as well as an NMR study of an RNH inhibitor interaction at the active site (25) were published. In contrast to these studies, which were focused on inhibitors chelating with Mg2+ ions at the active site, we recently characterized several acylhydrazone inhibitors that exhibit moderate antiviral activity; one of these, BHMP07, was found to bind the RNH domain at a novel allosteric site (1). This conclusion was based on chemical shift changes of backbone amide NMR signals, in the presence of BHMP07 at various concentrations, demonstrating that BHMP07 primarily recognizes the RNH substrate-handle region encompassing helices B and D (26) (i.e. residues 503 to 527 in RT). This observation was supported by a recent molecular docking study of hydrazone/hydrazine compounds on RT (27).
The RNH fragment used in our previous NMR study was a chimeric protein, created by replacing a small loop segment of HIV-1 RT-RNH with a 24 residue α-helical substrate-binding loop derived from Escherichia coli RNase H. Although this construct, termed p15-EC, has been widely used to screen RNH inhibitors and characterize protein-inhibitor interactions (2, 28, 29), it is unclear whether inhibitor binding sites obtained for the p15-EC construct are directly applicable to wild-type (WT) RT RNH. Addressing this issue is of critical importance because little structural information is available about the interaction mechanism of RNH inhibitors with WT RNH.
In this article, we describe the interaction between BHMP07 and the WT RNH fragment, evaluated using NMR spectroscopy. With this method, monitoring of backbone chemical shift perturbations (Δδ) can provide atomic resolution information for weakly interacting protein-ligand complexes, over the course of a titration series, allowing for localization of the ligand binding site. In general, magnitudes of chemical shift changes qualitatively correlate with the distance from the ligand. To gain insight into the allosteric inhibitor interaction with WT RNH, we monitored the impact of increasing concentrations of BHMP07 on both the monomeric WT RNH and a kinetically-trapped dimer form. First, we determined that the dimer interface encompasses the substrate-handle region and does not influence Mg2+ interaction at the active site. Thus, if BHMP07 binds at (or close to) the substrate-handle region, it will not be able to bind the dimer form of RNH. Our data indicate that this is the case: BHMP07 interacted with monomeric WT RNH but not with the dimeric form. Next, we demonstrated that Mg2+ prevents binding to both the monomeric and dimeric forms. Based on these results, we conclude that BHMP07 interacts with the monomeric RNH in a way that involves residues near both the substrate-handle region and the active site. Thus, while involvement of the substrate-handle region is consistent with our previous study of p15-EC, overlap of the BHMP07 interaction site with areas affected by Mg2+ binding is more pronounced in the WT RNH compared to p15-EC (1). This shift of the binding site is not surprising, as it is concomitant with a smaller number of hydrophobic amino acid side chains in the substrate-handle region of WT RNH compared to the chimeric construct. Finally, computational docking of BHMP07 to an RNH domain structure revealed three potential binding sites, one of which is located between the substrate-handle region and the active site and is consistent with the NMR data.
MATERIALS and METHODS
Sample preparation
We generated the isolated RNH domain (RT residues 427–560, with an additional N-terminal-peptide S–E–L) by expressing the domain in E. coli. In brief, the coding sequence was inserted into pET32a plasmid (EMD Chemicals Inc., San Diego, CA) as a thioredoxin fusion construct that included a Tobacco Etch Virus (TEV) protease recognition site at its C-terminus (30). For NMR experiments, the cells were cultured in modified M9 minimal media containing 15NH4Cl (1 g/L) as the sole nitrogen source in the absence or presence of [u-13C]-glucose as the sole carbon source (30). The protein was isolated from the cell lysate using a Hi-Trap SP ion-exchange column (GE Healthcare, Piscataway, NJ) at pH 7.5, using a 0 – 1 M NaCl gradient, followed by further purification by size exclusion chromatography on a Superdex 200 26/60 column (GE Healthcare, Piscataway, NJ), using buffer containing 25 mM sodium phosphate buffer (pH 7.0), 0.02% sodium azide, 100 mM NaCl and 1 mM DTT. The RNH protein was eluted at two positions (Fraction #1 and Fraction #2); both peaks were pooled and separately subjected to TEV digestion. Subsequently, each digested fraction was re-suspended in 20× volume of 25 mM Tris buffer at pH 8.5 containing 2 mM DTT, and subjected to a HiTrap TMQ SP column. Purity of the two protein species were confirmed by running 15% acrylamide gels in both SDS denatured and non-denaturing (native) conditions (PhastGel system, GE Healthcare, Piscataway, NJ). The final eluted protein fractions (~ 2 – 3 mg/mL) were stored separately in aliquots at −80°C. To prevent confusion due to the extra 3 residues in the N-terminus, in what follows, we report both the residue numbers reflecting the experimental construct used and, in parentheses, the RT numbering (see Figure S1).
Multi-angle static light scattering
Multi-angle static light scattering experiments were performed on a HELEOS instrument (Wyatt Technology) equipped with refractive index detectors (Optilab rEX, Wyatt Technology) and an analytical Superdex 200 column (1 × 30 cm, GE Healthcare), pre-equilibrated at room temperature with 25 mM sodium phosphate, 100 mM NaCl, 2 mM DTT and 0.02% (w/v) sodium azide at pH 7.0. 100 μL of thawed protein Fractions #1 and #2 (~ 3 mg/mL) were separately injected at a flow rate of 0.5 mL/min and eluted with the same buffer.
NMR experiments
Thawed aliquots of WT RNH Fraction #1 and Fraction #2 were dialyzed into 25 mM sodium phosphate containing 100 mM sodium chloride and 2 mM DTT, pH 7.0, and the proteins were concentrated, typically to ~200 μM, using Amicon Vivaspin concentrators (Millipore, Billerica, MA). NMR samples were prepared by combining ~380 μL protein solution with 20 μL D2O (to allow for deuterium locking) and stored in 5-mm NMR tubes susceptibility-matched to D2O (BMS-005B, Shigemi, Inc., Allison Park, PA).
