Abstract
This study documents the spatial and temporal distribution of Oct-4, Cdx-2 and acetylated H4K5 (H4K5ac) by immunocytochemistry staining using in-vivo-derived rabbit embryos at different stages: day-3 compact morulae, day-4 early blastocysts, day-4 expanded blastocysts, day-5 blastocysts, day-6 blastocysts and day-7 blastocysts. The Oct-4 signal was stronger in the inner cell mass (ICM)/epiblast cells than in the trophectoderm (TE) cells in all blastocyst stages except day-4 expanded blastocysts, where the signal was similarly weak in both the ICM and TE cells. The Cdx-2 signal was first detected in a small number of TE cells of day-4 early blastocysts, and became evident in the TE cells exclusively afterwards. A consistently strong H4K5ac signal was observed in the TE cells in all blastocyst stages examined. In particular, this signal was stronger in the TE than in the ICM cells in day-4 early blastocysts, day-4 expanded blastocysts and day-5 blastocysts. Double staining of H4K5ac with either Oct-4 or Cdx-2 on embryos at different blastocyst stages confirmed these findings. This work suggests that day 4 is a critical timing for lineage formation in rabbit embryos. A combination of Oct-4, Cdx-2 and H4K5ac can be used as biomarkers to identify different lineage cells in rabbit blastocysts.
Keywords: Cdx-2, embryos, H4K5ac, in vivo, Oct-4, rabbit
Introduction
Laboratory rabbits (Oryctolagus cuniculus) have been long used in biochemical research as valuable experimental models for various human diseases (Fan and Watanabe, 2003). Scientists have attempted to derive authentic rabbit embryonic stem cells and inducible pluripotent stem cells, but with little success (Honda et al., 2010; Honda et al., 2008; Honda et al., 2009; Intawicha et al., 2009; Wang et al., 2008; Wang et al., 2007). There are, in fact, very limited studies on key transcription factors and epigenetic programming events in preimplantation-stage rabbit embryos. Some fundamental knowledge, such as the expression profile of caudal-like transcription factor (Cdx-2) in different lineage cells in rabbit embryos, is not available as yet (Chen et al., 2012).
Recently, Chen et al. (2012) documented the spatial and temporal distribution of the Oct-4 protein and acetylated H4K5 (H4K5ac) using in-vitro-cultured rabbit embryos. The Oct-4 signal was detected in both the inner cell mass (ICM) and the trophectoderm (TE) cells, a pattern similar to that in human embryos, but different from that in mouse embryos, supporting the argument that rabbit could serve as a better model than mice for human embryology and stem cell studies. Chen et al. (2012) was the first work from this study centre, and one of the few reported works towards the understanding of the genetic and epigenetic events during early embryo development in rabbits.
As a continuation of the previous work, the aim of the present study was to determine the dynamics of several key genetic and epigenetic markers, such as Oct-4, Cdx-2 and H4K5ac using in-vivo-derived rabbit embryos.
The germline-specific protein, POU-domain transcription factor Oct-4 (also known as POU5F1), plays a critical role for early embryo development and pluripotent lineage formation (Scholer et al., 1990). It is regarded as the most important factor to maintain the pluripotency of stem cell lines, such as embryonic stem cells, inducible pluripotent stem cells, embryonic carcinoma cells and embryonic germ cells. Loss of its expression almost always results in differentiation (Pardo et al., 2010). In mouse blastocysts, Oct-4 protein appears to only be expressed in the ICM, but not in the TE (Palmieri et al., 1994; Yeom et al., 1996; Pesce et al., 1998; Ovitt and Schöler, 1998). The previous work found that Oct-4 protein is differently expressed in in-vitro-cultured rabbit embryos, showing detectable signals in both ICM and TE cells throughout the blastocyst stages (Chen et al., 2012). The expression pattern of Oct-4 in different lineage cells (i.e. of the ICM and the TE) of in-vivo-derived rabbit blastocysts is not available yet.
Cdx-2 is one of the earliest transcription factors and is essential for the maintenance of TE development in mammalian embryos, as demonstrated in mice, cattle and primates (Berg et al., 2011; Sritanaudomchai et al., 2009; Wu et al., 2010). It is restrictedly expressed in the TE lineage cells of pre- and early post-implantation embryos (Beck et al., 1995; Strumpf et al., 2005). Using Cdx-2 knockout and knockdown models, researchers found that although this protein might not be responsible for the initiation of TE lineage specification, lack of Cdx-2 expression caused low mitochondrial activity and abnormal ultrastructure of TE cells, which further impaired TE function and ceased embryo development (Meissner and Jaenisch, 2006; Sritanaudomchai et al., 2009; Strumpf et al., 2005; Wu et al., 2010). Surprisingly, as far as is known, no one has reported the expression pattern of Cdx-2 in rabbit embryos.
Epigenetic marks are involved in mammalian development by stabilizing gene expression and regulating cell lineage formation (Corry et al., 2009). Among epigenetic machinery, acetylated lysine residues of histone H4 play important roles in regulating gene expression by governing the access and binding activity of transcription factors and RNA polymerases to higher-order chromatin (Adenot et al., 1997; Lee et al., 1993; Tse et al., 1998). When histone H4 is hyperacetylated in an active genome, it often shows the ultimate acetylation at residue K5 (O’Neill and Turner, 1995; Thorne et al., 1990; Turner and Fellows, 1989). The acetylation of H4K5 hence reflects the hyperacetylation of histone H4 and correlates with the transcription activities of the associated genes (Grunstein, 1997; Turner, 1998). Chen et al. (2012) reported the dynamics of H4K5ac in in-vitro-cultured rabbit embryos. As far as is known, no group has reported the dynamics of H4K5ac in in-vivo-derived rabbit embryos.
The present study collected in-vivo-derived rabbit embryos 3–7 days post insemination (dpi). The immunostaining method was used to examine the temporal and spatial expression profiles of Oct-4 and Cdx-2 proteins, as well as the acetylation pattern of histone H4 at lysine 5 in these embryos. Furthermore, double staining of H4K5ac with either Oct-4 or Cdx-2 was conducted to investigate the temporal and spatial relationship of these three factors in rabbit blastocysts.
Materials and methods
All chemicals are purchased from Sigma Chemical Co. (St. Louis, MO, USA), unless otherwise indicated.
Animal maintenance and hormone administration
All animal maintenance, care and use procedures were reviewed and approved by the Institutional Animal Care and Use Committee of the National Taiwan University. Sexually mature (6–18 months old) New Zealand White female rabbits were maintained under a 12/12 light/dark cycle and superovulated with hormones using a routine regime (Du et al., 2009), consisting of two 0.3 mg, two 0.4 mg and two 0.6 mg injections of FSH (Folltropin-V; Bioniche Animal Health Canada, Belleville, Ontario, Canada) at intervals of 12 h, followed by 200 IU human chorionic gonadotrophin (HCG; Chorulon; IntervetInc, Millsboro, DE, USA). Superovulated rabbits were mated with fertile males and served as embryo donors.
Embryo collection
Dulbecco’s phosphate-buffered saline (DPBS; 15240-013; Gibco, Grand Island, NY, USA) containing 0.1% polyvinyl alcohol (P-8136) was used for flushing embryos from oviducts or uteri. Medium 199 with Earle’s salts, l-glutamine, 2.2 g/l sodium bicarbonate and 25 mmol/l HEPES (12340-014; Gibco) supplemented with 10% fetal bovine serum (SH0070.03; Hyclone, Logan, UT, USA) was used as the standard manipulation medium. In-vivo-derived embryos were collected on 3, 4, 5, 6 and 7 dpi, respectively.
