Abstract
Ricin A chain (RTA) depurinates the α-sarcin/ricin loop after it undergoes retrograde trafficking to the cytosol. The structural features of RTA involved in intracellular transport are not known. To explore this, we fused EGFP to precursor (preRTA-EGFP), containing a 35-residue leader, and mature RTA (matRTA-EGFP). Both were enzymatically active and toxic in S. cerevisiae. PreRTA-EGFP was localized in the endoplasmic reticulum (ER) initially and was subsequently transported to the vacuole, while mature RTA remained in the cytosol, indicating that ER localization is a prerequisite for vacuole transport. When the two glycosylation sites in RTA were mutated, mature form was fully active and toxic, suggesting that the mutations do not affect catalytic activity. However, nonglycosylated preRTA-EGFP had reduced toxicity, depurination and delayed vacuole transport, indicating that N-glycosylation affects transport of RTA out of the ER. Point mutations in the C-terminal hydrophobic region restricted RTA to the ER and eliminated toxicity and depurination, indicating that this sequence is critical for ER exit. These results demonstrate that N-glycosylation and the C-terminal hydrophobic region stimulate the toxicity of RTA by promoting ER export. The timing of depurination coincided with the timing of vacuole transport, suggesting that RTA may enter the cytosol during vacuole transport.
Keywords: Ricin, N-glycosylation, ribosome depurination, endoplasmic reticulum dislocation, vacuole transport
INTRODUCTION
Ricin, extracted from the castor bean (Ricinus communis), is a type 2 ribosome-inactivating protein (RIP) or an AB toxin, consisting of a catalytic A chain (RTA) linked to a cell binding B chain (RTB). RTA is an N-glycosidase, which specifically depurinates an adenine residue from the universally conserved α-sarcin/ricin loop (SRL) in the 28S rRNA, inhibiting protein synthesis (1). RTB is a lectin which specifically binds to cell surface glycolipids or glycoproteins with β-1,4-linked galactose residues and facilitates endocytosis of RTA (2). Ricin is one of the most toxic substances known and can cause severe morbidity and mortality. There are no specific protective measures or therapeutics effective against ricin intoxication and there is an urgent unmet need for therapy. In the castor bean, ricin is synthesized as a preproprotein with a 35-residue N-terminal leader, followed by the 267-residue mature RTA, which is connected to the 262-residue RTB by a 12-residue internal linker. Preproricin is translocated to the endoplasmic reticulum (ER) via the first 26-residue signal peptide in plants (3). After cleavage of the signal sequence in the ER, proricin is N-glycosylated, a disulfide bond is formed between RTA and RTB and within RTB (4). Proricin is then transported via the Golgi complex to the vacuole where the N-terminal 9-residue propeptide and the linker between RTA and RTB are processed to form mature ricin, which accumulates in protein storage vacuoles in the castor bean endosperm (5).
In mammalian cells, ricin is internalized either by clathrin-dependent or clathrin-independent mechanisms (2). After endocytosis, ricin is initially delivered to early endosomes. From there, it is either recycled back to the cell surface or delivered via late endosomes to lysosomes and is eventually degraded. Only a small portion of ricin follows the retrograde pathway from endosomes to the trans-Golgi network (TGN) (6). RTA is active only after it is separated from RTB in the ER by reduction of the disulfide bond (7). Reductive separation of RTA and RTB in the ER exposes a hydrophobic region at the C-terminus, which is proposed to mediate the association of RTA with the ER membrane (8, 9). RTA enters the cytosol by a process termed dislocation or retrotranslocation (10, 11). Dislocation is the most critical step in RTA trafficking, since RTA exerts its toxic effect on ribosomes after it enters the cytosol. RTA is thought to utilize the specific components of the ER associated degradation (ERAD) pathway for dislocation and has unique characteristics compared with other ERAD substrates (12). Since very few RTA molecules reach the Golgi and the ER in mammalian cells, RTA variants containing sulfation and glycosylation sites have been used to study retrograde transport (13). These studies identified components of the ERAD pathway, such as the ER degradation enhancing α–mannosidase I-like protein (EDEM), which participates in dislocation of RTA (14). Yeast has been used to probe the mechanism of dislocation of RTA by targeting enzymatically attenuated RTA variants to the ER lumen (15). A folding competent RTA variant required the Hrd1p (HMG-CoA reductase degradation-1 protein) for dislocation in yeast (15). Trafficking to the Golgi from the ER was also required for dislocation of the RTA variants in yeast (15). SEL1L, a regulator of the HRD complex, promoted dislocation of an enzymatically attenuated RTA variant to the cytosol in mammalian cells (16). Png1p, Htm1p and Yos9p were implicated in the degradation of a glycosylated structurally defective RTA variant, but were not required for efficient degradation of the nonglycosylated form (17). Some of these studies were carried out with enzymatically attenuated RTA variants, which may differ from wild type RTA in their requirements for dislocation, while others with structurally defective forms, which were unable to fold into an active conformation and appeared to act as bonafide ERAD substrates.
Structural features of wild type RTA involved in dislocation remain to be determined. Ricin contains four glycosylation sites, two on RTA and two on RTB. N-glycosylation occurs on asparagines 10 and 236 in the consensus Asn-X-Ser/Thr motifs in RTA (18) in the ER (13). The role of N-glycosylation in intracellular transport, in vivo depurination and cytotoxicity of RTA has not been examined. Recombinant nonglycosylated RTA is as active in inhibiting protein synthesis as plant derived glycosylated RTA in vitro (19), suggesting that glycosylation does not affect the catalytic activity of RTA. However, deglycosylated RTA is approximately one thousand fold less toxic than glycosylated RTA in mice and has been evaluated as a vaccine (20). Toxicity of ricin isoforms correlates with their glycosylation levels in immunotoxins targeted against cancer cells (21). Moreover, addition of a C-terminal glycosylation signal increases the toxicity of the recombinant RTA compared to the nonglycosylated form (13). These studies suggest a potential role for glycosylation in the toxicity of RTA. However, glycosylation was reported not to affect trafficking, since both glycosylated and nonglycosylated ricin precursor ultimately accumulated in the protein bodies in the castor bean (5).
In the present study, we examine the trafficking of wild type RTA in yeast using the EGFP tagged precursor, which contains the native N-terminal leader. We investigate the role of glycosylation in intracellular transport, depurination activity and cytotoxicity of wild type RTA. Our results show that RTA is transported to the ER initially and then to the vacuole. Mutation of the N-glycosylation sites does not affect the catalytic activity of RTA, but reduces toxicity and delays depurination and vacuole transport. These results demonstrate for the first time that N-glycosylation contributes to the toxicity of RTA by promoting more efficient transport out of the ER. Mutation of the C-terminal hydrophobic stretch prevents vacuole transport, depurination and cytotoxicity, indicating that this sequence is critical for ER exit.
RESULTS
The precursor form of RTA is transported from the ER to the vacuole in yeast
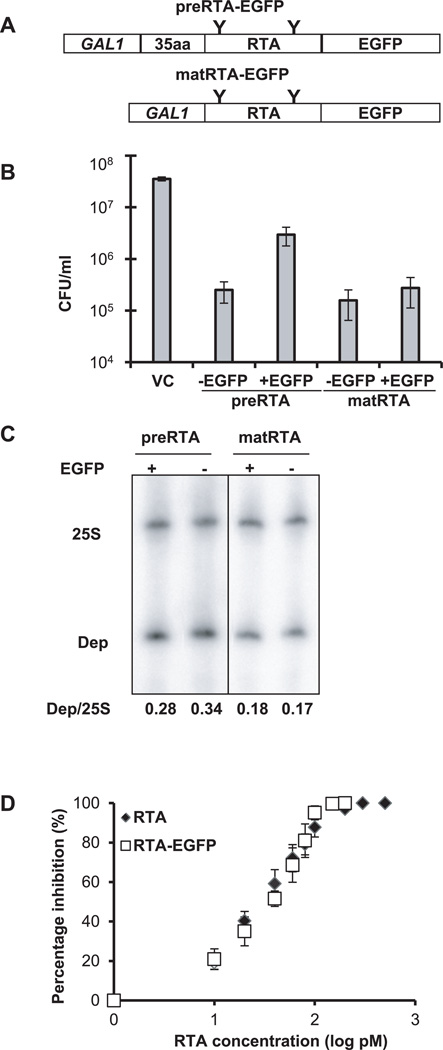
To examine the trafficking of RTA in vivo in yeast, we fused the enhanced green fluorescent protein (EGFP) to the C-terminal end of the precursor (preRTA-EGFP) or the mature form of RTA (matRTA-EGFP) (Figure 1A). The preRTA-EGFP contained the native 35-residue N-terminal leader followed by the mature RTA, while matRTA-EGFP did not contain the leader. To determine if the C-terminal EGFP fusions affected the toxicity of RTA, we examined the viability of yeast expressing preRTA or mature RTA with or without EGFP at 10 hours post induction (hpi) (Figure 1B). EGFP tagging reduced the toxicity of preRTA. However, preRTA-EGFP was still 12 times more toxic than the vector control (VC) (Figure 1B). EGFP tagging did not affect the toxicity of mature RTA. Both matRTA and matRTA-EGFP were about two orders of magnitude more toxic than VC. PreRTA and preRTA-EGFP were slightly less toxic than matRTA and matRTA-EGFP, respectively (Figure 1B). PreRTA and matRTA with the C-terminal EGFP tag had similar depurination activity as preRTA and matRTA without the EGFP tag by dual primer extension analysis (22) at 10 hpi (Figure 1C). Furthermore, recombinant matRTA-EGFP had similar activity (IC50 31.3 pM) as the recombinant matRTA without the EGFP tag (IC50 28.4 pM) by in vitro translation in the rabbit reticulocyte lysate (Figure 1D). Thus, the presence of the EGFP tag does not affect the depurination activity of RTA or its ability to inhibit translation, while causing some reduction in the toxicity of preRTA.