All NMR experiments were recorded at 20°C on a Bruker Avance 600 spectrometer (Bruker BioSpin, Billerica, MA) equipped with a 5 mm triple-resonance z-gradient cryogenic probe. Backbone 1H/15N signals were assigned via essentially the same approach used for the p15-EC RNH study (1) by recording HNCA, CBCACONH and HNCACB experiments using 13C /15N-labeled protein at ~500 μM concentration (31). To facilitate backbone assignment, a separate set of experiments was collected on a sample of 13C/15N-labeled protein at ~500 μM concentration in the presence of 40 mM MgCl2. NMR spectra were processed and analyzed using NMRPipe, NMRview, and CcpNmr Analysis 2.1.5 (32, 33)(34). After manual assignment of ~75% of the sequence, existing spin systems were frozen and further sequential links were identified with the aid of predictions generated independently by the PINE server 1.0 (35) and the Nexus automated assignment protocol that is included in the CcpNmr Analysis software. A series of 1H-15N HSQC experiments were recorded at different Mg2+ concentrations (0, 5, 10, 15 and 20 mM) to identify the Mg2+ interaction sites. Similarly, a series of 1H-15N HSQC experiments were recorded at different concentrations of BHMP07 (inhibitor:protein ratios ranging from 0 to ~1.5:1) to identify the inhibitor-interaction site in the presence and absence of 20 mM Mg2+. All titration series were conducted under the same conditions and each completed within a half day.
Combined chemical shift changes for each step i of the titration series are calculated as the quadratically weighed amplitudes of the 1H and 15N chemical shift differences:
| Eq. 1 |
Endpoints for the titrations, although falling short of complete ligand saturation, were chosen because higher concentrations of the inhibitor solution (in DMSO) resulted in significant precipitation. Hence, the maximal amplitude of chemical shift changes are shown at the 1.5:1 ratio, to facilitate comparison between monomer and dimer and to minimize the contribution of unspecific binding effects.
Determination of dissociation constants
Dissociation constants (KD) were derived from the chemical shift changes logged for discrete resonances by fitting the obtained titration curves individually via nonlinear regression analysis. Assuming single-site binding for a system in fast exchange, at each titration step i the theoretical is given based on the total protein concentration [P]total and the current ligand concentration [L]i as (36):
| Eq. 2 |
The unknown parameters, i.e. KD and the extent of chemical shift change at saturation, , were optimized by minimizing the difference between the experimentally measured Δδobs and the Δδcal for each titration curve, i.e., minimizing the χ2 function
| Eq.3 |
Uncertainties of the optimized parameters were estimated via Monte Carlo resampling, by generating 50 sets of synthetic data of Δδ with a Gaussian distribution whose standard deviation (σ) is given by an experimental chemical shift uncertainty, δnoise (37).
Computational docking experiments
Molecular docking calculations were carried out with the Schrödinger Suite 2011 programs. The structure of the wild type RNH fragment of HIV-1 RT, previously determined by X-Ray crystallography (PDB access code 3KP2), was downloaded from the Protein Data Bank. To prepare structures for further docking calculations, hydrogen atom and hydrogen bond assignment, side chain ionization, and energy minimization were done with the Protein Preparation Wizard Protocol (Schrödinger Suite 2011 Protein Preparation Wizard; Epik version 2.2, Schrödinger, LLC, New York, NY, 2011; Impact version 5.7, Schrödinger, LLC, New York, NY, 2011; Prime version 3.0, Schrödinger, LLC, New York, NY, 2011).
The geometry of BHMP07 was minimized with MacroModel (MacroModel, version 9.9, Schrödinger, LLC, New York, NY, 2011) and then the ligand was prepared for docking by force field assignment and ionization states prediction (at target pH = 7.5 ± 1.0) by LigPrep application (LigPrep, version 2.5, Schrödinger, LLC, New York, NY, 2011). Prepared structures were then used to generate conformers with ConfGen (version 2.3, Schrödinger, LLC, New York, NY, 2011) and conformers with Boltzmann populations greater than 5% were taken into further docking. The receptor grid box was centered on the residues that showed the largest chemical shifts upon NMR binding (see below) and made to accommodate a ligand within 20 Å of length (i.e. the approximate length of the most extended BHMP07 conformer). The potential of non-polar parts of the receptor was softened by applying a 0.8 scaling factor to van der Waals radii of atoms with partial charges less than 0.15 a.u. Docking computations were carried out using Glide XP in extra precision mode (Glide, version 5.7, Schrödinger, LLC, New York, NY, 2011).
RESULTS
Isolation of dimer and monomer fractions of RNH in solution
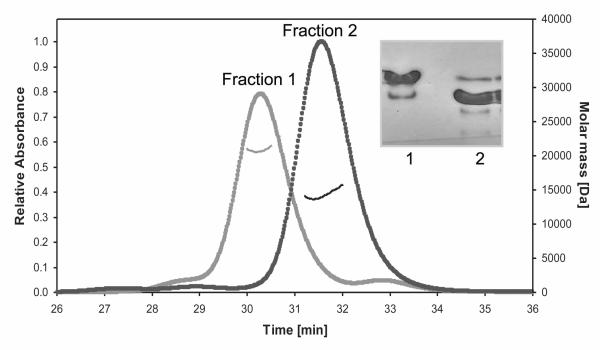
We previously synthesized and characterized acylhydrazones that inhibit RT RNH activity and exhibit moderate antiviral activity (1, 38). To further characterize the binding mechanism of acylhydrazones to the RT RNH fragment, we prepared WT RNH by expressing the domain in E. coli. Interestingly, the domain was eluted in two different fractions from gel filtration column chromatography, henceforth referred to as Fraction #1 and #2. The protein fractions exhibited similar migration on SDS PAGE and the same molecular masses as determined by mass spectroscopy (15094.8 Da and 15094.7 Da for non-isotope labeled Fractions #1 and #2, respectively; data not shown), confirming that both fractions contain the same polypeptide. However, the gel filtration profiles of the two fractions and the associated molecular masses, determined by MALS equipped with a refractive index detector, revealed that Fraction #1 is predominantly a dimer (Figure 1). Consistent with this observation, native gel profiles showed different migration patterns for Fraction #1 and Fraction #2 (Figure 1, inset). 1H-15N HSQC spectra of the two fractions, despite differing in ~20 distinct cross-peaks, exhibit overall similar resonance patterns, suggesting that the basic secondary and tertiary fold of the dimer is almost identical to that of the monomer (Figure 2A). No interconversion between the two forms was seen at 20°C or 40°C over three days. However, upon unfolding by adding concentrated urea and then re-folding, the biophysical characteristics of the protein in Fraction #1 became consistent with those of Fraction #2 (data not shown). Taken together, the data suggest that the RNH domain in Fraction #1 represents a kinetically trapped dimer. To our knowledge, no dimer of RNH in solution has been heretofore reported in the literature.
Figure 1.
Isolation of dimer and monomer fractions of RNH in solution. Overlay of elution profiles of the MALS for the WT RNH Fraction #1 (gray) and Fraction #2 (black). The molecular masses calculated from the refractive index were 22.47 kDa and 14.4 kDa for Fractions #1 and #2, respectively. An electrophoretic native gel for the two fractions is inserted in the graph, showing the different migration patterns in the two fractions. All experiments were performed in the absence of Mg2+.