Immunostaining of rabbit embryos
As described previously (Chen et al., 2012), the embryos were first fixed with fresh paraformaldehyde. After washing in DPBS for 10 min, permeabilization was achieved by treatment of 0.5% Triton-X 100 for 15–30 min and washing in 0.25% DPBS/Tween 20 (PBST) for 30 min at room temperature (20–25°C). Treatment with DPBS supplemented with 2% bovine serum albumin for 1 h at room temperature was used to block nonspecific binding sites. Immunostaining of Oct-4 and Cdx-2 was performed by incubation of the embryos with primary antibodies, i.e. Oct-4 (monoclonal, 1:150, MAB4401; Millipore, Billerica, MA, USA) or Cdx-2 (monoclonal, 1:200, CDX-2-88; BioGenex, San Ramon, CA, USA), at 4°C overnight followed by washing in PBST at room temperature. Embryos were incubated with the secondary antibody (Alexa Fluor 488 goat anti-mouse IgG, 1:500, A11029; Invitrogen, Carlsbad, CA, USA) for 1 h at 37°C. Immunostaining of H4K5ac was performed by incubation of the embryos with the primary antibody for H4K5ac (monoclone, 1:250, ab51997; Abcam, Cambridge Science Park, Cambridge, UK) at room temperature for 1.5 h. Incubation with the secondary antibody (Alexa Fluor 594 donkey anti-rabbit IgG, 1:500, A21207; Invitrogen) was performed for 1 h at 37°C. Double staining of Oct-4/H4K5ac or Cdx-2/H4K5ac was performed by first processing the staining of Oct-4 or Cdx-2, followed by processing the staining of H4K5ac on the same embryos. Finally, the embryos were washed and stained for DNA with 100 ng/ml 4(,6-diamidino-2-phenylindole) (DAPI; D9564) for 10 min and mounted on slides with 50% glycerol in DPBS.
Image processing and quantitation
Image sections of individual embryos stained with DAPI, Oct-4, Cdx-2 or H4K5ac were observed and captured by laser scanning confocal microscopy (Olympus IX71 with UltraVIEW confocal software; PerkinElmer, Covina, California, USA). Volocity version 5.3.1 (Improvision) and ImageJ version 1.45b (National Institutes of Health) were applied for intensity analysis of these images.
The immunofluorescent intensity of individual embryos was measured as described previously (Chen et al., 2012). For comparison of Oct-4 intensity between different regions in the different stages of embryos (i.e. inside cells versus outside cells at compact morula stage or ICM/epiblast versus TE at blastocyst stages), a representative single plane across both the ICM and the TE region of each embryo was selected. Images were first converted to 8-bit grey scale and the background value was eliminated by the background subtract function. The test areas (i.e. the ICM/epiblast and the TE regions) were visually identified and tested using the measure function of ImageJ. The same measurement procedures were applied for Cdx-2 and H4K5ac intensity analysis.
Statistical analysis
Statistical analyses were performed with PRISM version 5.0C (GraphPad Software, La Jolla, CA, USA). Differences of embryo size among different stages were subjected to one-way ANOVA. Paired t-test was used to compare the intensity difference of Oct-4 or H4K5ac between the ICM and the TE regions in the embryos as described previously (Chen et al., 2012). A P-value <0.05 was considered statistically significant.
Results
The morphology of rabbit embryos derived in vivo
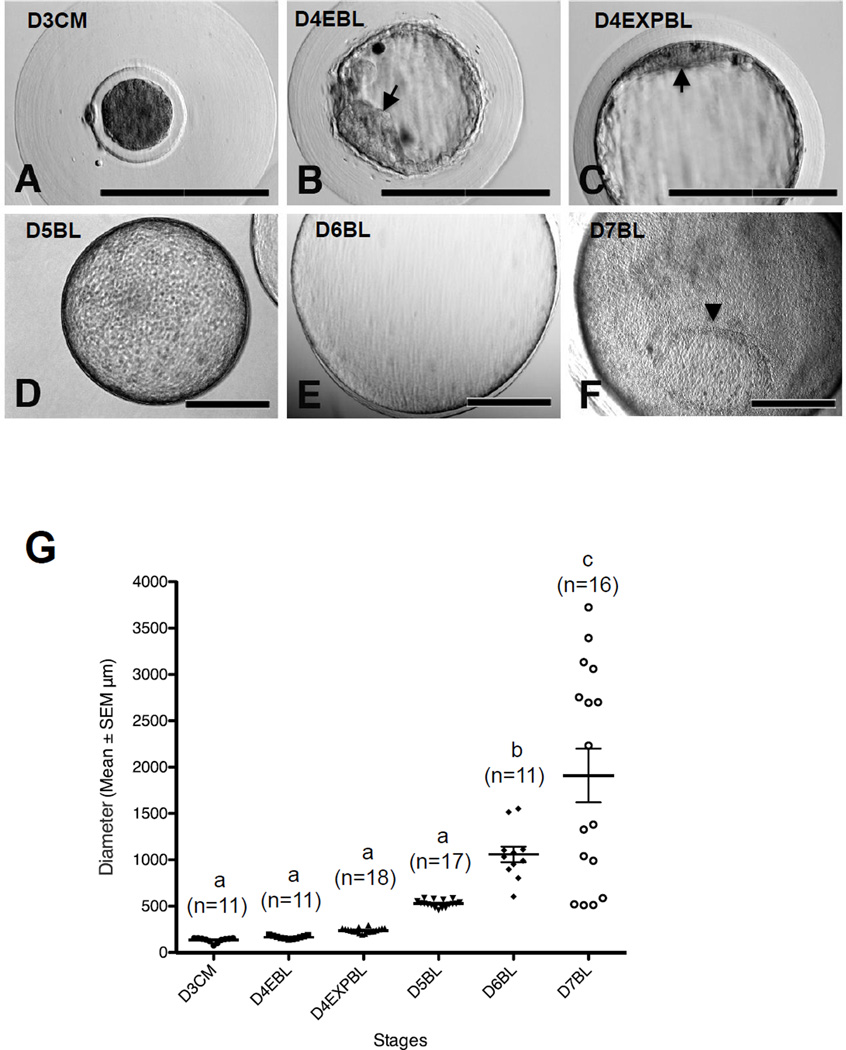
The embryos at 3 dpi (68–72 h), 4 dpi (92–96 h), 5 dpi (116–120 h), 6 dpi (140–144 h) and 7 dpi (164–168 h) were collected in the present study (Figure 1A–F). Embryos collected at 3 dpi (n = 11) showed the typical morphology of compact morulae and were surrounded with thick mucin coats (Figure 1A). The diameter (mean ± SEM) of day-3 embryos was 135 ± 8 µm, ranging from 77 to 156 µm (Figure 1G).
Figure 1.
The morphology of rabbit embryos derived in vivo. Rabbit embryos were flushed at 3–7 days post insemination (dpi). (A) Compact morulae were coated with a thick mucin layer at 3 dpi. (B) A typical early blastocyst with clear inner cell mass (ICM) structure (arrow) shown at 4 dpi. (C) Some of the blastocysts at 4 dpi had reached expanded blastocyst stage and a flattened ICM structure was seen (arrow). (D, E) Fully expanded blastocysts were observed at 5 dpi (D) and 6 dpi (E). (F) Embryonic disc (arrowhead) was seen in the blastocyst at 7 dpi. (G) Embryo diameter at each stage, measured and analysed by ImageJ and PRISM. Results marked with different letters are significantly different (P < 0.05). D3CM = day-3 compact morulae; D4EBL = day-4 early blastocysts; D4EXPBL = day-4 expanded blastocysts; D5BL = day-5 blastocysts; D6BL = day-6 blastocysts; D7BL = day-7 blastocysts. Bars = 200 µm.
There were two distinct categories of blastocysts for embryos collected at 4 dpi. The first group of embryos showed the typical morphology of early blastocysts, with small blastocoeles (Figure 1B). The other group of embryos already had enlarged blastocoeles and showed typical morphology of expanded blastocysts (Figure 1C). The mean diameter of day-4 early blastocysts (n = 11) was 165 ± 5 µm (140–191 µm) and of day-4 expanded blastocysts (n = 18) was 240 ± 6 µm (194–296 µm; Figure 1G).
This study observed fast embryo growth as indicated by the embryo size after 4 dpi. Embryos collected at 5 dpi (n = 17) became fully expanded (Figure 1D). The mean diameter of these embryos was 529 ± 10 µm (458–589 µm; Figure 1G). Embryos collected at 6 dpi (n = 11) had mean diameter of 1058 ± 84 µm (603–1551 µm; Figure 1E and G). Embryos collected at 7 dpi (n = 16) had a mean diameter of 1910 ± 290 µm (511–3725 µm; Figure 1F and G). Embryonic discs were observed in day-7 blastocysts (Figure 1F). The average size of the embryos on and beyond day 4 (i.e. day-4 expanded blastocysts and day-5, day-6 and day-7 blastocysts) showed linear correlation with the embryo age (i.e. dpi) (R2 = 0.95). Interestingly, the SEM of the embryo size of these stages also showed a linear relationship with the embryo age (R2 = 0.80).
Based on these morphology observations, the embryos were categorized into six groups for the immunostaining experiments: day-3 compact morulae, day-4 early blastocysts, day-4 expanded blastocysts, day-5 blastocysts, day-6 blastocysts and day-7 blastocysts.