Figure 1. Cytotoxicity and enzymatic activity of RTA-EGFP fusions.
(A) Schematic representation of the C-terminal EGFP fusions with the precursor (preRTA-EGFP) and mature form of RTA (matRTA-EGFP). PreRTA contains the 35-residue leader and the 267-residue mature RTA, while mature RTA contains only the 267-residue RTA downstream of the GAL1 promoter. “Y” indicates the glycosylation sites. (B) Viability of yeast harboring the empty vector (VC), preRTA, preRTA-EGFP, mature RTA and mature RTA-EGFP. The CFU/ml was calculated based on the analysis of eleven different transformants at 10 hpi. (C) Ribosome depurination in yeast expressing the precursor and mature form of RTA with and without the EGFP tag. Total RNA isolated after 10 h of growth on galactose was analyzed by dual primer extension analysis using two different end-labeled primers, the depurination primer (Dep), which was used to measure the extent of depurination, and the 25S rRNA primer (25S), which was used to measure the total amount of 25S rRNA (22). The depurination was quantified by calculating the ratio between the intensity of depurination product (Dep) relative to the intensity of the 25S rRNA (25S) and is shown below each lane. (D) Translation inhibition by recombinant RTA-EGFP and recombinant RTA without EGFP. Purified RTA-EGFP and RTA were added to the Flexi Rabbit Reticulocyte Lysate System at 0, 10, 20, 40, 60, 80, 100, 200, 500, 1000 pM. The reaction was incubated at 30 °C for 30 min and the luciferase activity was measured. Data are the mean ± SE of at least 3 individual experiments.
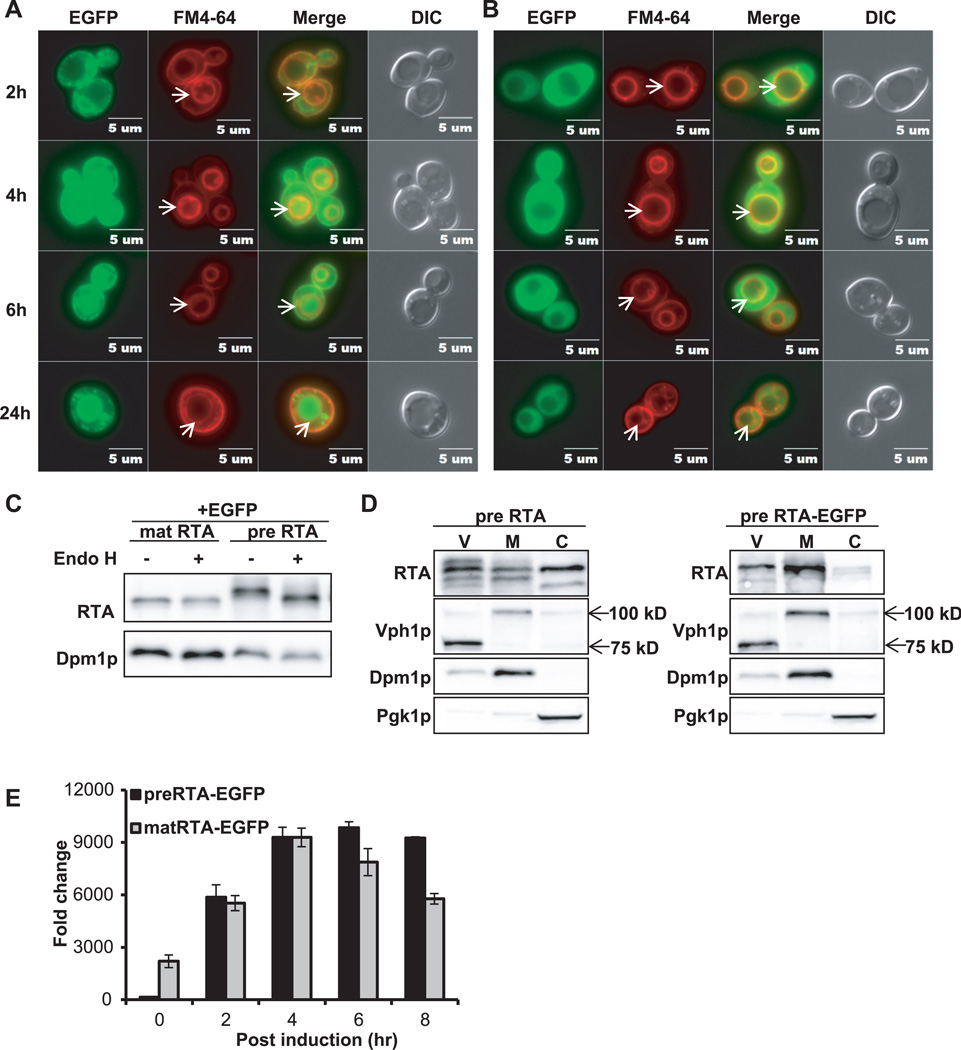
We examined the localization of preRTA-EGFP at various times after induction by epifluorescence microscopy. PreRTA-EGFP showed perinuclear localization at 2 hpi indicative of ER localization (Figure 2A). To determine if it co-localized with the ER, preRTA-EGFP and matRTA-EGFP were transformed into yeast harboring the ER marker, RFP-tagged Alg9 (Alg9-RFP) (23). Epifluorescence microscopy indicated that preRTA-EGFP co-localized with the ER marker, while matRTA-EGFP did not (Supplemental Figure S1). The perinuclear pattern was confirmed by indirect immunofluorescence with yeast harboring preRTA without the EGFP tag (Supplemental Figure S2). At 4 hpi, preRTA-EGFP co-localized with the vacuole in 40% of the cells (n=57) as indicated by co-localization with the red-fluorescent probe FM4-64, which stains the vacuole membranes (24) (Figure 2A). At 6 hpi, 88% (n=54) and at 24 hpi 100% (n=33) of preRTA-EGFP co-localized with the vacuole. In contrast, matRTA-EGFP remained in the cytosol at all time points (Figure 2B). Immunoblot analysis indicated that preRTA-EGFP migrated slower than matRTA-EGFP, suggesting that it was glycosylated (Figure 2C). In order to distinguish the glycosylated form from the nonglycosylated form, yeast membrane fraction was treated with Endoglycosidase H (Endo H), which cleaves the N-linked mannose-rich oligosaccharides (25). The slower migrating form changed in mobility after Endo H treatment (Figure 2C), indicating that it represented the N-glycosylated form of preRTA-EGFP. In contrast, matRTA-EGFP only had a single form, which did not change in mobility after EndoH treatment, indicating that it was not glycosylated (Figure 2C). These results confirmed that preRTA-EGFP was translocated into the ER and was glycosylated, while matRTA-EGFP remained in the cytosol and was not glycosylated. To confirm that vacuole localization was not an artifact of the EGFP tag, we isolated the vacuole fraction from cells expressing preRTA without EGFP and preRTA-EGFP at 14 hpi. Immunoblot analysis showed that both preRTA and preRTA-EGFP were present in the vacuole, as well as in the membrane and cytosol (Figure 2D). The vacuole fraction had some ER membrane contamination based on the signal from Dpm1p, the ER membrane marker. However, the amount of Dpm1p in the vacuole fraction was only 18% of that in the membrane fraction, while RTA expression level was similar in the two fractions (Figure 2D), suggesting that the presence of RTA in the vacuole fraction was not due to the ER contamination. The preRTA appeared as several forms in the vacuole fraction, which may be due to processing in the vacuole. The vacuole marker, Vph1p, encoding the Vacuolar H+-ATPase was 100-kDa in the membrane fraction, but 75-kDa in the vacuole fraction (Figure 2D), possibly due to proteolysis during vacuole isolation (26). These results demonstrate that the 35-residue N-terminal leader contains information for targeting RTA to the ER and from the ER to the vacuole in yeast and provide evidence that ER localization is a prerequisite for vacuolar transport.
Figure 2. Intracellular transport, protein expression and depurination activity of preRTA-EGFP and mature RTA-EGFP.
(A) Localization of preRTA-EGFP and (B) mature RTA-EGFP. Yeast cells were grown in SD medium with glucose and induced with galactose. The images were taken at 2, 4, 6 and 24 hpi with an Olympus BX41 fluorescence microscope. Yeast cells were treated with FM4-64 to stain the vacuole. Merged images show localization of each protein relative to the vacuole. The arrows indicate the vacuoles. (C) Immunoblot analysis of mature RTA-EGFP and preRTA-EGFP. Membrane fraction isolated at 6 hpi was treated with (+) or without (−) Endo H to cleave the glycans. The proteins (5 µg) were separated on a 10% SDS-polyacrylamide gel and probed with monoclonal anti-RTA (1:5000). The blot was reprobed with the ER membrane marker Dpm1p as a loading control. (D) Immunoblot analysis of preRTA and preRTA-EGFP in vacuole (V), membrane (M) and cytosol (C) fractions. The proteins (5 µg) were separated on a 10% SDS-polyacrylamide gel and probed with monoclonal anti-RTA (1:5000). The blot was reprobed with vacuole membrane marker Vph1p, ER membrane marker, Dpm1p and cytosol marker, Pgk1p. (E) Ribosome depurination by preRTA-EGFP and matRTA-EGFP in vivo by qRT-PCR. Yeast ribosomes were extracted at 0, 2, 4, 6 and 8 hpi. Two pairs of primers were designed to amplify the target amplicon (depurinated SRL) and the reference amplicon (25S rRNA) (27). The data was analyzed by the comparative ΔCT method (ΔΔCT). The y-axis indicates the fold change in depurination in yeast harboring the preRTA-EGFP and matRTA-EGFP over yeast harboring the empty vector. Data are mean ± SD from triplicates.