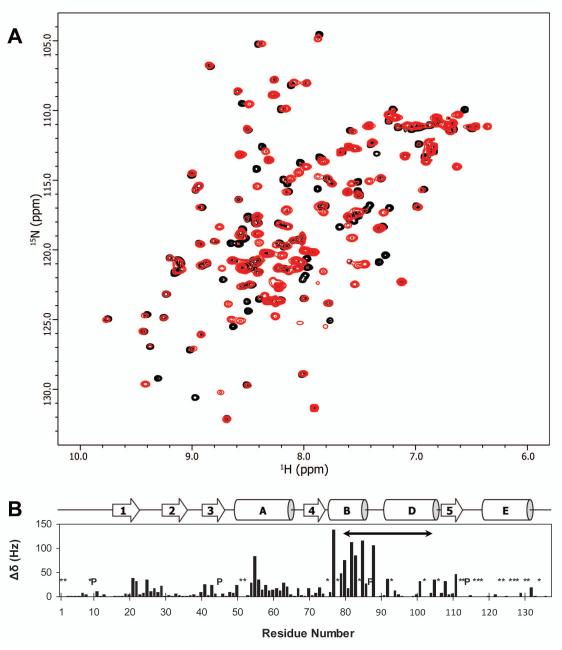
Figure 2.
Difference in NMR chemical shifts between dimer and monomer fractions of RNH. (A) Overlay of 1H-15N HSQC spectra of WT RNH dimer (Fraction #1, red) and the monomer (Fraction #2, black). Both experiments were performed in the absence of Mg2+. (B) Normalized quadratically weighed 1H, 15N backbone amide resonance shift differences (Δδ, cf. Eq. 1) between the monomer and dimer forms are shown relative to sequence residue. Prolines (P) and unassigned/undetected backbone NH amide groups (*) in either form are indicated; the “substrate-handle region” is denoted by a solid arrow (residues 80 to 103). A schema of secondary structure elements is included for reference [helix C is an insert that occurs only in E. Coli RNAse H (26)]. Data were recorded at 600 MHz (1H), at 20°C and pH 7.0.
Assignment of NMR spectra
Using spectra recorded in the absence of metal ions or in the presence of 40 mM MgCl2, we were able to assign amide resonances for 83.5% of the RNH residues in Fraction #2 (total 111 residues among 133 backbone amide. Note that there are 4 Pro residues). This represents a significant increase in completeness over published assignments (39) and, notably, includes most residues in the C-terminal helix. Thus, we were able to assign the amide resonances of 118 residues in Fraction #1 and directly compare 109 chemical shifts between Fractions #1 and #2 (Figure 2).
Characterization of the dimer interface in solution
Based on the NMR signal assignments, significant backbone chemical shift differences (, i.e. greater than one standard deviation from the average response) in the dimer compared to the monomer were found to be restricted to a relatively few residues, including Glu55(478) [Δδ ~ 83 Hz], Gln77(500) [Δδ ~ 138 Hz], Ala79(502) [Δδ ~ 49 Hz], Leu80(503) [Δδ ~ 75 Hz], Ile82(505) [Δδ ~ 113 Hz], Ile83(506) [Δδ ~ 86 Hz], Ala85(508) [Δδ ~ 116 Hz], Asp88(511) [Δδ ~ 106 Hz] and Ala111(534) [Δδ ~ 46 Hz] (Figure 2B). Thus, the interface helix is located at the substrate-handle region (residues 80 to 104 in our NMR sequence, see Figure S1 and Figure 3A). Crucially, this interface area is largely distinct from the active site (Figure 3A); this is confirmed by the fact that Mg2+ interacts with both, resulting in congruous effects in the monomer and dimer forms (Figure S2). Indeed, both amplitude (Figure S2, compare panels A and B) and spatial localization (Figure S2, compare panels C and D) of the chemical shift perturbation upon Mg2+ addition were almost identical for both protein forms.
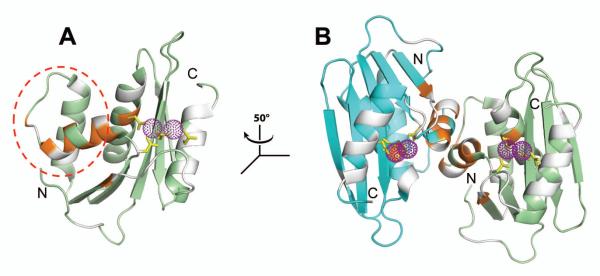
Figure 3.
Dimer interface mapped onto RNH crystal structures. Two RNH chains (PDB: 3K2P) are shown in ribbon representation and colored, respectively, in green and cyan. Residues exhibiting significant chemical shift changes (, i.e. greater than one standard deviation from the average response in (Figure 2) between the dimer and the monomer are highlighted in orange; undetected/unassigned residues are shaded in gray. Purple spheres, shown in dots, delineate the position of manganese ions in the active site; the side chains of the metal coordinating residues (D20, E55, D74, and D126) are indicated by yellow sticks. (A) Interface mapped onto one RNH subunit, the substrate-handle region (residues from 80 to 103) is outlined (red dashed circle). (B) Illustration of the kinetically trapped dimer interface via a coinciding (see text) crystallographic dimer. Relative to panel (A), the view in (B) is rotated by −50° around the z-axis (see inset). Note that although the crystal structure contains manganese ions as coordinating ions, magnesium was used for NMR experiments to avoid paramagnetic effects by manganese.
Interestingly, the interface mapped by NMR spectroscopy coincides with one identified in a previously reported crystallographic dimer structure (PDB=3K2P, (23)). Indeed, mapping of the chemical shift differences between the monomer and dimer in solution presents a high degree of overlap with the interface in the previously reported crystallographic dimer (Figure 3B). It must, however, be emphasized that the kinetically trapped RNH dimer form in solution, discussed here, is distinct from the crystallographic dimer which solely reflects packing effects. While further structural studies would be needed for an in-depth characterization, we propose that this stable dimer may involve domain swapping at the interfacial α-helix in the substrate-handle region. Since such a domain swap would retain most of the secondary and tertiary structure of the monomer, it would explain why the pattern of NMR chemical shifts of the dimer was similar to that of monomer, with only 24 resonances significantly altered between the two structures (Figure 2A).