Oct-4 expression patterns
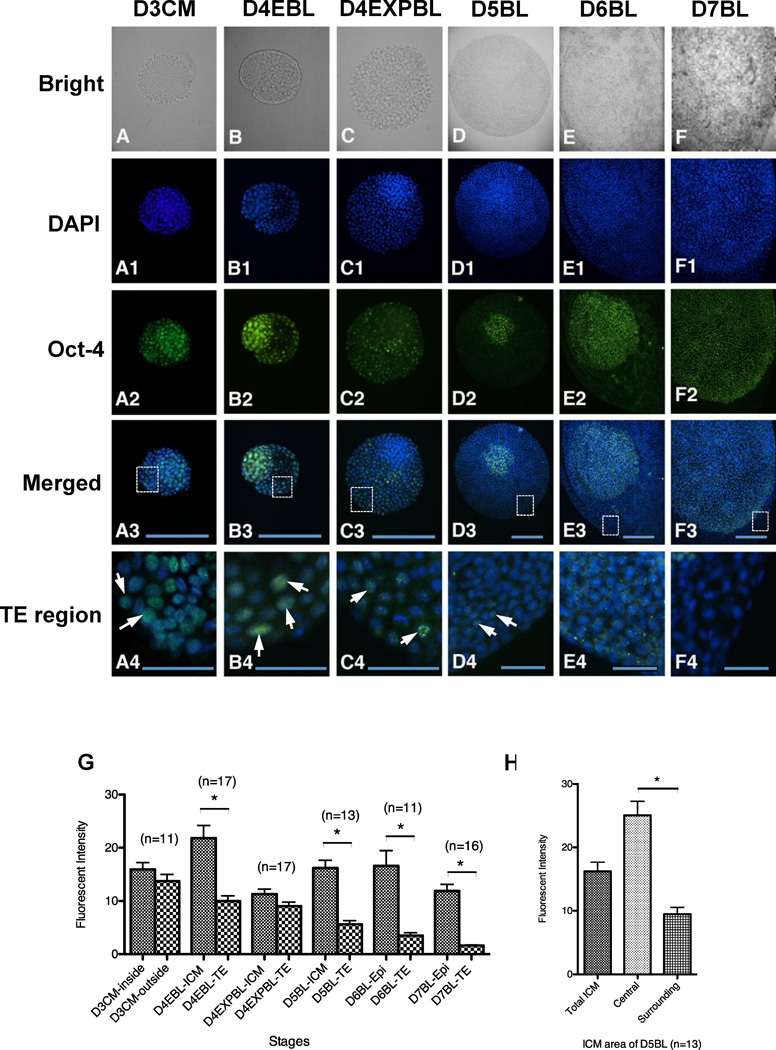
After collection, embryos were directly subjected to immunocytochemistry without any in-vitro culture. Strong nuclear Oct-4 staining was detected in day-3 compact morulae with an intensive signal in both the inside cells and the outside cells (Figure 2A2 and G).
Figure 2.
Oct-4 protein expression in in-vivo-derived rabbit embryos. Embryos were fixed for double staining of DNA with DAPI (blue) and Oct-4 with antibody (green) and the intensity of Oct-4 signals were quantitated. (A–A4) Day-3 compact morulae had an intense Oct-4 signal in inside cells in contrast to the mild intensity in outside cells (n = 11). (B–B4) Strong Oct-4 fluorescence was still present in the ICM of day-4 early blastocysts (n = 17). (C–C4) In day-4 expanded blastocysts, obvious Oct-4 diminishment was observed in the ICM cells rather than in the TE cells (n = 17). (D–D4) In day-5 blastocysts (n = 13), a wave of Oct-4 elevation was observed manifestly in some specified cells of ICM region, while weak Oct-4 expression was still detectable in TE. (E–F4) Clear Oct-4 expression was observed in the epiblast of day-6 blastocysts (E–E4, n = 11) and day-7 blastocysts (F–F4, n = 16). (G) Comparison of the Oct-4 signal intensity of different regions in embryos collected at different stages. The Oct-4 images of in-vivo-derived embryos were used for intensity analysis and different regions of embryo were manually selected for comparison: in merged figures (A3–F3), the TE regions in rectangles were magnified and the arrows (A4–F4) indicate the representative TE cells with Oct-4 signals. Embryos at all stages examined, with the exception of day-3 compact morulae and day-4 expanded blastocysts, revealed a significant difference in Oct-4 intensity, showing stronger Oct-4 intensity in ICM and epiblast regions rather than in TE. (H) Two groups of cells (central and surrounding cells) in the ICM area of day-5 blastocysts showed different Oct-4 intensity (P < 0.05). D3CM = day-3 compact morulae; D4EBL = day-4 early blastocysts; D4EXPBL = day-4 expanded blastocysts; D5BL = day-5 blastocysts; D6BL = day-6 blastocysts; D7BL = day-7 blastocysts; Epi = epiblast; ICM = inner cell mass; TE = trophectoderm. Asterisks indicate statistically significant differences (P < 0.05). Bars = 200 µm (A3–F3) and 50 µm (A4–F4).
A unique transition point was observed in blastocysts collected at 4 dpi. In day-4 early blastocysts, both ICM and TE cell nuclei expressed Oct-4 (Figure 2B2), while the signal in the ICM was significantly stronger than that in the TE (Figure 2G; P < 0.05). In day-4 expanded blastocysts, the Oct-4 signal in the ICM cells diminished to a strength indistinguishable from that of the TE cells (Figure 2C2 and G). The Oct-4 signal in the ICM cells was soon significantly elevated, as shown in day-5 blastocysts (P < 0.05). In contrast, Oct-4 signal remained weak in the TE cells (Figure 2D2 and G) at this stage.
Notably, the cells located in the centre of the ICM region of day-5 blastocysts displayed higher Oct-4 signal intensity than the surrounding cells (Figure 2H), although they were both located within the ICM boundary defined by the observations of bright field and DAPI staining. In the following embryonic stages, the study did not observe two such distinct groups of cells inside the embryos.
In day-6 and day-7 blastocysts, most cells in the embryonic disc showed a strong Oct-4 signal in the nuclei (Figure 2E2 and F2). Cytoplasmic, but not nuclear, weak Oct-4 staining was observed in the TE cells in day-6 blastocysts and soon disappeared in the TE cells in day-7 blastocysts (Figure 2E4 and F4). In fact, the average signal intensity in the TE cells continuously decreased after the day-4 early blastocyst stage and was lowest at the day-7 blastocyst stage (R2 = 0.96).
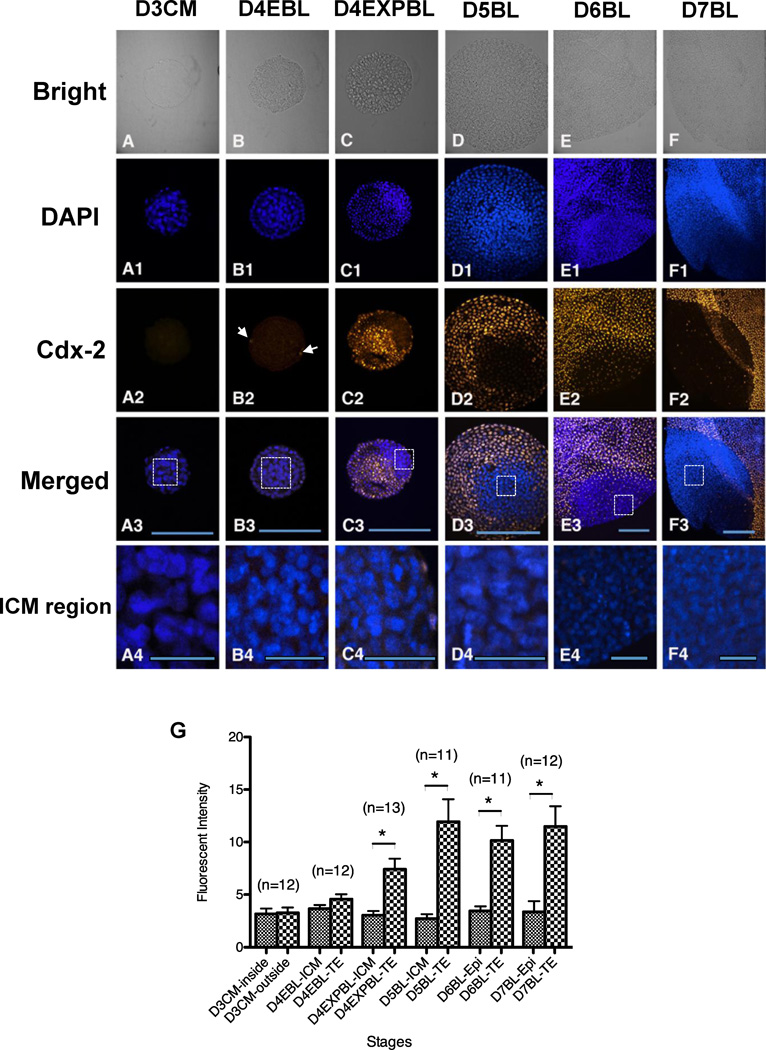
Cdx-2 expression patterns
In order to delineate TE lineage formation during rabbit blastulation, the present study examined the spatial and temporal profiles of Cdx-2 protein of in-vivo-derived blastocysts by immunocytochemistry. No Cdx-2 staining was detected in any of the 12 day-3 compact morulae examined in this study (Figure 3A2). In all embryos, the Cdx-2 signal was restricted to the nuclei of TE cells and absent in the ICM cells (Figure 3C2–F2 and C4–F4). Notably, the Cdx-2 was only detectable in the TE cells in three out of 12 (25%) day-4 early blastocysts examined. Furthermore, only a few TE cells displayed positive Cdx-2 signal in these three day-4 early blastocysts (Figure 3B2). Cdx-2 signal became evident in most TE cells in day-4 expanded blastocysts and continued so in the day 5, day-6 and day-7 blastocysts examined (Figure 3C2–F2 and G).