Ribosome depurination is a sensitive indicator of RTA transport to the cytosol. We examined the level of depurination over time using qRT-PCR (Figure 2E) (27). In this assay, two pairs of primers are used to amplify the target amplicon (depurinated SRL) and the reference amplicon (25S rRNA) and the data are analyzed by the comparative ΔCT method (ΔΔCT) (27). Depurination level of preRTA-EGFP was negligible at 0 hpi, but matRTA-EGFP had detectable depurination activity, which was 15-fold higher than preRTA-EGFP. However, at 2 hpi preRTA-EGFP had similar depurination level as matRTA-EGFP, suggesting that preRTA-EGFP dislocated to the cytosol at 2 hpi. The depurination level for both proteins reached a peak level at 4 hpi. At 6 and 8 hpi, the depurination level of preRTA-EGFP was higher than that of matRTA-EGFP. These results indicate that the depurination by preRTA-EGFP was delayed compared to matRTA-EGFP, possibly because preRTA-EGFP was transported to the ER. The delayed depurination of preRTA-EGFP was consistent with its lower cytotoxicity compared to matRTA-EGFP (Figure 1B).
In tobacco, the first 26 amino acids of the 35-residue leader of RTA act as the signal peptide (3), while the following 9 residues affect co-translational import and the extent of glycosylation (28). To determine if the N-terminal 26 and 9 amino acids function similarly in yeast, we deleted the first 26 residues (RTAΔ26-EGFP) and the subsequent 9 residues (RTAΔ9-EGFP) separately and constructed C-terminal EGFP fusions with each protein (Figure 3A). Epifluorescence microscopy showed that RTAΔ26-EGFP remained in the cytosol at all time points after induction (Figure 3B) as matRTA-EGFP (Figure 2B), indicating that the first 26 residues are required for ER targeting in yeast as in plants.
Figure 3. Intracellular transport, protein expression, cytotoxicity and depurination activity of RTAΔ26-EGFP and RTAΔ9-EGFP.
(A) Schematic representation of RTAΔ26-EGFP and RTAΔ9-EGFP. RTAΔ26-EGFP contains a deletion of the 26-residue signal peptide, but contains the 9-residue propeptide upstream of the mature RTA. RTAΔ9-EGFP contains the 26-residue signal peptide, but does not contain the 9-residue propeptide upstream of the mature RTA. (B) Intracellular transport of RTAΔ26-EGFP. Yeast cells expressing RTAΔ26-EGFP were grown in SD medium with glucose and induced with galactose. The images were taken at 2, 4, 6, 8 and 24 hpi with an Olympus BX41 fluorescence microscope. Cells were treated with FM4-64 to stain the vacuole. Merged images show localization of each protein relative to the vacuole. The arrows indicate the vacuoles. (C) Immunoblot analysis of membrane fraction isolated from cells expressing each protein at 6 hpi after treatment with (+) or without (−) Endo H to cleave the glycans. The proteins (5 µg) were separated on a 10% SDS-polyacrylamide gel and probed with monoclonal anti-RTA (1:5000). The blot was reprobed with the ER membrane marker Dpm1p as a loading control. (D) Viability of yeast expressing each protein and the vector control (VC). (E) Intracellular transport of RTAΔ9-EGFP was analyzed as in B. (F) Ribosome depurination by RTAΔ26-EGFP and matRTA-EGFP by qRT-PCR. (G) Ribosome depurination by RTAΔ9-EGFP and preRTA-EGFP by qRT-PCR. Data are mean ± SD from triplicates.
RTAΔ26-EGFP migrated as a single band on SDS-PAGE, which did not change in mobility after Endo H treatment (Figure 3C), indicating that it was not glycosylated. RTAΔ26-EGFP reduced viability about 50-fold compared to the vector control at 10 hpi, and was almost as toxic as matRTA-EGFP (Figure 3D). Comparison of the level of depurination in yeast expressing RTAΔ26-EGFP with matRTA-EGFP (Figure 3F) showed that RTAΔ26-EGFP caused lower level of depurination than matRTA-EGFP at 4 hpi, but similar level of depurination at later time points. These results suggested that the presence of the 9-residue propeptide in RTAΔ26-EGFP did not have a major effect on the depurination activity and toxicity.
RTAΔ9-EGFP co-localized with the ER at 2 and 4 hpi (Figure 3E), which was confirmed by transforming RTAΔ9-EGFP into yeast harboring an ER marker (Supplemental Figure S1). RTAΔ9-EGFP accumulated in the vacuole in 42% of the cells (n=78) at 6 hpi (Figure 3E), indicating that its vacuole transport was delayed. These results demonstrated that the 9-residue propeptide affected the transport of RTA to the vacuole. Immunoblot analysis (Figure 3C) showed that RTAΔ9-EGFP migrated as a single band and did not change in mobility after Endo H treatment, indicating that it was not glycosylated. RTAΔ9-EGFP migrated slower than RTAΔ26-EGFP on SDS-PAGE (Figure 3C), suggesting incomplete cleavage of the signal peptide. Viability analysis showed that the cytotoxicity of RTAΔ9-EGFP was similar to preRTA-EGFP (Figure 3D). RTAΔ9-EGFP and preRTA-EGFP were more viable than RTAΔ26-EGFP and matRTA-EGFP at 10 hpi, suggesting that trafficking of RTAΔ9-EGFP and preRTA-EGFP to the ER delays depurination and reduces toxicity.
RTAΔ9-EGFP depurinated 1.7–fold less than preRTA-EGFP at 2 hpi, but reached a similar level at 4 hpi (Figure 3G). Subcellular fractionation at 6 hpi indicated that RTAΔ26-EGFP was present in the cytosol and membrane fraction and did not change in mobility after Endo H treatment (Supplemental Figure S3A). In contrast, almost all of RTAΔ9-EGFP was associated with the membrane fraction and did not change in mobility after Endo H treatment (Supplemental Figure S3A). These results demonstrate that the first 26 residues of the N-terminal leader mediate ER import, while the remaining 9 residues influence ER import, glycosylation and vacuole transport of RTA.
Glycosylation of preRTA in the ER affects trafficking beyond the ER
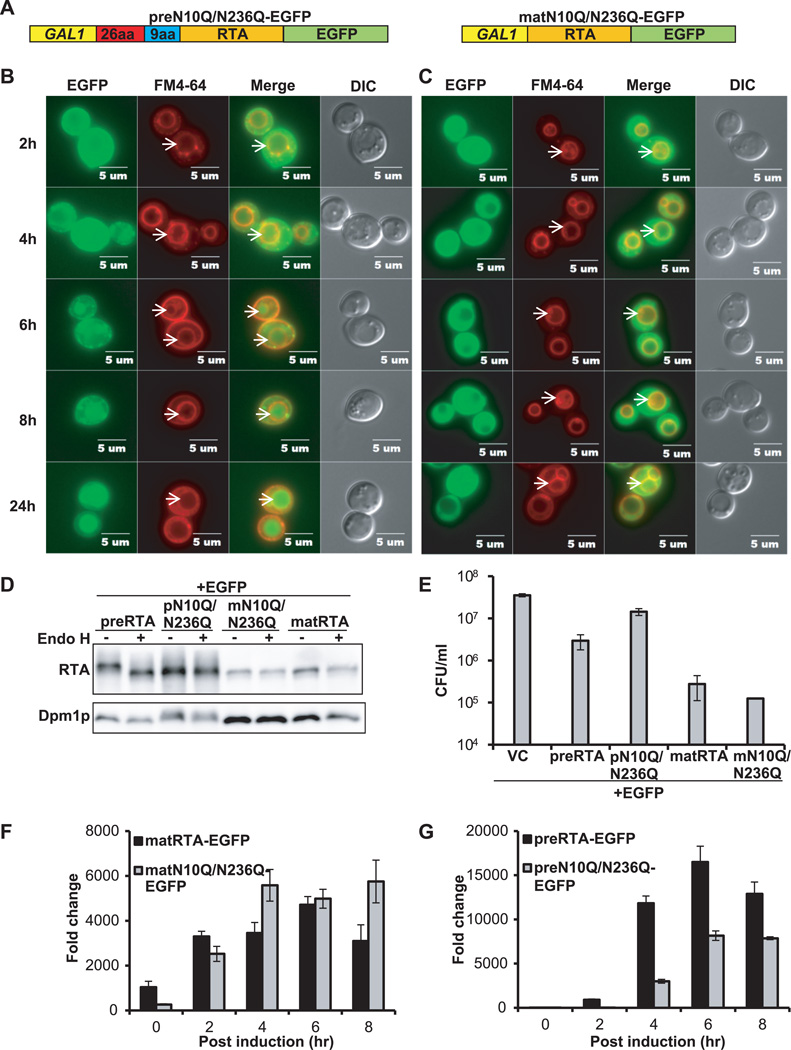
The correlation observed between the absence of glycosylation and delayed vacuole transport of RTAΔ9-EGFP suggested that glycosylation might be important for transport of RTA out of the ER. To address this, we mutated the two glycosylation sites in RTA by substituting Asn10 and Asn236 with Gln (N10Q/N236Q). The C-terminal EGFP fusions were constructed with the precursor (preN10Q/N236Q-EGFP) and the mature (matN10Q/N236Q-EGFP) form (Figure 4A). As shown in Figure 4B, preN10Q/N236Q-EGFP was localized in the ER at 2 hpi and was detected in the vacuole in 45% of the cells (n=51) at 6 hpi, two hours later than wild type preRTA-EGFP (Figure 2A). Discrete foci were observed in cells expressing preN10Q/N236Q-EGFP (Figure 4B), possibly due to aggregation of the nonglycosylated preN10Q/N236Q-EGFP in the ER. The matN10Q/N236Q-EGFP remained in the cytosol at all time points (Figure 4C). Immunoblot analysis (Figure 4D) showed that preN10Q/N236Q-EGFP did not change in mobility after Endo H treatment, demonstrating the lack of glycosylation. The matN10Q/N236Q-EGFP comigrated with wild type matRTA-EGFP and did not change in mobility after EndoH treatment, consistent with cytosolic localization.