BHMP07 binds to the RNH monomer in the absence of Mg2+
Upon addition of BHMP07 to the RNH monomer, in absence of metal, a number of 1H-15N HSQC cross-peaks shifted monotonically with increasing ligand concentration, implying a fast exchange régime for the protein/ligand interaction (Figure S3A). The most strongly affected polypeptide backbone amide NH resonances were from residues located at the dimer interface, notably Glu55(478) [Δδ ~ 47 Hz], Ser76(499) [Δδ ~ 46 Hz] and Ala79(502) [Δδ ~ 87 Hz]; or in the vicinity of the active site, such as Ala23(446) [Δδ ~ 58 Hz], Asn24(447) [Δδ ~ 33 Hz], Arg25(448) [Δδ ~ 43 Hz], Gly30(453) [Δδ ~ 47 Hz], Ala32(455) [Δδ ~ 40 Hz] and Gly118(541) [Δδ ~ 67 Hz]. Additional strong perturbations were observed for C-terminal residues, including Ile133(556) [Δδ ~ 42 Hz], Lys135(558) [Δδ ~ 48 Hz] and Val136(559) [Δδ ~ 50 Hz] (Figure 4A). Overall, the sequence-averaged perturbation was ~ 17.3 ± 15.4 Hz, the large σ reflecting the site-specificity of the binding interaction. The equilibrium dissociation constant (KD) between BHMP07 and RNH monomer was ~ 49 ± 25 μM in the absence of Mg2+, determined by nonlinearly fitting the ligand titration profiles for 26 individual residues that exhibited significant changes compared to the average perturbation (i.e. , Table S1). It must be noted that although complete saturation could not be observed experimentally (vide supra), the measured titration profiles are clearly hyperbolic (Table S1 and Figure S3B). Here, the relatively small uncertainty of the KD, given as a standard deviation, indicates that the binding is specific and cooperative. Although this in vitro KD for the RNH fragment is rather large, it nevertheless compares favorably to the KD ~ 142 ± 36 μM that was previously derived for the p15-EC RNH construct (1).
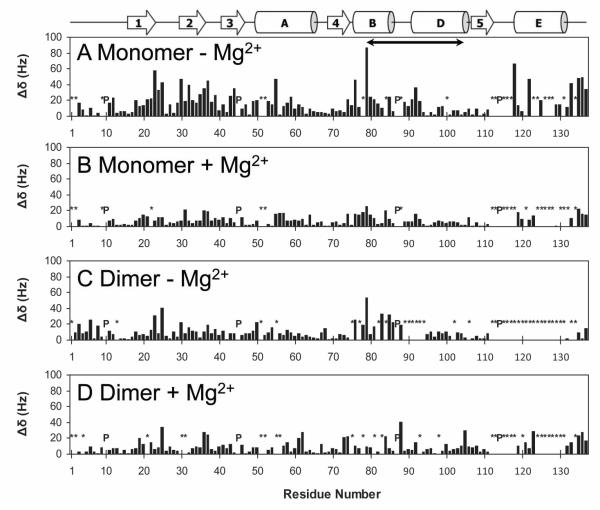
Figure 4.
NMR chemical shift perturbation induced by BHMP07 interaction with the RNH monomer in the absence (A) or presence (B) of 20 mM Mg2+; and with the RNH dimer in the absence (C) or presence (D) of 20 mM Mg2+. Normalized, quadratically weighed 1H, 15N backbone amide resonance shifts induced by BHMP07 (Δδ, cf. Eq. 1) are shown relative to sequence residue. Prolines (P) and backbone NH amide groups whose chemical shift changes could not be tracked (*) are indicated. A schema of secondary structure elements (26) is included for easy reference; the “substrate-handle region” (58) is denoted by a solid arrow (residues 80 to 103). Data were recorded at 600 MHz (1H), at 20°C and pH 7.0.
Mg2+ negatively impacts BHMP07 binding in the RNH monomer
Pre-addition of 20 mM MgCl2 to the RNH monomer resulted in significantly reduced perturbation effects during titration with BHMP07 (Figure S4A), with a sequence-averaged perturbation of ~ 7.8 ± 5.8 Hz (Figure 4B) and with no residue exhibiting Δδ > 30 Hz. The minute amplitude of the observed chemical shift changes forms compelling evidence that, in essence, BHMP07 does not bind to RNH monomer in the presence of Mg2+. Furthermore, the observed small shifts are clearly independent of any conformational changes caused by the Mg2+ interaction: indeed, the Δδ for the latter interaction do not correlate with the perturbations induced by BHMP07 (Figure S2). Further inspection of chemical shift changes of the individual 1H (Figure S5) and 15N (Figure S6) chemical shift differences support these results. This result is in agreement with our previous study on p15-EC. However, it is rather idiosyncratic that the present distribution of small chemical shift perturbations closely mirrors the one recorded without Mg2+. Qualitatively, these tiny Δδs yield KD ~ 53 ± 129 μM from nonlinear fitting of 15 curves (Figure S4B), a value that is similar to the one obtained in the absence of Mg2+. Since interaction of Mg2+ with WT RNH is known to be weak (39), an appreciable population (~5%) of metal-free RNH is expected to exist in the current condition. Hence, our tentative interpretation is that the observed BHMP07-induced Δδ in the presence of 20 mM Mg2+ (Figure 4B) were due to inhibitor interaction with the Mg2+-free form of RNH in solution.
BHMP07 does not specifically interact with the RNH dimer
As discussed above, the RNH dimer interface is located at the substrate-handle region (Figure 3). Indeed, chemical shift differences between the monomer and the dimer (Figure 2B) overlap with the shift perturbations of the monomer caused by BHMP07 interaction (Figure 4A). Thus, any interaction with BHMP07 involving the substrate-handle region would likely be impeded in the dimer form of RNH due to its reduced accessibility upon dimerization. To test this hypothesis, analogous titrations with BHMP07 were performed on the RNH dimer.
Clearly, the addition of BHMP07 to the dimer resulted in weaker chemical shift changes ( ~10.2 ± 7.5 Hz) when compared to the monomer (Figure 4, compare panels A and C). In particular, most titration curves did not show a consistent hyperbolic profile but were rather erratic (not shown), implying that the corresponding Δδs were not indicative of a specific binding interaction. The significant reduction in chemical shift changes in the dimeric RNH demonstrates that the substrate-handle region is involved in the BHMP07 interaction. Pre-addition of Mg2+ did not lead to an increased BHMP07 perturbation for most residues either (Figure 4D), with the sequence-averaged perturbations being ~ 9.8 ± 8.0 Hz. Thus, the above titration results using the RNH dimer verify that the substrate-handle region is involved in the BHMP07 interaction.