Figure 3.
Cdx-2 protein expression in in-vivo-derived rabbit embryos. Embryos were used for double labelling of DNA with DAPI (blue) and Cdx-2 with antibody (yellow, pseudocolour assigned by Volocity). A representative single section of individual embryo across the major ICM and TE areas was selected for analysis and the intensity of Cdx-2 signals were quantitated. Cdx-2 (A–A4) Cdx-2 was undetectable in day-3 compact morulae (n = 12). (B–B4) In some of day-4 early blastocysts (3/12), Cdx-2 signals were shown in few TE cells (arrows). (C–C4) The Cdx-2 signal became evident in day-4 expanded blastocysts (n = 13), co-localized with the TE cells, but not with the ICM cells. (D–F4) Thereafter, Cdx-2 was expressed only in TE lineage of in day-5 blastocysts (D–D4, n = 11), day-6 blastocysts (E–E4, n = 11) and day-7 blastocysts (F–F4, n = 12). (G) Comparison of the Cdx-2 signal intensity of different regions in embryos collected at different stages. Significantly different Cdx-2 intensity was found at the day-4 expanded blastocyst stage (P < 0.05), showing stronger Cdx-2 intensity in the TE regions compared with the ICM. Because the Cdx-2 signals were not visually observed in both ICM and TE regions of day-3 compact morulae and the ICM/epiblast regions of day-4–7 blastocysts, the measured Cdx-2 intensity can be considered as a background value. In merged figures (A3–F3), the ICM regions in rectangles were magnified and Cdx-2 signals were not detected by the current method (A4–F4). D3CM = day-3 compact morulae; D4EBL = day-4 early blastocysts; D4EXPBL = day-4 expanded blastocysts; D5BL = day-5 blastocysts; D6BL = day-6 blastocysts; D7BL = day-7 blastocysts; Epi = epiblast; ICM = inner cell mass; TE = trophectoderm. Asterisks indicate statistically significant differences (P < 0.05). Bars = 200 µm (A3–F3) and 50 µm (A4–F4).
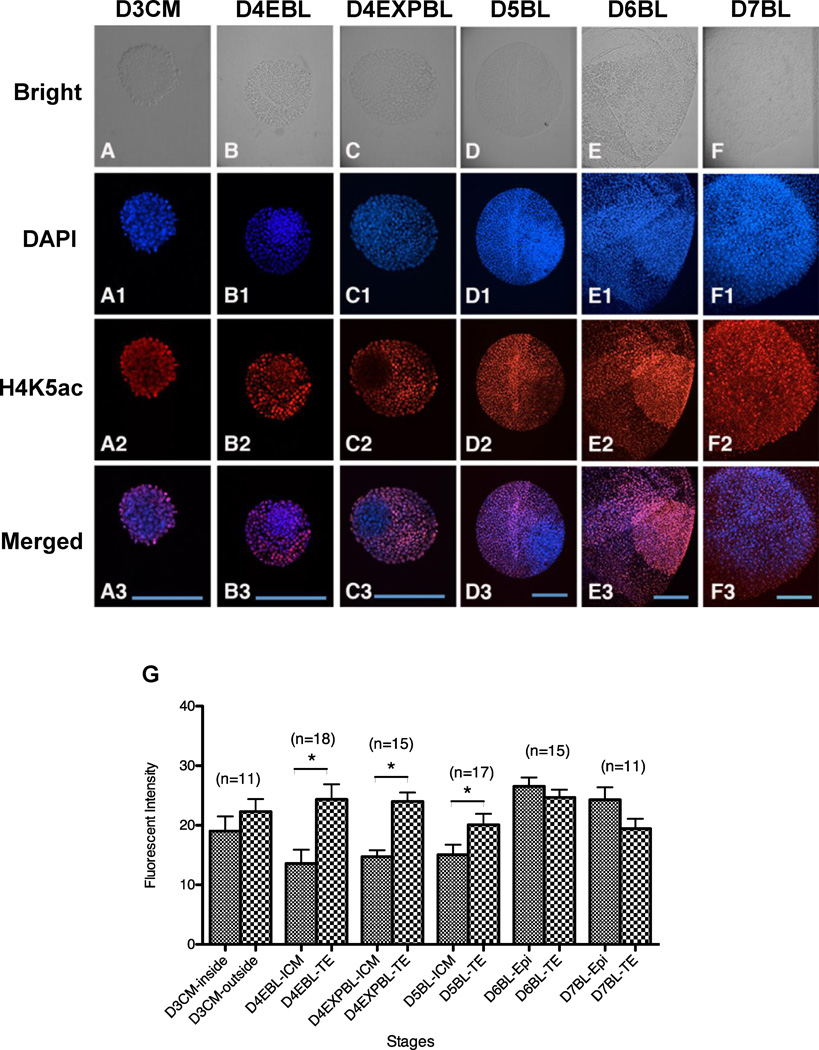
H4K5ac patterns
A consistently strong H4K5ac signal was observed in the TE cells of embryos at different stages, from day-3 compact morulae to day-7 blastocysts (Figure 4G). The H4K5ac signal in the ICM cells, however, was weaker in day-4 early blastocysts, day-4 expanded blastocysts and day-5 blastocyst-stage embryos (Figure 4B2–D2), compared with embryos at earlier stages (i.e. day-3 compact morulae, Figure 4A2) or later (i.e. day-6 or day-7 blastocysts, Figure 4E2 and F2). Consequently, the H4K5ac signal was similar between the ICM and the TE cells in all stages examined, except the day-4 early blastocysts, day-4 expanded blastocysts and day-5 blastocyst stages, where the H4K5ac signal in the TE cells was significantly stronger than that in the ICM cells (Figure 4G; P < 0.05).
Figure 4.
Acetylation pattern of H4K5 in in-vivo-derived rabbit embryos. Embryos were fixed for double staining of DNA with DAPI (blue) and acetylated H4K5 with antibody (red) and the intensity of H4K5ac signals were quantitated. (A–D3) A stronger H4K5ac signal was shown in the outside area of day-3 compact morulae (A–A3, n = 11), in the TE cells of day-4 early blastocysts (B–B3, n = 18), day-4 expanded blastocysts (C–C3, n = 15) and day-5 blastocysts (D–D3, n = 17), compared with the inside area of day-3 compact morulae and ICM of day-4 early blastocysts, day-4 expanded blastocysts and day-5 blastocysts, respectively. (E–F3) A reverse of this trend was observed starting in day-6 blastocysts. The H4K5ac intensity in the epiblast appeared higher than that in the TE cells of day-6 (E–E3, n = 15) and day-7 blastocysts (F–F3, n = 11). (G) Analysis of H4K5ac revealed that its signal was significantly weaker in the ICM than in the TE cells in day-4 early blastocysts, day-4 expanded blastocysts and day-5 blastocysts (P < 0.05). On the contrary, H4K5ac intensity was similar between the ICM and TE cells in day-6 and day-7 blastocysts, as well as between the outside and inside regions of day-3 compact morulae. D3CM = day-3 compact morulae; D4EBL = day-4 early blastocysts; D4EXPBL = day-4 expanded blastocysts; D5BL = day-5 blastocysts; D6BL = day-6 blastocysts; D7BL = day-7 blastocysts; Epi = epiblast; ICM = inner cell mass; TE = trophectoderm. Asterisks indicate statistically significant differences (P < 0.05). Bars = 200 µm.