Figure 4. Intracellular transport, protein expression, cytotoxicity and depurination activity of the glycosylation mutants.
(A) Schematic representation of preN10Q/N236Q-EGFP and matN10Q/N236Q-EGFP. Glycosylation sites at Asn10 and Asn236 were replaced with glutamine in both preRTA-EGFP and mature RTA-EGFP. (B) Trafficking of preN10Q/N236Q-EGFP and (C) matN10Q/N236Q-EGFP. Yeast cells expressing each construct were grown in SD medium with glucose and induced with galactose. The images were taken at 2, 4, 6, 8 and 24 hpi with an Olympus BX41 fluorescence microscope. Vacuoles were stained with FM4-64. Merged images show localization of each protein relative to the vacuole. The arrows indicate the vacuoles. (D) Immunoblot analysis of membrane fraction isolated from cells expressing each protein at 6 hpi after treatment with (+) or without (−) Endo H to cleave the glycans. Protein (5 µg) was separated on a 10% SDS-polyacrylamide gel and probed with monoclonal anti-RTA (1:5000). The blot was reprobed with the ER membrane marker Dpm1p as a loading control. (E) Viability of yeast expressing each protein and the vector control (VC). (F) Ribosome depurination in yeast expressing matN10Q/N236Q-EGFP compared to matRTA-EGFP by qRT-PCR. (G) Ribosome depurination in yeast expressing preN10Q/N236Q-EGFP compared to preRTA-EGFP by qRT-PCR. Data are mean ± SD from triplicates.
The viability analysis (Figure 4E) indicated that matN10Q/N236Q-EGFP had similar viability as wild type matRTA-EGFP. In contrast, preN10Q/N236Q-EGFP was more viable than wild type preRTA-EGFP. A similar increase in viability was observed when viability of untagged preN10Q/N236Q was compared to untagged wild type preRTA (Supplemental Figure S4A). These results confirmed that the increase in viability of the preN10Q/N236Q-EGFP relative to wild type preRTA-EGFP was not due to the EGFP tag. Since transport of preN10Q/N236Q-EGFP to the vacuole was delayed in the absence of glycans, we further determined if the depurination activity was also affected. The depurination level of matN10Q/N236Q-EGFP and matRTA-EGFP was similar (Figure 4F). In contrast, depurination level of preN10Q/N236Q-EGFP was considerably lower than wild type preRTA-EGFP at all time points (Figure 4G). Depurination by preN10Q/N236Q-EGFP could not be detected until 4 hpi and the level was about one fourth of that observed with wild type preRTA-EGFP (Figure 4G). To confirm that this was not due to the EGFP tag, we examined depurination by the glycosylation mutants without the EGFP tag. Untagged matN10Q/N236Q depurinated at a similar level as untagged wild type mature RTA (Supplemental Figure S5A), whereas depurination by untagged preN10Q/N235Q was inhibited and reached only about half the depurination level of the untagged wild type preRTA at 4 hpi (Supplemental Figure S5B). These results demonstrate that the mutation of the glycosylation sites does not reduce the catalytic activity of RTA. However, lack of glycosylation reduces cytotoxicity by impairing depurination.
Subcellular fractionation indicated that only the glycosylated form of wild type preRTA-EGFP was observed in the ER, since it migrated faster after the Endo H treatment (Supplemental Figure S3A). The glycosylated form and a less abundant, faster migrating form were detected in the cytosol. Since RTA may be deglycosylated during dislocation (16), the less abundant form may represent the deglycosylated RTA. In contrast, preN10Q/N236Q-EGFP was primarily associated with the membrane fraction (Supplemental Figure S3A). A very low level of this mutant was detected in the cytosol and it migrated faster than the membrane associated fraction, suggesting that the 9-residue propeptide was processed. Since this processing step occurs in the vacuole (5), the faster migrating form might be released from the vacuole during the fractionation. The immunoblot analysis suggested that reduced depurination of preN10Q/N236Q-EGFP was not due to lower level of expression, since expression level of preRTA-EGFP and preN10Q/N236Q-EGFP was similar in the ER membrane (Figure 4D). These results indicate that glycosylation promotes transport of RTA out of the ER.
The C-terminal hydrophobic region affects transport of RTA beyond the ER
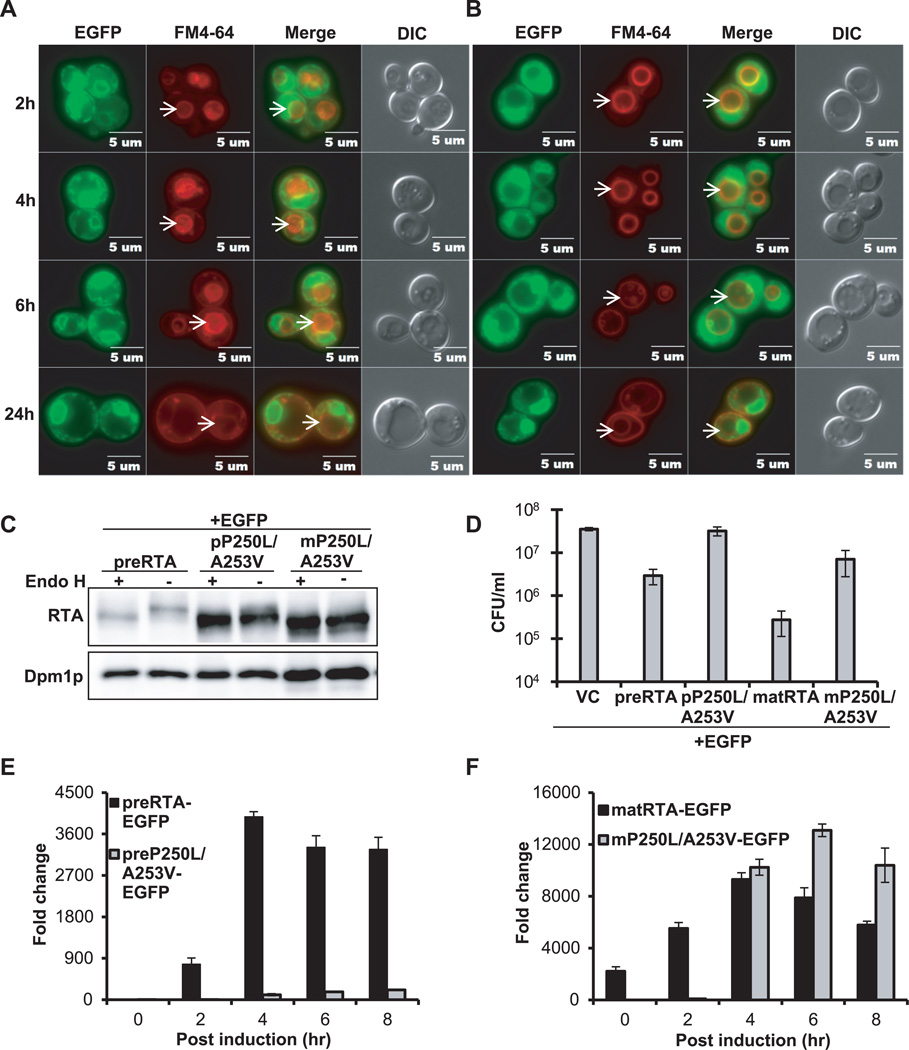
Reduction of the disulfide bond between RTA and RTB in the ER exposes a hydrophobic sequence at the C-terminus of RTA (Val245-Val256), which was proposed to mediate the association of RTA with the ER membrane (29). Mutations in this region do not affect catalytic activity of RTA in vitro, but reduce toxicity when the mutated RTA is reassociated with RTB (29). We previously showed that a double mutation, P250L/A253V, in this region eliminated depurination activity and cytotoxicity of RTA in yeast (30). To examine the trafficking of this mutant, we constructed C-terminal EGFP fusions with the precursor and the mature form of P250L/A253V and examined their localization using epifluorescence microscopy. The preP250L/A253V-EGFP showed only perinuclear localization at all time points up to 24 hpi and did not accumulate in the vacuole (Figure 5A). The matP250L/A253V-EGFP was observed in the ER and cytosol at 2 hpi and accumulated in the ER after 2 hpi. At 24 hpi it showed only perinuclear localization (Figure 5B). Both preP250L/A253V-EGFP and matP250L/A253V-EGFP co-localized with the ER marker at 6 hpi (Supplemental Figure S1). ER localization of preP250L/A253V was confirmed by indirect immunofluorescence (Supplemental Figure S2).
Figure 5. Intracellular transport, protein expression, cytotoxicity and depurination activity of preP250L/A253V-EGFP and mature P250L/A253V-EGFP.