Molecular docking
The NMR data above indicate that BHMP07 interacts with the monomer RNH at a region near the active site and the substrate-handle region. Using this information, we carried out theoretical molecular docking studies. NMR chemical shift perturbations not only map direct binding effects but are also sensitive to indirect, long-range effects that may result from conformational changes. Thus, for our docking calculations, we designed a receptor grid to encompass residues with strongly perturbed chemical shifts upon BHMP07 interaction, namely Ala23(446), Asp24(447), Asn25(448), Gly30(453), Ala32(455), Ser76(499), and Ala79(502) (see Figure 4A). We did not include the residues at the loop or hinge regions that may undergo significant chemical shift changes when the torsion angles vary or slight conformational adjustments occur; for the same reason, we also did not include the residues at the C-terminus, which are dynamic in solution (39, 40).
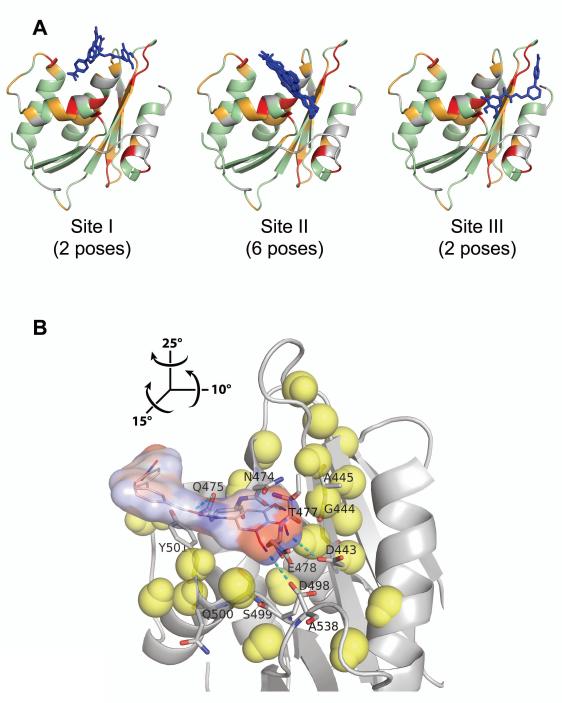
The docking results revealed three potential interaction sites: Site I involves predominantly the substrate-handle region; Site II is located in a region that bridges the substrate-handle and the active site regions, and Site III is located at the active site region (Figure 5A). Residues that interact with BHMP07 are explicitly indicated in Figure S7. Based on calculated docking scores BHMP07 shows strong affinity to Site I and II, while binding into Site III is less favorable. It can be argued that an interaction involving exclusively Site I would not fully satisfy the experimental NMR results as it cannot explain the observed perturbations near the active site. On the other hand, an interaction similarly dependent on Site III exclusively would not cause significant chemical shift changes in the substrate-handle region. By contrast, Site II is located proximal to the substrate-handle region and involves active site residues. Furthermore, inclusion of manganese ions at the active site prevents in silico docking of BHMP07 at Site II due to steric clash (data not shown), consistent with our NMR results that showed suppression of BHMP07 binding in the presence of Mg2+ (Figure 4).
Figure 5.
Interaction of BHMP07 with RNH monomer mapped onto the crystallographic structure (PDB code, 3K2P, chain A). (A) Ten BHMP07 conformers with the highest docking scores at Site I, Site II, and Site III (see text). RNH residues (ribbon cartoon) that exhibit significant NMR chemical shift changes upon addition of 1.5× excess of BHMP07 (Figure 4A) are highlighted: ; ; ; undetected/unassigned residues are colored gray. The sampled BHMP07 poses (blue sticks, nonpolar hydrogens are omitted) were predicted via NMR-based molecular docking in the absence of divalent ions (see text). (B) Minimum energy conformer of BHMP07 docked at Site II on RNH (gray ribbon). The inhibitor pose is presented as the skeletal formula with superposed van der Waals contact surface, colored by partial atomic charges from −0.4 a.u. (red) to +0.4 a.u. (blue). Interacting RNH residues (Figure S7) are labeled and shown in stick representation with carbon, nitrogen and oxygen atoms denoted in gray, blue and red, respectively; predicted intermolecular hydrogen bonds are indicated (cyan dashes). Neighboring (< 11 Å) RNH backbone H and N atoms that exhibit significant chemical shift changes () are shown (yellow van der Waals spheres). Relative to panel (A), the view in (B) is rotated by 15°, −10° and −25° about the x, y and z-axes, respectively (see inset).
The Site II position suggests that BHMP07 is partially sandwiched between two secondary structural units, β-strand 1 and the α-helix B. In this conformation, residues near BHMP07 include Asp20(443), Gly21(444), Ala24(445) in the β-strand 1; and Ser76(499), Gln77(500), Tyr(501), Ser76(499) in the α-helix B (Figures S7 and S8). These residues overlap with the residues included in the molecular docking [Ala23(446), Ala24(447), Asn25(448) in the β-strand 1; and Asp76(499), Ala79(502) in the α-helix B]. There is a slight difference between the NMR-observed Δδs and the BHMP07 interacting residues, presumably because chemical shift perturbations can also reflect indirect effects such as conformational shift induced by ligand interaction with residue side chains. In addition, since docking was done into a rigid receptor, possible slight conformational changes of protein backbone and side chain orientations, which might be caused by ligand binding, were not taken into account. Overall, while the binding configurations of Site I and Site III may not be discarded out of hand, a BHMP07 interaction mechanism that predominantly involves Site II correlates the best with our NMR results.
DISCUSSION
In the protein energy landscape, slight changes in the balance of energetics can result in significant effects in protein folding and molecular interaction (41-43). Thus, protein crystals may show inhibitors trapped at interaction sites in a manner different from that predicted by biochemical experiments, because the energetics in a crystal are different from those in solution. For example, in the crystal structure of RT with an N-acylhydrazone analogue, the inhibitor DHBNH, while specifically affecting RNH function, was found to bind to the non-nucleoside RT inhibitor binding pocket in the polymerase domain and not to the RNH site (38). In another crystallographic study, the non-nucleoside inhibitor nevirapine was soaked into RT crystals to block the polymerase site and to allow an RNH active-site inhibitor to bind at the RNH site rather than the polymerase (24). Dynamic structural changes of RT in solution have been observed by computational and experimental studies (44-46). Inspection of inhibitor interactions using RNH in solution, as a complement strategy of the RT crystal, is, therefore, of paramount importance (1). However, in cases where the binding is weak, particularly if the inhibitor is not highly soluble in aqueous solution, structure determination of protein-inhibitor complexes by NMR becomes challenging. Accordingly, the present study focuses on a hybrid approach, combining NMR binding site identification with computational prediction of the binding site. Since BHMP07 has moderate antiviral activity (1), understanding the mechanism of inhibition at an atomic level is critically important for future design of RT RNH inhibitors.