Relationship of H4K5ac with Oct-4 at days 3–5 dpi
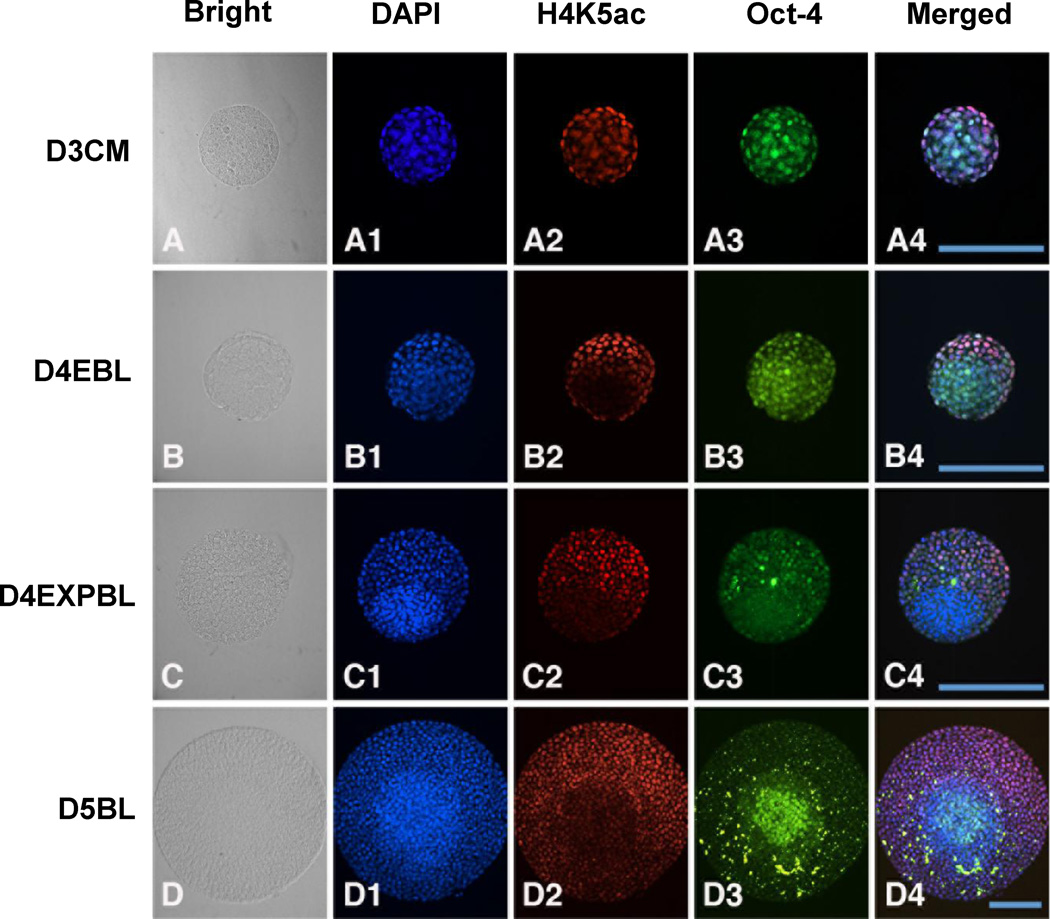
This study performed double staining with H4K5ac and Oct-4 on rabbit embryos on day-3 compact morulae, day-4 early blastocysts, day-4 expanded blastocysts and day-5 blastocyst stages (Figure 5). In the ICM cells, the H4K5ac signal was consistently weak, while the Oct-4 signal was intensive except at the day-4 expanded blastocyst stage. Consequently, the H4K5ac signal appeared to be inversely related with the Oct-4 signal except at the day-4 expanded blastocyst stage in the ICM cells. In the TE cells, the H4K5ac signal was intensive but the Oct-4 signal was weak throughout all blastocyst stages examined. As a result, the H4K5ac signal appeared to be inversely related with the Oct-4 signal in the TE cells.
Figure 5.
Double staining of Oct-4 and H4K5ac in in-vivo-derived rabbit embryos. A representative section across most of the area of ICM and TE was chosen for evaluation. Images of DNA (blue), H4K5ac (red) and Oct-4 (green) at different stages are shown. (A–A4) In day-3 compact morulae, Oct-4 signal was shown in both the outside and inside cells and a more intense H4K5ac signal was located outside of the embryos. (B–B4) In day-4 early blastocysts, intensive Oct-4 staining and hypoacetylated H4K5 was observed in the ICM region and a mild Oct-4 signal with hyperacetylated H4K5 in the TE region, revealing the inverse relationship between Oct-4 and H4K5ac expression. (C–C4) In day-4 expanded blastocysts, diminished Oct-4 signal and the continued hypo-acetylation of H4K5 was observed in the ICM. (D–D4) In day-5 blastocysts, the hypo-acetylated H4K5 pattern was kept in ICM region, while a small group of ICM cells regained a highly intensive Oct-4 signal. D3CM = day-3 compact morulae; D4EBL = day-4 early blastocysts; D4EXPBL = day-4 expanded blastocysts; D5BL = day-5 blastocysts; D6BL = day-6 blastocysts; D7BL = day-7 blastocysts; ICM = inner cell mass; TE = trophectoderm. Bars = 200 µm.
Relationship of H4K5ac with Cdx-2 at days 3–5
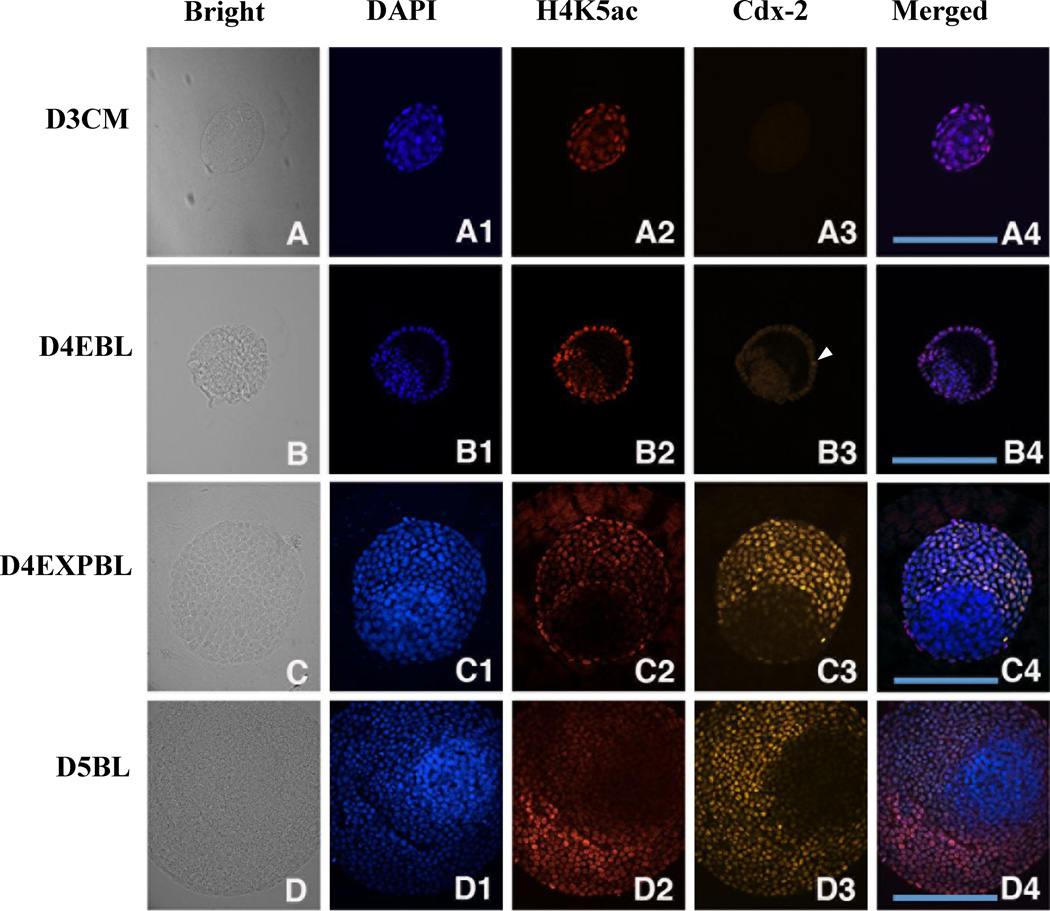
This study performed double staining with H4K5ac and Cdx-2 on rabbit embryos at day-3 compact morulae, day-4 early blastocysts, day-4 expanded blastocysts and day-5 blastocyst stages (Figure 6). In the ICM cells, the Cdx-2 and H4K5ac signals were both weak throughout the blastocysts examined (i.e. day-4 early blastocysts, day-4 expanded blastocysts and day-5 blastocysts). Consequently, the Cdx-2 signal and H4K5ac signal were concordant with each other in the ICM cells in the stages examined. In the TE cells, the H4K5ac signal was concordant with the DAPI signal.