(A) Trafficking of preP250L/A253V-EGFP and (B) matP250L/A253V-EGFP in yeast. The images were taken at 2, 4, 6 and 24 hpi with Olympus BX41 fluorescence microscope. Merged images show localization of each protein relative to the vacuole. The arrows indicate the vacuoles. (C) Immunoblot analysis of membrane fraction isolated from cells expressing each protein at 6 hpi were treated with (+) or without (−) Endo H to cleave the glycans. The proteins (5 µg) were separated on a 10% SDS-polyacrylamide gel and probed with monoclonal anti-RTA (1:5000). The blot was reprobed with the ER membrane marker Dpm1p as a loading control. (D) Viability of yeast expressing each protein and the vector control (VC). (E) Ribosome depurination by preP250L/A253V-EGFP and preRTA-EGFP by qRT-PCR. (F) Ribosome depurination by matP250L/A253V-EGFP and matRTA-EGFP by qRT-PCR. Data are mean ± SD from triplicates.
Immunoblot analysis (Figure 5C) showed that preP250L/A253V-EGFP migrated as a single band with a weak slower migrating band, which disappeared after Endo H treatment, suggesting that it was glycosylated. The matP250L/A253V-EGFP migrated as a single band, and did not change in mobility after Endo H treatment, indicating that it was not glycosylated.
The viability of cells expressing preP250L/A253V-EGFP was similar to the viability of cells harboring the vector (Figure 5D). Yeast expressing matP250L/A253V-EGFP were slightly less viable than preP250L/A253V-EGFP, but were over 25-fold more viable than yeast expressing wild type matRTA-EGFP. The depurination level of preP250L/A253V-EGFP was barely detectable throughout the time course and 36-fold lower than that of the wild type preRTA-EGFP at 4 hpi (Figure 5E). Depurination by matP250L/A253V-EGFP was not detectable at 0 and 2 hpi, but the depurination level increased at 4 hpi and reached the highest level at 6 hpi (Figure 5F). Since matP250L/A253V-EGFP depurinated ribosomes, it was slightly more toxic than preP250L/A253V-EGFP (Figure 5D). However, it was less toxic than wild type matRTA-EGFP, possibly due to the delay in depurination during the early stages of induction. Cell fractionation analysis confirmed that preP250L/A253V-EGFP was associated primarily with the membrane fraction and migrated slightly faster after Endo H treatment (Supplemental Figure S3B). In contrast, matP250L/A253V-EGFP was equally distributed between the membrane and the cytosolic fraction, as wild type matRTA-EGFP (31) and did not change in mobility after Endo H treatment. These results demonstrate that the C-terminal hydrophobic stretch is critical for ER exit.
DISCUSSION
RTA is transported from the ER to the vacuole in yeast
In this study, we used EGFP tagged wild type preRTA to examine the time course of intracellular transport and carried out mutation analysis to reveal structural features important for trafficking to the cytosol. We show that plant derived native N-terminal leader sequence directs RTA to the ER membrane in yeast. PreRTA was transported from the ER to the vacuole, while mature RTA without the N-terminal leader stayed in the cytosol. These results demonstrate that RTA is transported from the ER to the vacuole and ER import is necessary for vacuole transport.
To rule out possible artifacts of ER and vacuole localization due to the EGFP tag, we confirmed ER localization by indirect immunofluorescence using untagged preRTA in fixed cells (Supplemental Figure S2). Vacuole localization was confirmed by subcellular fractionation (Figure 2D), since vacuoles did not maintain balanced osmolarity and were ruptured during fixation in indirect immunofluorescence (32). These results demonstrated that the EGFP tag does not alter the localization of RTA. The viability of preRTA-EGFP was slightly higher than preRTA, while mature RTA had similar viability with or without the EGFP tag (Figure 1B), possibly because the C-terminal EGFP tag caused a delay in ER export. Similarly, the viability of EGFP tagged preN10Q/N236Q was slightly higher than the untagged form, while the EGFP tagged or untagged mature forms had similar viability (Supplemental Figure S4B). Nevertheless, preRTA-EGFP reduced viability of yeast compared to the vector control 12-fold (Figure 1B) and depurinated ribosomes as preRTA without the EGFP tag in vivo (Figure 1C), indicating that EGFP tagging is a useful strategy to study the time course of intracellular transport of RTA.
Since very little RTA gets into the cytosol, and is impossible to visualize, we used a very sensitive qRT-PCR assay we developed to quantify ribosome depurination (27) as an indicator of RTA trafficking to the cytosol. Ribosomes are depurinated and growth is severely inhibited when wild type preRTA-EGFP is transported to the cytosol in yeast. Mature form of RTA-EGFP was used as a control to detect changes in catalytic activity in vivo. Since mature form is localized in the cytosol, changes observed in depurination would be due to its catalytic activity, and not to trafficking. Deletion analysis indicated that only the first 26 residues of the 35-residue N-terminal leader were necessary and sufficient for ER targeting of RTA in yeast as in plants (3). RTA without the 9-residue propeptide was unprocessed, nonglycosylated and was not imported to the ER in tobacco protoplasts (28). However, although in tobacco protoplasts the 9-residue propeptide did not influence vacuolar targeting or the rate of dislocation of RTA to the cytosol (28), we show here that deletion of the 9-residue propeptide delayed vacuole transport and ribosome depurination. These results suggested that N-glycosylation may promote ER export.
Glycosylation promotes transport of RTA out of the ER
Analysis of the glycosylation mutants indicated that vacuole transport was delayed when the glycosylation sites were mutated in preRTA-EGFP, suggesting that N-glycosylation affected transport of RTA from the ER to the vacuole. While mature form of the glycosylation mutant was fully active, ribosome depurination by precursor form was reduced 4-fold compared to wild type at 4 hpi, suggesting that the ability to traffic to the cytosol was also impaired in nonglycosylated preN10Q/N236Q-EGFP. Consistent with the depurination data, toxicity of nonglycosylated preN10Q/N236Q-EGFP was reduced over 5-fold compared to wild type preRTA-EGFP, while the mature form had similar toxicity as wild type matRTA-EGFP (Figure 4E). Similar results were obtained using the precursor and mature forms of non EGFP-tagged glycosylation mutants (Supplemental Figure S4A). Subcellular fractionation indicated that preRTA-EGFP lacking N-glycans was associated primarily with the membrane fraction. These results demonstrate that N-glycosylation stimulates the cytotoxicity of RTA by promoting its transport to the cytosol.
A previous study concluded that glycosylation is not required for intracellular trafficking, since nonglycosylated proricin ultimately accumulated in the protein bodies as the N-glycosylated precursor in the castor bean (5). However, this study did not examine the time course of transport. We demonstrate here that although both N-glycosylated and nonglycosylated RTA are transported to the vacuole, the transport of the nonglycosylated RTA is delayed. This delay appears to be significant, since it results in markedly reduced cytotoxicity. These results suggest that glycosylation is not required for correct targeting of RTA, but allows more efficient intracellular transport to the cytosol and to the vacuole. The timing of trafficking to the cytosol, measured by depurination, coincides with the timing of vacuole transport, indicating that the two processes are coupled. These results suggest that RTA may enter the cytosol during transport through the secretory pathway to the vacuole. This finding is significant because up to now the ERAD pathway is thought to be the only pathway for dislocation of RTA from the ER to the cytosol. We present the first evidence here that RTA may enter the cytosol during transport to the vacuole, suggesting that this may be an alternative mechanism to dislocation from the ER.
Glycosylation in the ER increases hydrophilicity and promotes protein folding (33–35). Discrete foci were observed in yeast expressing the nonglycosylated preN10Q/N236Q-EGFP, but not wild type preRTA-EGFP, suggesting that the nonglycosylated preN10Q/N236Q-EGFP likely aggregated in the ER. Inhibition of glycosylation activates the unfolded protein response (UPR) and triggers ERAD (33, 36, 37). Activation of ERAD prevents toxicity of ricin by reducing its dislocation (14). These results suggest that RTA may need to be properly folded to exit the ER. N-glycosylation may promote folding of RTA in the ER to allow efficient transport out of the ER. However, N-glycosylation is not only important for folding of RTA in the ER, since fully folded deglycosylated RTA is less toxic than glycosylated RTA in mice (20).
The pathway that leads to degradation of RTA in the cytosol is unclear. ER import was shown to be a prerequisite for the degradation of a folding competent RTA variant (15). However, this variant was not degraded by the proteasome core and its disappearance in a yeast strain lacking vacuolar proteinase A was similar to that in the wild type (15). These results do not exclude the possibility that vacuolar turnover may be important for degradation of RTA. Our results suggest that ER to vacuole trafficking may reduce cytotoxicity of RTA, possibly by promoting its degradation. Hence, the vacuole may be an alternative compartment for RTA turnover.
C-terminal hydrophobic sequence is critical for transport of RTA out of the ER
Our previous results showed that a double mutation in a hydrophobic sequence at the C-terminus of RTA, P250L/A253V, eliminated toxicity and depurination activity of RTA (30). The P250A mutation alone affected the secondary structure of RTA, reducing its interaction with EDEM1, and transport from the ER to the cytosol in mammalian cells (38). We show here that preP250L/A253V-EGFP was restricted to the ER and was not transported to the vacuole. Ribosome depurination and toxicity of preP250L/A253V-EGFP were almost completely inhibited compared to wild type preRTA-EGFP, suggesting that it was not able to dislocate to the cytosol. Subcellular fractionation confirmed that this mutant was mainly localized in the membrane fraction. These results indicate that the C-terminal hydrophobic region allows ER exit.
The matP250L/A253V-EGFP was also associated with the ER membrane, even though it did not contain the signal sequence. The matP250L/A253V-EGFP was not glycosylated and thus had not reached the ER by translocation. Toxicity of matP250L/A253V-EGFP was reduced compared to wild type matRTA-EGFP. Depurination was delayed, possibly because the C-terminal mutations allowed insertion of matP250L/A253V-EGFP into the ER membrane, but prevented the release of this mutant from the membrane and inhibited its folding to the native conformation. This mutant likely depurinated ribosomes on the ER membrane, thereby delaying depurination and reducing toxicity.