Our identification of a previously unreported dimeric form of RNH in solution is not surprising; indeed, proteins are not static entities and undergo various conformational fluctuations that can result in various quaternary structures, including oligomerization and domain-swapping (47-54). For RNase H, although dimers have been detected during purification for some RNHs (55, 56), so far, only crystallographic dimers have been observed. The dimer that we observed in solution most likely results from domain swapping at the helix region (Figure 3). Previously, the helix region was observed to be unstable in the folding intermediate of the RNH fragment (57), suggesting that the helix can be swapped during the folding process. Indeed, unfolding of the WT RNH monomer in 6 M urea followed by re-folding at very high protein concentration in NMR buffer (> 10 mM) yielded almost exclusively Fraction #1 (data not shown), in line with earlier observations for other domain swapped dimers (52).
Here, we demonstrate that BHMP07 binds to the monomer RNH but has little effect on the dimer (Figure 4), strongly supporting our earlier hypothesis that the substrate-handle region is involved in the inhibitor interaction (1). From combined in silico molecular docking results and in vitro NMR data, we predict that for WT RNH the preferred BHMP07 binding site is located at the juncture between the active site and the substrate-handle region, i.e., Site II (Figure 5B). This result, thus, refines our earlier predictions using p15-EC, in which BHMP07 was seen to bind to the substrate-handle region itself (1). Indeed, compared to the WT RNH, the p15-EC construct contains 24 additional residues, including three additional Trp, in the substrate-handle region. We speculate that these surplus hydrophobic residues distort the interaction site of p15-EC for BHMP07 and lead it to bind at the elongated substrate binding handle. This hypothesis is supported by the fact that BHMP07 binds to WT RNH with a KD approximately threefold lower than the value obtained for p15-EC, demonstrating that the binding mode discussed in the present study is the more favorable of the two.
In striking contrast to RNH inhibitors that exhibit NMR chemical shift changes in the presence of Mg2+, we find that BHMP07 causes significant perturbation of RNH signals only in the absence of Mg2+. Consequently, the BHMP07 binding mode that we propose in this and our previous study is clearly different from that of RNH active-site inhibitors, which requires metal coordination (22-25). As revealed by the observed chemical shift changes (Figure 4), BHMP07 also structurally affects residues in areas remote from the likely binding site (Figure 5), clearly demonstrating the allosteric modulation effect on the structure. Combined with the fact that BHMP07 was previously shown to present moderate anti-HIV activity (1), knowing this allosteric mechanism is critically important to future design of RT RNH inhibitors.
CONCLUSIONS
Although extensive screening has identified a variety of small molecular RNH inhibitors, little information about the structural basis of their interaction with RNH is known. Here, we report on a kinetically trapped dimer form of RNH in which the substrate-handle region provides the dimer interface. We also demonstrate that the substrate-handle region and the active site of a wild type RNH fragment are conjointly involved in the interaction with an acylhydrazone inhibitor, BHMP07. Our results support the presence of an inhibitor binding site between the substrate-handle region and the active site of RNH. This binding mechanism is significantly different from active site metal-mediated inhibitors, promoting an alternative line of attack for future RNH inhibitor design.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by grants from the National Institutes of Health (AI077424 to R.I. and M.A.P., and AI073975 to M.A.P.), the National Science Foundation (MCB 0814905 to R.I.), and funds from the University of Pittsburgh. We thank Wazo Myint and Maria DeLucia for technical assistance in this work and Teresa Brosenitsch for critical reading of the manuscript.
Abbreviations
- BHMP07
a trihydroxy acylhydrazide compound (1)
- Δδ
chemical shift perturbations (Hz)
sequence-averaged backbone chemical shift perturbation (Hz)
- HSQC
heteronuclear single-quantum correlated spectroscopy
- KD
protein – inhibitor equilibrium dissociation constant
- MALS
multi-angle light-scattering
- p15-EC
a catalytically active RNH chimeric construct (2)
- RNH
HIV-1 RT ribonuclease H domain
- RT
reverse transcriptase
- WT
wild type
Footnotes
SUPPORTING INFORMATION Supporting Information is available: Eight figures, including (1) the amino acid sequence of WT RT RNH used in this study, (2) Mg2+ effects on both monomer and dimer RNH, (3) overlays of 1H-15N HSQC spectra of RNH monomer at different BHMP07 concentrations and the titration curves in absence of 20 mM Mg+, (4) overlays of 1H-15N HSQC spectra of RNH monomer at different BHMP07 concentrations and the titration curves in presence of 20 mM Mg+, (5) histograms for the 1H chemical shift perturbations induced by BHMP07, Mg2+, or both, on the RNH monomer, (6) histograms for the 15N chemical shift perturbations induced by BHMP07, Mg2+, or both, on the RNH monomer, (7) diagram of the RNH residues predicted to surround BHMP07 in each of the three distinct docking sites, (8) per-residue interactions with BHMP07, as predicted for the minimum energy BHMP07 pose at Site II, and (9) a table of individual KD values for the interaction of RNH monomer with BHMP07.