Figure 6.
Double staining of Cdx-2 and H4K5ac in in-vivo-derived rabbit embryos. A representative section across most of the area of ICM and TE was chosen for evaluation. The images of DNA (blue), H4K5ac (red) and Oct-4 (green) at different stages are shown. (A–A4) Cdx-2 signal was not detected in day-3 compact morulae, but hyperacetylated H4K5 was shown in the outside cells. (B–B4) Few Cdx-2 positive TE cells were initially found in day-4 early blastocysts (B3, arrowhead). Hyperacetylation of H4K5ac was continually shown in the TE lineage in contrast to hypo-acetylation in the ICM lineage. (C–C4) Cdx-2 signal exclusively appeared in the TE cells of day-4 expanded blastocysts accompanied with hyperacetylated H4K5 signal. (D–D4) A similar relationship between Cdx-2 and H4K5ac was found day-5 blastocysts. The expression patterns of Cdx-2 and H4K5ac across the TE and ICM regions were similar and overlapped in day-4 expanded blastocysts and day-5 blastocysts. D3CM = day-3 compact morulae; D4EBL = day-4 early blastocysts; D4EXPBL = day-4 expanded blastocysts; D5BL = day-5 blastocysts; D6BL = day-6 blastocysts; D7BL = day-7 blastocysts; ICM = inner cell mass; TE = trophectoderm. Bars = 200 µm.
The Cdx-2 signal became evident from day-4 expanded blastocyst stage and the signal strength was concordant with the DAPI signal. As a result, the Cdx-2 signal and H4K5ac signal were concordant with each other in day-4 expanded blastocysts and day-5 blastocysts.
Discussion
This study, as far as is known for the first time, reports the protein expression patterns of Cdx-2 in rabbit morulae and blastocysts. Cdx-2 is specifically expressed in the TE cells in mouse embryos (Nishioka et al., 2009; Yagi et al., 2007). It plays a vital role in TE development and commitment and is often used as a biomarker to identify TE lineage cells (Beck et al., 1995; Ralston and Rossant, 2008; Sritanaudomchai et al., 2009; Strumpf et al., 2005). The present results revealed that the Cdx-2 protein was also exclusively expressed in the TE cells but not in the ICM cells in rabbit blastocysts. However such distinction was very weak at the day-4 early blastocyst stage, where only very few TE cells showed Cdx-2 signals. Such signal became strongly detectable in most TE cells at the day-4 expanded blastocyst stage. Consequently, this study does not recommend the use of Cdx-2 to identify TE cells in early blastocysts. Rather, it should only be used to mark TE cells at or beyond the day-4 expanded blastocyst stage in rabbits.
This work reveals a different temporal expression pattern of Cdx-2 protein in the rabbit embryos from that in the mouse embryos. In mouse embryos, the onset of embryonic Cdx-2 gene expression was found as early as the 8-cell stage (Dietrich and Hiiragi, 2007; Sritanaudomchai et al., 2009; Strumpf et al., 2005; Wu et al., 2010). In rabbits, the protein signal of Cdx-2 was first detected in day-4 early blastocysts. Because the heterogeneous expression of Cdx-2 in blastomeres takes place prior to compaction, it has been postulated as one factor contributing to cell lineage determination in mouse embryos (Jedrusik et al. 2008). In the present work, although the initial onset of Cdx-2 expression in rabbit embryos was also heterogeneous, it did not happen until after embryo compaction, making it an unlikely causal factor for cell lineage formation in this species. In primate embryos (Sritanaudomchai et al., 2009), interestingly, the onset of Cdx-2 expression was also after compaction (at morula stage), but such expression is homogeneous in all embryonic cells, contrasting what this study observed in rabbit embryos, and previously reported in mouse embryos (Dietrich and Hiiragi, 2007; Strumpf et al., 2005). These results suggest that Cdx-2 expression patterns are species specific. Future work is needed to examine Cdx-2 expression patterns among different species.
The previous study using in-vitro-cultured rabbit embryos suggested that day 4 might be the critical time point for lineage formation in rabbit embryos (Chen et al., 2012). The findings in the present work further strengthen this speculation. First, the Cdx-2 signal started to emerge in very few TE cells of the day-4 early blastocysts and became strongly detectable exclusively in TE cells in the day-4 expanded blastocysts and beyond. Second, there is a unique wave of Oct-4 signal change in the ICM cells, which bottomed at the day-4 expanded blastocyst stage in both in-vitro-cultured and in-vivo-derived embryos. Thirdly, the present study observed a wave of hypoacetylation of H4K5 in the ICM cells, which started at the day-4 early blastocyst stage and ended at the day-5 blastocyst stage.
The Oct-4 signal change in the ICM cells on day 4 may be associated with the switch of the distal enhancer (DE) of Oct-4 to the proximal enhancer. The distal enhancer is responsible for driving Oct-4 expression in the ICM and primordial germ cells as well as the in-vitro derivatives (e.g. embryonic stem cells and embryonic germ cells). By contrast, the proximal enhancer is active and directs Oct-4 expression in the epiblast and their in-vitro counterpart, epiblast stem cells. Thus, the regulatory machinery to express Oct-4 is a useful tool to distinguish the pluripotent cell property and its origins (Bao et al., 2009). Based on the observation that the Oct-4 bottomed at the expanded blastocyst stage in both in-vitro-cultured and in-vivo-derived embryos, it is postulated that such down-regulation is caused by the enhancer switching from the distal enhancer to the proximal enhancer, which takes place at the expanded blastocyst stage, and that day-4 early blastocyst-stage embryos may serve better for authentic embryonic stem cell derivation in rabbits, as the the distal enhancer driving Oct-4 expression is associated with ICM and embryonic stem cells (Bao et al., 2009).
The two different groups of cells (i.e. the cells in the centre with stronger Oct-4 signal, and the surrounding cells with relatively weaker Oct-4 signal) in the ICM of the day-5 blastocysts might signal the emergence of the epiblast and hypoblast cells in ICM derivatives. In mouse, the separation of the epiblast and the hypoblast takes place in peri-implantation stage (embryonic day 4.5), driven by a series of processes, such as asymmetric division during compaction, expression of lineage-specific transcription factors, cell position, cell sorting and apoptosis (Chazaud et al., 2006; Morris et al., 2010; Plusa et al., 2008; Rossant et al., 2003). Very recently, using immunostaining markers for epiblast (NANOG) and hypoblast (GATA-6), it is shown that the segregation of these two lineages in primate ICM may take place before implantation (Tachibana et al., 2012). Although the underlying mechanism for epiblast and hypoblast commitments in rabbits is undetermined, this present work may have found the starting point of such separation (i.e. day 5, before implantation). It is speculated that the centre cells with very bright Oct-4 signals are the emerging epiblast cells, and the surrounding cells with relatively weaker Oct-4 signals are the hypoblast cells. Further work is needed to characterize these two groups of cells to determine whether they are truly epiblast and hypoblast cells. The present work was not able to do so because of the lack of a suitable antibody for rabbit hypoblast cells.