Glycosylation stimulates toxicity by increasing the rate of transport of RTA into the cytosol
We previously showed that ribosome depurination alone is not sufficient for the cytotoxicity of RTA (30). Factors that contribute to cytotoxicity other than depurination have not yet been identified. We show here that cytotoxicity depends on the timing of depurination. A high depurination level during the early stages of induction implies a high rate of depurination, inducing extensive cell death and degradation of damaged ribosomes. Thus, very little depurinated rRNA accumulates in yeast expressing highly active RTA. Yeast expressing matRTA or matRTA-EGFP were less viable than yeast expressing preRTA or preRTA-EGFP, possibly due to a higher level of depurination during the early stages of induction. A low level of depurination at the early stages of induction indicates a low rate of depurination. The depurinated rRNA accumulates with time and a higher level of depurination is detected at the later stages of induction since cells do not die. Therefore, a high level of depurination at the later stages of induction does not suggest high catalytic activity.
Depurination is a dynamic process regulated by the rate of dislocation of RTA and its enzymatic activity. The rate of depurination is determined by the rate of dislocation to the cytosol, since dislocation allows exposure of RTA to cytosolic ribosomes. Our results indicate that glycosylation does not affect the catalytic activity of RTA, but affects cytotoxicity by increasing the rate of transport to the cytosol. Although the lack of glycosylation delayed transport to the cytosol by only two hours, it significantly reduced toxicity. Our results are consistent with other studies, which showed that glycosylation is required for efficient intracellular transport of carboxypeptidase Y to the vacuole in yeast (39) and the rate of transport of barley lectin to vacuoles in plants (40).
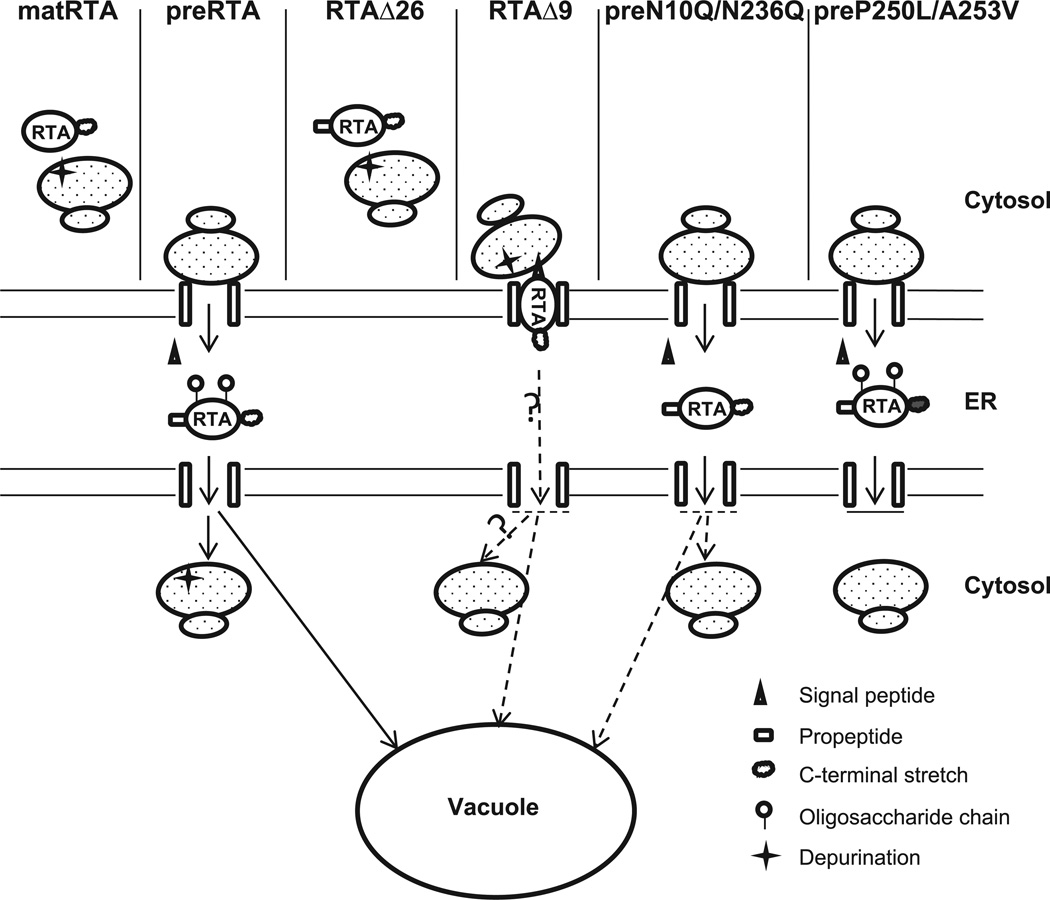
We propose a model that explains our findings (Figure 6). The mature RTA without the 35-residue leader remains in the cytosol and depurinates ribosomes in the cytosol. The preRTA is translocated to the ER, the signal sequence is cleaved and it is N-glycosylated. The N-glycosylated RTA is transported to the vacuole. Some of it enters the cytosol and depurinates ribosomes. Since RTAΔ26 lacks the signal sequence, it remains in the cytosol and depurinates cytosolic ribosomes. RTAΔ9 is targeted to the ER after synthesis, but remains on the cytosolic face of the ER membrane and depurinates membrane-bound ribosomes. Some of this protein might be translocated into the ER, but it is not glycosylated and vacuole transport is delayed. The preN10Q/N236Q is translocated to the ER and the signal sequence is processed. It is not glycosylated in the ER. It causes a delay in depurination because ER-cytosol and ER-vacuole transport are delayed. The preP250L/A253V is targeted to the ER after synthesis, and some of it is glycosylated. However, it is not able to exit the ER due to point mutations in the C-terminal hydrophobic sequence. Consequently, it causes a markedly lower level of depurination and is not toxic.
Figure 6. A model describing intracellular transport of wild type and mutant forms of RTA.
Wild type mature RTA does not contain a 35-residue leader and stays in the cytosol and depurinates ribosomes. Wild type preRTA is targeted to the ER after synthesis, where the signal peptide is cleaved. The preRTA is transported from the ER to the vacuole. Some of it gets into the cytosol and depurinates ribosomes. RTAΔ26 does not contain a signal peptide and remains in the cytosol and depurinates ribosomes. RTAΔ9 is targeted to the ER after synthesis. It may tether to the cytosolic face of the ER membrane and depurinate membrane-bound ribosomes. Some of it may be translocated into the ER lumen. However, it is not glycosylated and the vacuole transport is delayed. The preN10Q/N236Q is targeted to the ER where the signal peptide gets processed. It is not glycosylated in the ER. The depurination in the cytosol is markedly lower because its transport to the cytosol is impaired and its transport to the vacuole is delayed. PreP250L/A253V is targeted to the ER after synthesis. The signal peptide gets processed and some of it gets glycosylated. However, its transport beyond the ER is inhibited.
In summary, our results provide the first evidence that N-glycosylation stimulates toxicity by increasing the rate of transport of RTA to the cytosol. N-glycosylation also affects vacuole transport of RTA, suggesting that it appears to be the driving force for transport of RTA out of the ER. Point mutations in the C-terminal hydrophobic stretch prevent the ability of RTA to get out of the ER and eliminate toxicity and depurination, indicating that this sequence is necessary for ER exit. Our results suggest that RTA may enter the cytosol during transport to the vacuole. These results contribute to our mechanistic understanding of the intracellular transport of ricin and suggest that altering this pathway may have implications for the development of antidotes and vaccines to combat ricin intoxication.
MATERIALS AND METHODS
Plasmids and yeast strains
The following cDNAs were fused with EGFP tag at the 3’ end and cloned into yeast vector containing the LEU2 marker (NT198) downstream of the galactose-inducible GAL1 promoter. Wild type preRTA contains a 35-residue signal sequence, followed by 267-residue RTA (preRTA-EGFP, NT1205); wild type mature RTA consists of 267-residue RTA (matRTA-EGFP, NT1206). The first 26 residues of the 35-residue leader was deleted using primer pairs 5’-CCTCTAGAGTCGAGGATGTGGTCTTTCACATTAGAGG-3’ and 5’-TAAGAATTCAAACTGTGACGATGGTGGAGG-3’(RTAΔ26-EGFP, NT1493). The 9-residue propeptide was deleted by site directed mutagenesis using the primer 5’-GGATCCACCTCAGGGATATTCCCCAAACAATACC-3’ (RTAΔ9-EGFP, NT1494). The glycosylation mutants were generated by substituting Gln at Asn10 and Asn236 using 5’-CCAAACAATACCCAATTATACAATTTACCACAGCGGGTGCC-3' and 5'-CCAATTCAACTGCAAAGACGTCAAGGTTCCAAATTCAGTGTG-3'(preN10Q/N236Q-EGFP, NT1497; matN10Q/N236Q-EGFP, NT1500). The C-terminal mutants contained cDNA with point mutations P250L and A253V (preP250L/A253V-EGFP, NT1253; matP250L/A253V-EGFP, NT1259). The cDNA corresponding to precursor and mature RTA was cloned into NT198 downstream of the GAL1 promoter to generate preRTA and matureRTA without the EGFP tag (preRTA, NT849; matRTA, NT1456) (30). The plasmids were then transformed into Saccharomyces cerevisiae strain W303 (MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100), and transformants were selected on SD-Leu medium containing 2% glucose. The yeast strain, PMY147, containing the ER marker Alg9-RFP (MATa trp1-Δ 901 leu2-3,112 his3-Δ200 ura3-52 lys2-801 suc2-Δ9 can1∷hisG ALG9∷RFP, TRP1) was a gift from Dr. Peter Mayinger. PreRTA-EGFP, mature RTA-EGFP, RTAΔ9-EGFP, preN10Q/N236Q-EGFP, preP250L/A253V-EGFP and matP250L/A253V-EGFP were transformed into PMY147 and transformants were selected on SD-Leu,Trp medium containing 2% glucose.