REFERENCES
- 1.Gong Q, Menon L, Ilina T, Miller LG, Ahn J, Parniak MA, Ishima R. Interaction of HIV-1 Reverse Transcriptase Ribonuclease H with an Acylhydrazone Inhibitor. Chem Biol Drug Des. 2011;77:39–47. doi: 10.1111/j.1747-0285.2010.01052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keck JL, Marqusee S. Substitution of a highly basic helix/loop sequence into the RNase H domain of human immunodeficiency virus reverse transcriptase restores its Mn(2+)-dependent RNase H activity. Proc Natl Acad Sci U S A. 1995;92:2740–4. doi: 10.1073/pnas.92.7.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarafianos SG, Das K, Hughes SH, Arnold E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr Opin Struct Biol. 2004;14:716–30. doi: 10.1016/j.sbi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 4.De Clercq E. Non-nucleoside reverse transcriptase inhibitors (NNRTIs): past, present, and future. Chem Biodivers. 2004;1:44–64. doi: 10.1002/cbdv.200490012. [DOI] [PubMed] [Google Scholar]
- 5.Basavapathruni A, Anderson KS. Reverse transcription of the HIV-1 pandemic. FASEB J. 2007;21:3795–808. doi: 10.1096/fj.07-8697rev. [DOI] [PubMed] [Google Scholar]
- 6.Sluis-Cremer N, Tachedjian G. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Res. 2008;134:147–56. doi: 10.1016/j.virusres.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jochmans D. Novel HIV-1 reverse transcriptase inhibitors. Virus Res. 2008;134:171–85. doi: 10.1016/j.virusres.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Ilina T, Parniak MA. Inhibitors of HIV-1 reverse transcriptase. Adv Pharmacol. 2008;56:121–67. doi: 10.1016/S1054-3589(07)56005-9. [DOI] [PubMed] [Google Scholar]
- 9.Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–8. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 10.DeStefano JJ, Bambara RA, Fay PJ. Parameters that influence the binding of human immunodeficiency virus reverse transcriptase to nucleic acid structures. Biochemistry. 1993;32:6908–15. doi: 10.1021/bi00078a014. [DOI] [PubMed] [Google Scholar]
- 11.Cirino NM, Cameron CE, Smith JS, Rausch JW, Roth MJ, Benkovic SJ, Le Grice SF. Divalent cation modulation of the ribonuclease functions of human immunodeficiency virus reverse transcriptase. Biochemistry. 1995;34:9936–43. doi: 10.1021/bi00031a016. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh M, Williams J, Powell MD, Levin JG, Le Grice SF. Mutating a conserved motif of the HIV-1 reverse transcriptase palm subdomain alters primer utilization. Biochemistry. 1997;36:5758–68. doi: 10.1021/bi963045e. [DOI] [PubMed] [Google Scholar]
- 13.Loya S, Hizi A. The interaction of illimaquinone, a selective inhibitor of the RNase H activity, with the reverse transcriptases of human immunodeficiency and murine leukemia retroviruses. J Biol Chem. 1993;268:9323–8. [PubMed] [Google Scholar]
- 14.Palaniappan C, Fay PJ, Bambara RA. Nevirapine alters the cleavage specificity of ribonuclease H of human immunodeficiency virus 1 reverse transcriptase. J Biol Chem. 1995;270:4861–9. doi: 10.1074/jbc.270.9.4861. [DOI] [PubMed] [Google Scholar]
- 15.Borkow G, Fletcher RS, Barnard J, Arion D, Motakis D, Dmitrienko GI, Parniak MA. Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry. 1997;36:3179–85. doi: 10.1021/bi9624696. [DOI] [PubMed] [Google Scholar]
- 16.Budihas SR, Gorshkova I, Gaidamakov S, Wamiru A, Bona MK, Parniak MA, Crouch RJ, McMahon JB, Beutler JA, Le Grice SF. Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 2005;33:1249–56. doi: 10.1093/nar/gki268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw-Reid CA, Feuston B, Munshi V, Getty K, Krueger J, Hazuda DJ, Parniak MA, Miller MD, Lewis D. Dissecting the effects of DNA polymerase and ribonuclease H inhibitor combinations on HIV-1 reverse-transcriptase activities. Biochemistry. 2005;44:1595–606. doi: 10.1021/bi0486740. [DOI] [PubMed] [Google Scholar]
- 18.Tramontano E, Esposito F, Badas R, Di Santo R, Costi R, La Colla P. 6-[1-(4-Fluorophenyl)methyl-1H-pyrrol-2-yl)]-2,4-dioxo-5-hexenoic acid ethyl ester a novel diketo acid derivative which selectively inhibits the HIV-1 viral replication in cell culture and the ribonuclease H activity in vitro. Antiviral Res. 2005;65:117–24. doi: 10.1016/j.antiviral.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Schultz SJ, Champoux JJ. RNase H activity: structure, specificity, and function in reverse transcription. Virus Res. 2008;134:86–103. doi: 10.1016/j.virusres.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wendeler M, Lee HF, Bermingham A, Miller JT, Chertov O, Bona MK, Baichoo NS, Ehteshami M, Beutler J, O’Keefe BR, Götte M, Kvaratskhelia M, Le Grice S. Vinylogous ureas as a novel class of inhibitors of reverse transcriptase-associated ribonuclease H activity. ACS Chem Biol. 2008;3:635–44. doi: 10.1021/cb8001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirschberg TA, Balakrishnan M, Squires NH, Barnes T, Brendza KM, Chen X, Eisenberg EJ, Jin W, Kutty N, Leavitt S, Liclican A, Liu Q, Liu X, Mak J, Perry JK, et al. RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information. J Med Chem. 2009;52:5781–4. doi: 10.1021/jm900597q. [DOI] [PubMed] [Google Scholar]
- 23.Himmel DM, Maegley KA, Pauly TA, Bauman JD, Das K, Dharia C, Clark ADJ, Ryan K, Hickey MJ, Love RA, Hughes SH, Bergqvist S, Arnold E. Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site. Structure. 2009;17:1625–35. doi: 10.1016/j.str.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su HP, Yan Y, Prasad GS, Smith RF, Daniels CL, Abeywickrema PD, Reid JC, Loughran HM, Kornienko M, Sharma S, Grobler JA, Xu B, Sardana V, Allison TJ, Williams PD, et al. Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors. J Virol. 2010;84:7625–33. doi: 10.1128/JVI.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan J, Wu H, Tom T, Brodsky O, Maegley K. Targeting Divalent Metal Ions at the Active Site of the HIV-1 RNase H Domain: NMR Studies on the Interactions of Divalent Metal Ions with RNase H and Its Inhibitors. Am J Anal Chem. 2011;2:639–49. [Google Scholar]
- 26.Davies J.F.n., Hostomska Z, Hostomsky Z, Jordan SR, Matthews DA. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 27.Felts AK, Labarge K, Bauman JD, Patel DV, Himmel DM, Arnold E, Parniak MA, Levy RM. Identification of Alternative Binding Sites for Inhibitors of HIV-1 Ribonuclease H Through Comparative Analysis of Virtual Enrichment Studies. J Chem Inf Model. 2011;51:1986–98. doi: 10.1021/ci200194w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stahl SJ, Kaufman JD, Vikić-Topić S, Crouch RJ, Wingfield PT. Construction of an enzymatically active ribonuclease H domain of human immunodeficiency virus type 1 reverse transcriptase. Protein Eng. 1994;7:1103–8. doi: 10.1093/protein/7.9.1103. [DOI] [PubMed] [Google Scholar]