Different from mouse but similar to the human, the TE cells of rabbit blastocysts expressed both Oct-4 and Cdx-2 (Berg et al., 2011; Hansis et al., 2000; Kirchhof et al., 2000). In mouse embryos (Dietrich and Hiiragi, 2007; Ovitt and Scholer, 1998; Palmieri et al., 1994; Pesce et al., 1998; Yeom et al., 1996), Oct-4 and Cdx-2 displayed a reciprocal expression pattern with Cdx-2 localized exclusively in the TE, and Oct-4 in the ICM. When Cdx-2 was mutated, the Oct-4 expression was found high in the TE cells of mouse embryos (Strumpf et al., 2005). Overexpression of Cdx-2 in mouse embryonic stem cells, on the other hand, leads to the down-regulation of Oct-4 and, ultimately, the differentiation of these cells towards TE-like cells (Niwa and Miyazaki, 2000; Niwa et al., 2005). At the molecular level, the Cdx-2 protein was in fact co-operating with transcription factor Tcfap2 to bind the Oct-4 distal autoregulatory enhancer CR4 to achieve Oct-4 repression in mice (Berg et al., 2011; Kuckenberg et al., 2010; Niwa et al., 2005). Interestingly, the Tcfap2 binding sites in the Oct-4 promoter sequence is absent in human, cattle and rabbits (Berg et al., 2011). This is likely the reason why Cdx-2-mediated Oct-4 down-regulation does not take place in rabbit TE cells, as observed in the present study.
It has been suggested that epigenetic factors may be involved in mammalian embryo development to stabilize gene expression and regulate cell lineage formation (Corry et al., 2009). The present study observed a maintained hyperacetylation status of H4K5 in the TE cells from day-4 early blastocysts to day-7 blastocysts. The H4K5 in the ICM cells, on the other hand, showed a wave of hypo-acetylation from day-4 early blastocysts to day-5 blastocysts. During the initiation of blastocyst formation (day-4 early blastocysts to day-5 blastocysts), the H4K5ac signal was significantly stronger in the TE than in the ICM cells, implying that a wave of genes, likely those associated with blastocyst formation and lineage segregation, are more active in TE cells during this time window. Moreover, the present study suggests that H4K5ac might be involved in the regulation of the Cdx-2 expression in the TE cells and the Oct-4 expression in the ICM cells. First, the TE lineage was associated with hyperacetylation of H4K5 immediately after compaction (day-4 early blastocysts), a time point when only very few cells in the TE displayed a detectable Cdx-2 signal. In other words, the hyperacetylation of H4K5 in the TE cells was a preceding event of the TE-specific Cdx-2 expression. It is not until day-4 expanded blastocyst stage that the majority of TE cells strongly expressed Cdx-2. On the other hand, when looking at the relationship between H4K5ac and Oct-4 in the ICM cells, the hypo-acetylation of H4K5 in the ICM cells started earlier (day-4 early blastocysts) and ended later (day-5 blastocysts) than the Oct-4 down-regulation event (day-4 expanded blastocysts).
It is noted that different lineage cells displayed different signature profiles of Oct-4, Cdx-2 and H4K5ac in in-vivo-derived rabbit embryos, and one single biomarker is not sufficient to mark the cell lineage (i.e. ICM or TE) in rabbit blastocysts. While Cdx-2 signal was only detected in the TE cells, it did not become robust until day-4 expanded blastocyst stage. So it cannot be reliably used for lineage identification on early blastocysts. Oct-4 signal, on the other hand, was present in both ICM and TE cells, and the signal strength was similar between the ICM and TE cells at the day-4 expanded blastocyst stage. Therefore, it cannot be used to identify cell lineage in all stages of rabbit embryos either. The unique H4K5ac patterns in different blastocyst stages complemented the information provided by Cdx-2 and Oct-4. The H4K5 showed differential acetylation patterns between the ICM and TE cells as early as the early blastocyst stage. H4K5ac signal was stronger in the TE cells than in the ICM cells in day-4 early blastocysts, day-4 expanded blastocysts and day-5 blastocysts. Taking these together, the present results demonstrat that a combination of Oct-4, Cdx-2 and H4K5ac can be used to accurately identify different lineages of cells in blastocyst-stage rabbit embryos (Table 1). Briefly, day-4 early blastocysts are characterized with high Oct-4 in the ICM and high H4K5ac in the TE. Day-4 expanded blastocysts are characterized with high Cdx-2 and H4K5ac in the TE, and low Oct-4 in the ICM. Day-5 blastocysts are characterized with high Cdx-2 in the TE and low H4K5ac in the ICM. For embryos at or beyond day 5, they are characterized with high Oct-4 in the epiblast and high Cdx-2 in the TE cells.
Table 1.
Oct-4, Cdx-2 and H4K5ac profiles in different cell lineages of in-vivo-derived rabbit embryos.
| Cell lineage | Marker | Stage of development | |||||
|---|---|---|---|---|---|---|---|
| D3CM | D4EBL | D4EXPBL | D5BL | D6BL | D7BL | ||
| Inside/ICM/Epi | Oct-4 | +++ | ++++ | ++ | +++ | +++ | ++ |
| Cdx-2 | − | − | − | − | − | − | |
| H4K5ac | ++++ | ++ | ++ | ++ | ++++ | ++++ | |
| Outside/TE | Oct-4 | +++ | ++ | ++ | + | +/− | +/− |
| Cdx-2 | − | +/− | +++ | ++++ | +++++ | +++++ | |
| H4K5ac | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | |
++++ = ; +++ = ; ++ = ; +/− = ; − =.
Epi = epiblast; ICM = inner cell mass; TE = trophectoderm; D3CM = day-3 compact morulae; D4EBL = day-4 early blastocysts; D4EXPBL = day-4 expanded blastocysts; D5BL = day-5 blastocysts; D6BL = day-6 blastocysts; D7BL = day-7 blastocysts.
The present study used in-vivo-derived embryos derived from superovulated rabbits. It is noted that the in-vivo endocrine environment may be altered by superovulation treatment and influence the lineage formation in subsequent embryo development. In the mouse study, superovulation appears to affect cell allocation to the embryonic or abembryonic pole in blastocysts compared with that in unstimulated animals (Katayama and Roberts, 2010). Thus, the effects of FSH stimulation given to the rabbits on the embryo development and lineage formation need to be determined in future studies.
In-vitro-cultured embryos may differ from in-vivo-derived embryos in several ways, such as gene expression profiles (Mamo et al., 2007; Niemann and Wrenzycki, 2000), embryo morphology, cell number, embryo size and development speed (Hyttel and Niemann, 1990; Machaty et al., 1998). This present work confirms that there is some difference between in-vitro-cultured and in-vivo-derived rabbit embryos. Comparing to the in-vitro counterparts, in-vivo-derived rabbit embryos were insulated by a unique coating of mucin layer on zona pellucida; they developed more rapidly (as indicated by the average size of a given stage), displayed well-structured morphology and did not show hatching in all stages examined (versus hatching as early as day 5 in in-vitro-cultured embryos). They also differed in the pattern of H4K5ac at day 5. The H4K5ac signal in the TE cells was stronger than that in the ICM cells in day-5 in-vivo-derived embryos; it was the opposite in day-5 in-vitro-cultured embryos. Interestingly, at the same stage (day-5 blastocysts), the present study observed two distinct groups of cells in the ICM cells characterized by the location and differential strengths of the Oct-4 signal using in-vivo-derived embryos. This phenomenon was not revealed in the previous report using in-vitro-cultured embryos (Chen et al., 2012). Nevertheless, despite some difference observed, most findings, such as the unique Oct-4 pattern in the ICM cells and the hyperacetylation status of H4K5 in the TE cells, were consistent between in-vitro-cultured and in-vivo-derived embryos.
In summary, this study documents the spatial and temporal distribution of Oct-4, Cdx-2 and H4K5ac using in-vivo-derived rabbit blastocysts. The data pinpoint day 4 as the critical timing for lineage formation in rabbit embryos and demonstrates that a combination of Oct-4, Cdx-2 and H4K5ac could be used as biomarkers to identify different lineage cells in rabbit blastocysts. The present work provides novel information on key transcription factors and epigenetic events during rabbit embryo development.
Acknowledgements
The authors would like to gratefully acknowledge the confocal resources supplied from Dr Song-Kun Shyueat Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan. This study was supported by the National Institutes of Health (grant number 5R44RR023774 to JX) and the National Science Council, Taiwan, ROC (grant number 100-2313-B-002-045-MY3 to L-YS).