Analysis of protein expression
Yeast cells were grown in SD-Leu supplemented with 2% glucose overnight and then transferred to SD-Leu supplemented with 2% galactose at OD600 of 0.3 to induce RTA expression. Cells were collected at 6 hpi and membrane fractions were isolated as previously described (41). Membrane fractions were treated with Endo H using the manufacturer’s protocol (New England Biolabs, Ipswich, MA). The protein samples were separated on a 10% SDS-polyacrylamide gel and the blot was probed with monoclonal anti-RTA (1:5000), a gift of Dr. Nicholas J. Mantis. The blot was stripped with 8M guanidine hydrochloride and reprobed with antibody against the ER maker, dolichol phosphate mannose synthase (Dpm1p; Invitrogen, Eugene, Oregon) (1:1000) and developed using ChemiDoc MP imaging system (Bio-Rad, Philadelphia, PA).
Subcellular fractionation
Subcellular fractionation was carried out as previously described (42). Membrane fraction was collected at 6 or 14 hpi by centrifuging at 16,000 × g for 15 min. The supernatant was further centrifuged at 200,000 × g for 1 h to pellet the ribosomes. The membrane fraction and the post ribosomal supernatant were treated with or without Endo H and analyzed by immunoblot analysis using monoclonal anti-RTA followed by anti-Dpm1p as described above. The blot was reprobed using anti-3-phosphoglycerate kinase (Pgk1p; Invitrogen, Eugene, Oregon), as a marker for the cytosol. The vacuole fraction was isolated as described (43). Briefly, cells harvested at 14 hpi were washed with DTT solution (0.1 M Tris pH9.4, 10mM DTT) and lysed by lyticase at 20 U/OD in spheroplasting buffer (0.16 × YPD, 0.4 M sorbitol, 50 mM potassium phosphate, pH 7.5). The spheroplasts were treated with DEAE-Dextran for gentle lysis. Purified vacuoles were obtained by flotation in a 0, 4, 8, 15% Ficoll step gradient. The gradients were centrifuged at 110,000 × g for 90 min in a Beckman L8-70M Ultracentrifuge (Beckman Coulter, Brea, CA) and vacuoles were collected from the 0%–4% interface. The vacuole marker was anti-H+-ATPase (V-ATPase; Invitrogen, Eugene, Oregon) (1:500). The band intensity was quantified using Quantity One software (Bio-Rad, Philadelphia, PA).
Viability analysis
Yeast were induced as described above and 8 µL of a series of 10-fold dilutions (OD600 of 10−1 to 10−5) were plated on SD-Leu plates containing 2% glucose at 0, 4, 6 and 10 hpi. Plates were incubated at 30°C for 48 h.
Depurination analysis
For dual primer extension, total yeast RNA from W303 expressing precursor and mature forms of RTA with and without EGFP tag was isolated at 10 hpi and hybridized using a depurination primer and a 25S control primer, as described previously (22). Extension products were separated on a 7M urea-5% polyacrylamide gel and quantified using a PhosphorImager.
The qRT-PCR analysis of depurination was carried out as previously described (27, 44). Cells were harvested at 0, 2, 4, 6 and 8 hpi and total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA). Total RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). The 25S rRNA was detected using (5’-AGA CCG TCG CTT GCT ACA AT-3’ and 5’- ATG ACG AGG CAT TTG GCT AC- 3’). The depurinated rRNA was detected using the forward primer (5’- CTA TCG ATC CTT TAG TCC CTC-3’) and the reverse primer (5’- CCG AAT GAA CTG TTC CAC A-3’). Real time PCR was performed using an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Carlsbad, CA). The data was analyzed by the comparative ΔCT (ΔΔCT) method for quantification (27).
Live cell imaging
Time course of RTA localization was carried out using yeast harboring the precursor and mature RTA with EGFP tag at 2, 4, 6, 8 and 24 hpi. The cells were directly added to 2% agar pads on slides. For ER co-localization, yeast harboring the ER marker, Alg9-RFP was visualized at 6hpi. For vacuole stain, yeast cells were harvested at 2, 4, 6, 8 and 24 hpi. FM4-64 (Invitrogen, Carlsbad, CA) dissolved in dimethyl sulfoxide (DMSO) was added at a final concentration of 80 µM and cells were incubated in the dark at 30°C for 60 min. Cells were pelleted and washed with YPD media and resuspended in YPD media to chase for 40 min at 30°C. The cell culture was applied to the agar pad and visualized using an Olympus BX41 fluorescence microscope equipped with a CCD camera (Hamamatsu, Bridgewater, NJ) and a 100X oil objective (1.45 N.A. Plan Apo, Olympus). Image acquisition and processing were performed using Metamorph Image Software (7.0; MDS Analytical Technologies). Data analysis was done by pooling images of expressed cells from 3 individual experiments.
Indirect immunofluorescence
Yeast cells harboring preRTA, preN10Q/N236Q and preP250L/A253V were grown in SD-Leu medium supplemented with 2% glucose and induced for RTA production with SD-Leu supplemented with 2% galactose. The cells were collected at 6 hpi and fixed with 4% paraformaldehyde by adding directly to the culture medium and incubated at 30°C for 30 min. The cells were spheroplasted by lyticase and incubated at 30°C for 30 min. The cells were then attached to the 0.1% poly-L-lysine coated Teflon slides. The cells were incubated with primary monoclonal antibody against RTA and then incubated with Alexa Fluor 488 goat anti-mouse antibody (Invitrogen, Carlsbad, CA). DAPI was applied at a concentration of 1µg/ml for 1 min at room temperature to stain the nuclei. The slides were mounted and sealed for observation under an Olympus BX41 fluorescence microscope equipped with a CCD camera (Hamamatsu, Bridgewater, NJ).
In vitro translation
Purified recombinant mature RTA-EGFP and recombinant mature RTA from E. coli (45) were added to the Flexi rabbit reticulocyte lysate system (Promega, Madison, WI) at 0, 10, 20, 40, 60, 80, 100, 200, 500 and 1000 pM according to the manufacturer’s protocol. The reactions were incubated at 30°C for 30 min and luciferase activity was measured using a luminometer. Data were collected from at least 3 individual experiments.
Supplementary Material
ACKNOWLEGEMENTS
We would like to thank Dr. Peter Mayinger for providing yeast strains harboring the ER marker, Dr. Nicholas Mantis for providing monoclonal antibody against RTA, Dr. Michael Pierce for constructing NT1456, Drs. Barth Grant, Jennifer Nielsen Kahn, Kerrie May and John McLaughlin for helpful comments. This work was supported by National Institutes of Health Grants AI072425 and AI077805 to NET.