- 29.Shaw-Reid CA, Munshi V, Graham P, Wolfe A, Witmer M, Danzeisen R, Olsen DB, Carroll SS, Embrey M, Wai JS, Miller MD, Cole JL, Hazuda DJ. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J Biol Chem. 2003;278:2777–80. doi: 10.1074/jbc.C200621200. [DOI] [PubMed] [Google Scholar]
- 30.Ahn J, Byeon IJ, Dharmasena S, Huber K, Concel J, Gronenborn AM, Sluis-Cremer N. The RNA binding protein HuR does not interact directly with HIV-1 reverse transcriptase and does not affect reverse transcription in vitro. Retrovirology. 2010;7:40. doi: 10.1186/1742-4690-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavanagh J, Fairbrother WJ, Palmer AGI, Skelton NJ. Protein NMR Spectroscopy. Academic Press; San Diego: 1996. [Google Scholar]
- 32.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 34.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinás M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN Data Model for NMR Spectroscopy: Development of a Software Pipeline. Proteins: Structure, Function, and Bioinformatics. 2005;59:687–96. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 35.Bahrami A, Assadi AH, Markley JL, Eghbalnia HR. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Computational Biology. 2009;5:e1000307. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fielding L. NMR methods for the determination of protein–ligand dissociation constants. Prog NMR Spec. 2007;51:219–42. doi: 10.2174/1568026033392705. [DOI] [PubMed] [Google Scholar]
- 37.Webb C, Upadhyay A, Giuntini F, Eggleston I, Furutani-Seiki M, Ishima R, Bagby S. Structural features and ligand binding properties of tandem WW domains from YAP and TAZ, nuclear effectors of the Hippo pathway. Biochemistry. 2011;50:3300–9. doi: 10.1021/bi2001888. [DOI] [PubMed] [Google Scholar]
- 38.Himmel DM, Sarafianos SG, Dharmasena S, Hossain MM, McCoy-Simandle K, Ilina T, Clark ADJ, Knight JL, Julias JG, Clark PK, Krogh-Jespersen K, Levy RM, Hughes SH, Parniak MA, Arnold E. HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. ACS Chem Biol. 2006;1:702–12. doi: 10.1021/cb600303y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pari K, Mueller GA, DeRose EF, Kirby TW, London RE. Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry. 2003;42:639–50. doi: 10.1021/bi0204894. [DOI] [PubMed] [Google Scholar]
- 40.Mueller GA, Pari K, DeRose EF, Kirby TW, London RE. Backbone dynamics of the RNase H domain of HIV-1 reverse transcriptase. Biochemistry. 2004;43:9332–42. doi: 10.1021/bi049555n. [DOI] [PubMed] [Google Scholar]
- 41.Rejto PA, Verkhivker GM. Unraveling principles of lead discovery: from unfrustrated energy landscapes to novel molecular anchors. Proc Natl Acad Sci USA. 1996;93:8945–50. doi: 10.1073/pnas.93.17.8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCammon JA. Theory of biomolecular recognition. Curr Opin Struct Biol. 1998;8:245–9. doi: 10.1016/s0959-440x(98)80046-8. [DOI] [PubMed] [Google Scholar]
- 43.Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Freer ST, Rose PW. Complexity and simplicity of ligand-macromolecule interactions: the energy landscape perspective. Curr Opin Struct Biol. 2002;12:197–203. doi: 10.1016/s0959-440x(02)00310-x. [DOI] [PubMed] [Google Scholar]
- 44.Termiz NA, Bahar I. Inhibitor binding alters the directions of domain motions in HIV-1 reverse transcriptase. Proteins. 2002;49:61–70. doi: 10.1002/prot.10183. [DOI] [PubMed] [Google Scholar]
- 45.Seckler JM, Howard KJ, Barkley MD, Wintrode PL. Solution structural dynamics of HIV-1 reverse transcriptase heterodimer. Biochemistry. 2009;48:7646–55. doi: 10.1021/bi900790x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seckler JM, Barkley MD, Wintrode PL. Allosteric suppression of HIV-1 reverse transcriptase structural dynamics upon inhibitor binding. Biophys J. 2011;100:144–53. doi: 10.1016/j.bpj.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arico-Muendel CC, Patera A, Pochapsky TC, Kuti M, Wolfson AJ. Solution structure and dynamics of a serpin reactive site loop using interleukin 1beta as a presentation scaffold. Protein Eng. 1999;12:189–202. doi: 10.1093/protein/12.3.189. [DOI] [PubMed] [Google Scholar]
- 48.Ishima R, Torchia DA, Lynch SM, Gronenborn AM, Louis JM. Solution structure of the mature HIV-1 protease monomer: insight into the tertiary fold and stability of a precursor. J Biol Chem. 2003;278:43311–9. doi: 10.1074/jbc.M307549200. Epub 2003 Aug 21. [DOI] [PubMed] [Google Scholar]
- 49.Libonati M, Gotte G. Oligomerization of bovine ribonuclease A: structural and functional features of its multimers. Biochem J. 2004;380:311–27. doi: 10.1042/BJ20031922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 2005;437:266–9. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 51.Baek JH, Im H, Kang UB, Seong KM, Lee C, Kim J, Yu MH. Probing the local conformational change of alpha1-antitrypsin. Protein Sci. 2007;16:1842–50. doi: 10.1110/ps.072911607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gronenborn AM. Protein acrobatics in pairs--dimerization via domain swapping. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller KH, Karr JR, Marqusee S. A hinge region cis-proline in ribonuclease A acts as a conformational gatekeeper for C-terminal domain swapping. J Mol Biol. 2010;400:567–78. doi: 10.1016/j.jmb.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonnen AF, Yu C, Evans EJ, Stuart DI, Davis SJ, Gilbert RJ. Domain metastability: a molecular basis for immunoglobulin deposition? J Mol Biol. 2010;399:207–13. doi: 10.1016/j.jmb.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rong YW, Carl PL. On the molecular weight and subunit composition of calf thymus ribonuclease H1. Biochemistry. 1990;29:383–9. doi: 10.1021/bi00454a012. [DOI] [PubMed] [Google Scholar]
- 56.Lim D, Gregorio GG, Bingman C, Martinez-Hackert E, Hendrickson WA, Goff SP. Crystal structure of the moloney murine leukemia virus RNase H domain. J Virol. 2006;80:8379–89. doi: 10.1128/JVI.00750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kern G, Handel T, Tracy M, Marqusee S. Characterization of a folding intermediate from HIV-1 ribonuclease H. Protein Sci. 1998;7:2164–74. doi: 10.1002/pro.5560071014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanaya S, Katsuda-Nakai C, Ikehara M. Importance of the positive charge cluster in Escherichia coli ribonuclease HI for the effective binding of the substrate. J Bio Chem. 1991;266:11621–7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.