Biography

Chien-Hong Chen was awarded an MSc in animal science in 2003 from the National Chung-Hsing University, Taichung, Taiwan. From 2003 to 2008, he worked as an assistant research fellow at the Animal Technology Institute in Taiwan. From 2010 to 2012, he worked as an associate scientist at Renova Life, USA. He was awarded his PhD in 2012 from the National Taiwan University, Taipei, Taiwan. His primary interests concern embryology and nuclear reprogramming through somatic cell nuclear transfer. At present, he is focusing on the establishment of germline competent embryonic stem cells in rabbits.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration: The authors report no financial or commercial conflicts of interest.
References
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck F, Erler T, Russell A, James R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 1995;204:219–227. doi: 10.1002/aja.1002040302. [DOI] [PubMed] [Google Scholar]
- Berg DK, Smith CS, Pearton DJ, Wells DN, Broadhurst R, Donnison M, Pfeffer PL. Trophectoderm lineage determination in cattle. Dev. Cell. 2011;20:244–255. doi: 10.1016/j.devcel.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Chen CH, Chang WF, Liu CC, Su HY, Shyue SK, Cheng WTK, Chen YE, Wu SC, Du F, Sung LY, Xu J. Spatial and Temporal Distribution of Oct-4 Expression and H4K5 Acetylation in Rabbit Embryos. Reprod. Biomed. Online. 2012 doi: 10.1016/j.rbmo.2012.01.001. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry GN, Tanasijevic B, Barry ER, Krueger W, Rasmussen TP. Epigenetic regulatory mechanisms during preimplantation development. Birth Defects Res. C Embryo Today. 2009;87:297–313. doi: 10.1002/bdrc.20165. [DOI] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Fan J, Watanabe T. Transgenic rabbits as therapeutic protein bioreactors and human disease models. Pharmacol. Ther. 2003;99:261–282. doi: 10.1016/s0163-7258(03)00069-x. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Hansis C, Grifo JA, Krey LC. Oct-4 expression in inner cell mass and trophectoderm of human blastocysts. Mol. Hum. Reprod. 2000;6:999–1004. doi: 10.1093/molehr/6.11.999. [DOI] [PubMed] [Google Scholar]
- Honda A, Hirose M, Hatori M, Matoba S, Miyoshi H, Inoue K, Ogura A. Generation of induced pluripotent stem cells in rabbits: potential experimental models for human regenerative medicine. J. Biol. Chem. 2010;285:31362–31369. doi: 10.1074/jbc.M110.150540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Hirose M, Inoue K, Ogonuki N, Miki H, Shimozawa N, Hatori M, Shimizu N, Murata T, Hirose M. Stable embryonic stem cell lines in rabbits: potential small animal models for human research. Reprod. Biomed. online. 2008;17:706–715. doi: 10.1016/s1472-6483(10)60320-3. [DOI] [PubMed] [Google Scholar]
- Honda A, Hirose M, Ogura A. Basic FGF and Activin/Nodal but not LIF signaling sustain undifferentiated status of rabbit embryonic stem cells. Exp. Cell Res. 2009;315:2033–2042. doi: 10.1016/j.yexcr.2009.01.024. [DOI] [PubMed] [Google Scholar]
- Hyttel P, Niemann H. Ultrastructure of porcine embryos following development in vitro versus in vivo. Mol. Reprod. Dev. 1990;27:136–144. doi: 10.1002/mrd.1080270208. [DOI] [PubMed] [Google Scholar]
- Intawicha P, Ou YW, Lo NW, Zhang SC, Chen YZ, Lin TA, Su HL, Guu HF, Chen MJ, Lee KH, Chiu YT, Ju JC. Characterization of embryonic stem cell lines derived from New Zealand white rabbit embryos. Cloning and Stem Cells. 2009;11:27–38. doi: 10.1089/clo.2008.0040. [DOI] [PubMed] [Google Scholar]
- Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 2008;22:2692–2706. doi: 10.1101/gad.486108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama M, Roberts RM. The effect of superovulation on the contributions of individual blastomeres from 2-cell stage CF1 mouse embryos to the blastocyst. Int. J. Dev. Biol. 2010;54:675–681. doi: 10.1387/ijdb.092942mk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhof N, Carnwath JW, Lemme E, Anastassiadis K, Scholer H, Niemann H. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol. Reprod. 2000;63:1698–1705. doi: 10.1095/biolreprod63.6.1698. [DOI] [PubMed] [Google Scholar]
- Kuckenberg P, Buhl S, Woynecki T, van Furden B, Tolkunova E, Seiffe F, Moser M, Tomilin A, Winterhager E, Schorle H. The transcription factor TCFAP2C/AP-2gamma co-operates with CDX2 to maintain trophectoderm formation. Mol. and Cell. Biol. 2010;30:3310–3320. doi: 10.1128/MCB.01215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Machaty Z, Day BN, Prather RS. Development of early porcine embryos in vitro and in vivo. Biol. Reprod. 1998;59:451–455. doi: 10.1095/biolreprod59.2.451. [DOI] [PubMed] [Google Scholar]
- Mamo S, Gal AB, Bodo S, Dinnyes A. Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev. Biol. 2007;7:14. doi: 10.1186/1471-213X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Jaenisch R. Generation of nuclear transfer-derived pluripotent ES cells from cloned Cdx2-deficient blastocysts. Nature. 2006;439:212–215. doi: 10.1038/nature04257. [DOI] [PubMed] [Google Scholar]
- Morris SA, Teo RTY, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc. Natl. Acad. Sci. 2010;107:6364–6369. doi: 10.1073/pnas.0915063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann H, Wrenzycki C. Alterations of expression of developmentally important genes in preimplantation bovine embryos by in vitro culture conditions: implications for subsequent development. Theriogenology. 2000;53:21–34. doi: 10.1016/s0093-691x(99)00237-x. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K-i, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H. The hippo signaling pathway components lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 Determines Trophectoderm Differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- O’Neill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovitt CE, Scholer HR. The molecular biology of Oct-4 in the early mouse embryo. Mol. Hum. Reprod. 1998;4:1021–1031. doi: 10.1093/molehr/4.11.1021. [DOI] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce M, Wang X, Wolgemuth DJ, Scholer H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech. Dev. 1998;71:89–98. doi: 10.1016/s0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Rossant J, Chazaud C, Yamanaka Y. Lineage allocation and asymmetries in the early mouse embryo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:1341–1348. doi: 10.1098/rstb.2003.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sritanaudomchai H, Sparman M, Tachibana M, Clepper L, Woodward J, Gokhale S, Wolf D, Hennebold J, Hurlbut W, Grompe M, Mitalipov S. CDX2 in the formation of the trophectoderm lineage in primate embryos. Dev. Biol. 2009;335:179–187. doi: 10.1016/j.ydbio.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao C-A, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Ramsey C, Ma H, Lee HS, Penedo MC, Mitalipov S. Generation of Chimeric Rhesus Monkeys. Cell. 2012;148:285–295. doi: 10.1016/j.cell.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne AW, Kmiciek D, Mitchelson K, Crane-Robinson C. Patterns of histone acetylation. Eur. J. Biochem. 1990;193:701–713. doi: 10.1111/j.1432-1033.1990.tb19390.x. [DOI] [PubMed] [Google Scholar]
- Tse C, Sera T, Wolffe AP, Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol. Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM, Fellows G. Specific antibodies reveal ordered and cell-cycle-related use of histone-H4 acetylation sites in mammalian cells. Eur. J. Biochem. 1989;179:131–139. doi: 10.1111/j.1432-1033.1989.tb14530.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Shen Y, Yuan X, Chen K, Guo X, Chen Y, Niu Y, Li J, Xu R-H, Yan X, Zhou Q, Ji W. Dissecting signaling pathways that govern self-renewal of rabbit embryonic stem cells. J. Biol. Chem. 2008;283:35929–35940. doi: 10.1074/jbc.M804091200. [DOI] [PubMed] [Google Scholar]
- Wang S, Tang X, Niu Y, Chen H, Li B, Li T, Zhang X, Hu Z, Zhou Q, Ji W. Generation and characterization of rabbit embryonic stem cells. Stem Cells. 2007;25:481–489. doi: 10.1634/stemcells.2006-0226. [DOI] [PubMed] [Google Scholar]
- Wu G, Gentile L, Fuchikami T, Sutter J, Psathaki K, Esteves TC, Arauzo-Bravo MJ, Ortmeier C, Verberk G, Abe K, Scholer HR. Initiation of trophectoderm lineage specification in mouse embryos is independent of Cdx2. Development. 2010;137:4159–4169. doi: 10.1242/dev.056630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, Depamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]