Abbreviations used
- EDEM1
ER degradation enhancing α-mannosidase I-like protein
- EGFP
enhanced green fluorescent protein
- Endo H
Endoglycosidase H
- ER
endoplasmic reticulum
- ERAD
ER associated degradation
- hpi
hour post-induction
- matRTA
mature form of ricin A chain
- preRTA
precursor form of ricin A chain
- RTA
ricin A chain
- RTB
ricin B chain
- SRL
α-sarcin/ricin loop
REFERENCES
- 1.Endo Y, Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 2.Sandvig K, Grimmer S, Lauvrak SU, Torgersen ML, Skretting G, van Deurs B, Iversen TG. Pathways followed by ricin and Shiga toxin into cells. Histochem Cell Biol. 2002;117:131–141. doi: 10.1007/s00418-001-0346-2. [DOI] [PubMed] [Google Scholar]
- 3.Halling KC, Halling AC, Murray EE, Ladin BF, Houston LL, Weaver RF. Genomic cloning and characterization of a ricin gene from Ricinus communis. Nucleic Acids Res. 1985;13:8019–8033. doi: 10.1093/nar/13.22.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lappi DA, Kapmeyer W, Beglau JM, Kaplan NO. The disulfide bond connecting the chains of ricin. Proc Natl Acad Sci USA. 1978;75:1096–1100. doi: 10.1073/pnas.75.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lord JM. Precursors of ricin and Ricinus communis agglutinin. Glycosylation and processing during synthesis and intracellular transport. Eur J Biochem. 1985;146:411–416. doi: 10.1111/j.1432-1033.1985.tb08667.x. [DOI] [PubMed] [Google Scholar]
- 6.Van Deurs B, Tonnessen TI, Petersen OW, Sandvig K, Olsnes S. Routing of internalized ricin and ricin conjugates to the Golgi complex. J Cell Biol. 1986;102:37–47. doi: 10.1083/jcb.102.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright HT, Robertus JD. The intersubunit disulfide bridge of ricin is essential for cytotoxicity. Arch Biochem Biophys. 1987;256:280–284. doi: 10.1016/0003-9861(87)90447-4. [DOI] [PubMed] [Google Scholar]
- 8.Bellisola G, Fracasso G, Ippoliti R, Menestrina G, Rosen A, Solda S, Udali S, Tomazzolli R, Tridente G, Colombatti M. Reductive activation of ricin and ricin A-chain immunotoxins by protein disulfide isomerase and thioredoxin reductase. Biochem Pharmacol. 2004;67:1721–1731. doi: 10.1016/j.bcp.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Spooner RA, Watson PD, Marsden CJ, Smith DC, Moore KA, Cook JP, Lord JM, Roberts LM. Protein disulphide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem J. 2004;383:285–293. doi: 10.1042/BJ20040742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wesche J, Rapak A, Olsnes S. Dependence of ricin toxicity on translocation of the toxin A-chain from the endoplasmic reticulum to the cytosol. J Biol Chem. 1999;274:34443–34449. doi: 10.1074/jbc.274.48.34443. [DOI] [PubMed] [Google Scholar]
- 11.Gillece P, Pilon M, Romisch K. The protein translocation channel mediates glycopeptide export across the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2000;97:4609–4614. doi: 10.1073/pnas.090083497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spooner RA, Lord JM. How ricin and Shiga toxin reach the cytosol of target cells: retrotranslocation from the endoplasmic reticulum. Current Topics in Microbiology and Immunology. 2012;357:19–40. doi: 10.1007/82_2011_154. [DOI] [PubMed] [Google Scholar]
- 13.Rapak A, Falnes PO, Olsnes S. Retrograde transport of mutant ricin to the endoplasmic reticulum with subsequent translocation to cytosol. Proc Natl Acad Sci USA. 1997;94:3783–3788. doi: 10.1073/pnas.94.8.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slominska-Wojewodzka M, Gregers TF, Walchli S, Sandvig K. EDEM is involved in retrotranslocation of ricin from the endoplasmic reticulum to the cytosol. Mol Biol Cell. 2006;17:1664–1675. doi: 10.1091/mbc.E05-10-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Spooner RA, Allen SC, Guise CP, Ladds G, Schnoder T, Schmitt MJ, Lord JM, Roberts LM. Folding-competent and folding-defective forms of ricin A chain have different fates after retrotranslocation from the endoplasmic reticulum. Mol Biol Cell. 2010;21:2543–2554. doi: 10.1091/mbc.E09-08-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redmann V, Oresic K, Tortorella LL, Cook JP, Lord M, Tortorella D. Dislocation of ricin toxin A chains in human cells utilizes selective cellular factors. J Biol Chem. 2011;286:21231–21238. doi: 10.1074/jbc.M111.234708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosomi A, Tanabe K, Hirayama H, Kim I, Rao H, Suzuki T. Identification of an Htm1 (EDEM)-dependent, Mns1-independent Endoplasmic Reticulum-associated Degradation (ERAD) pathway in Saccharomyces cerevisiae: application of a novel assay for glycoprotein ERAD. J Biol Chem. 2010;285:24324–24334. doi: 10.1074/jbc.M109.095919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rutenber E, Katzin BJ, Ernst S, Collins EJ, Mlsna D, Ready MP, Robertus JD. Crystallographic refinement of ricin to 2.5 Å. Proteins. 1991;10:240–250. doi: 10.1002/prot.340100308. [DOI] [PubMed] [Google Scholar]
- 19.Schlossman D, Withers D, Welsh P, Alexander A, Robertus J, Frankel A. Role of glutamic acid 177 of the ricin toxin A chain in enzymatic inactivation of ribosomes. Mol Cell Biol. 1989;9:5012–5021. doi: 10.1128/mcb.9.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soler-Rodriguez AM, Uhr JW, Richardson J, Vitetta ES. The toxicity of chemically deglycosylated ricin A-chain in mice. Int J Immunopharmacol. 1992;14:281–291. doi: 10.1016/0192-0561(92)90041-i. [DOI] [PubMed] [Google Scholar]
- 21.Sehgal P, Kumar O, Kameswararao M, Ravindran J, Khan M, Sharma S, Vijayaraghavan R, Prasad GB. Differential toxicity profile of ricin isoforms correlates with their glycosylation levels. Toxicology. 2011;282:56–67. doi: 10.1016/j.tox.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Parikh BA, Coetzer C, Tumer NE. Pokeweed antiviral protein regulates the stability of its own mRNA by a mechanism that requires depurination but can be separated from depurination of the α-sarcin/ricin loop of rRNA. J Biol Chem. 2002;277:41428–41437. doi: 10.1074/jbc.M205463200. [DOI] [PubMed] [Google Scholar]
- 23.Faulhammer F, Konrad G, Brankatschk B, Tahirovic S, Knodler A, Mayinger P. Cell growth-dependent coordination of lipid signaling and glycosylation is mediated by interactions between Sac1p and Dpm1p. J Cell Biol. 2005;168:185–191. doi: 10.1083/jcb.200407118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 26.Kane PM, Kuehn MC, Howald-Stevenson I, Stevens TH. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H+-ATPase. J Biol Chem. 1992;267:447–454. [PubMed] [Google Scholar]
- 27.Pierce M, Kahn JN, Chiou J, Tumer NE. Development of a quantitative RT-PCR assay to examine the kinetics of ribosome depurination by ribosome inactivating proteins using Saccharomyces cerevisiae as a model. RNA. 2011;17:201–210. doi: 10.1261/rna.2375411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolliffe NA, Di Cola A, Marsden CJ, Lord JM, Ceriotti A, Frigerio L, Roberts LM. The N-terminal ricin propeptide influences the fate of ricin A-chain in tobacco protoplasts. J Biol Chem. 2006;281:23377–23385. doi: 10.1074/jbc.M602678200. [DOI] [PubMed] [Google Scholar]
- 29.Simpson JC, Lord JM, Roberts LM. Point mutations in the hydrophobic C-terminal region of ricin A chain indicate that Pro250 plays a key role in membrane translocation. Eur J Biochem. 1995;232:458–463. doi: 10.1111/j.1432-1033.1995.tb20831.x. [DOI] [PubMed] [Google Scholar]
- 30.Li XP, Baricevic M, Saidasan H, Tumer NE. Ribosome depurination is not sufficient for ricin-mediated cell death in Saccharomyces cerevisiae. Infect Immun. 2007;75:417–428. doi: 10.1128/IAI.01295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parikh BA, Tortora A, Li XP, Tumer NE. Ricin inhibits activation of the unfolded protein response by preventing splicing of the HAC1 mRNA. J Biol Chem. 2008;283:6145–6153. doi: 10.1074/jbc.M707981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulholland J, Botstein D. Immunoelectron microscopy of aldehyde-fixed yeast cells. In: Guthrie C, Fink GR, editors. Methods Enzymol. Vol. 351. Elsevier Inc. Press; 2002. pp. 50–81. [DOI] [PubMed] [Google Scholar]
- 33.Lehrman MA. Oligosaccharide-based information in endoplasmic reticulum quality control and other biological systems. J Biol Chem. 2001;276:8623–8626. doi: 10.1074/jbc.R100002200. [DOI] [PubMed] [Google Scholar]
- 34.Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 36.Spiro RG. Glucose residues as key determinants in the biosynthesis and quality control of glycoproteins with N-linked oligosaccharides. J Biol Chem. 2000;275:35657–35660. doi: 10.1074/jbc.R000022200. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee S, Vishwanath P, Cui J, Kelleher DJ, Gilmore R, Robbins PW, Samuelson J. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci USA. 2007;104:11676–11681. doi: 10.1073/pnas.0704862104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokolowska I, Walchli S, Wegrzyn G, Sandvig K, Slominska-Wojewodzka M. A single point mutation in ricin A-chain increases toxin degradation and inhibits EDEM1-dependent ER retrotranslocation. Biochem J. 2011;436:371–385. doi: 10.1042/BJ20101493. [DOI] [PubMed] [Google Scholar]
- 39.Winther JR, Stevens TH, Kielland-Brandt MC. Yeast carboxypeptidase Y requires glycosylation for efficient intracellular transport, but not for vacuolar sorting, in vivo stability, or activity. Eur J Biochem. 1991;197:681–689. doi: 10.1111/j.1432-1033.1991.tb15959.x. [DOI] [PubMed] [Google Scholar]
- 40.Wilkins TA, Bednarek SY, Raikhel NV. Role of propeptide glycan in post-translational processing and transport of barley lectin to vacuoles in transgenic tobacco. Plant Cell. 1990;2:301–313. doi: 10.1105/tpc.2.4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parikh BA, Baykal U, Di R, Tumer NE. Evidence for retro-translocation of pokeweed antiviral protein from endoplasmic reticulum into cytosol and separation of its activity on ribosomes from its activity on capped RNA. Biochemistry. 2005;44:2478–2490. doi: 10.1021/bi048188c. [DOI] [PubMed] [Google Scholar]
- 42.Baykal U, Tumer NE. The C-terminus of pokeweed antiviral protein has distinct roles in transport to the cytosol, ribosome depurination and cytotoxicity. Plant J. 2007;49:995–1007. doi: 10.1111/j.1365-313X.2006.03012.x. [DOI] [PubMed] [Google Scholar]
- 43.Cabrera M, Ungermann C. Purification and in vitro analysis of yeast vacuoles. In: Klionsky DJ, editor. Methods Enzymol. Vol. 451. Elsevier Inc. Press; 2008. pp. 177–196. [DOI] [PubMed] [Google Scholar]
- 44.Chiou JC, Li XP, Remacha M, Ballesta JP, Tumer NE. Shiga toxin 1 is more dependent on the P proteins of the ribosomal stalk for depurination activity than Shiga toxin 2. Int J Biochem & Cell Biol. 2011;43:1792–1801. doi: 10.1016/j.biocel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li XP, Chiou JC, Remacha M, Ballesta JP, Tumer NE. A two-step binding model proposed for the electrostatic interactions of ricin A chain with ribosomes. Biochemistry. 2009;48:3853–3863. doi: 10.1021/bi802371h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.