Abstract
Paramagnetic Cu(II) ions enhance nuclear spin relaxation in a distance-dependent fashion and can be used as a structural probe of proteins. Cu(II) can also serve as functionally important ligands in proteins. Here we investigate the structural basis of Cu(II) inhibition of the influenza M2 proton channel through Cu(II)-induced paramagnetic relaxation enhancement (PRE). 13C T1 relaxation rates of the central residues of the transmembrane (TM) domain of M2 are significantly enhanced by Cu(II), and pronounced spectral broadening is observed for the proton-selective residue, His37. These data yielded quantitative distances of 13C spins to the Cu(II) center, and identified the Cu(II) binding site to be Nε2 of His37. This binding site is surrounded by four imidazole rings from the top and four indole rings of Trp41 from the bottom, thus explaining the high affinity of Cu(II) binding. Bound at this location, Cu(II) can inhibit proton currents by perturbing histidine-water proton exchange, preventing histidine conformational dynamics, and interfering with His-Trp cation-π interaction. The Cu(II) binding site is distinct from the binding site of the hydrophobic drug amantadine, which is about 10 Å N-terminal to His37. Consistently, Cu(II) and amantadine induce distinct conformational changes at several key residues, suggesting the possibility of designing new drugs that target the His37 site to inhibit amantadine-resistant mutant M2 proteins. In addition to the high-affinity His37 binding site, we also examined the weaker and non-specific binding of Cu(II) to membrane-surface lipid phosphates and the extent of the resulting PRE to surface-proximal protein residues. This study demonstrates the feasibility of NMR studies of paramagnetic-ion complexed membrane proteins, where the ion serves as both a functional ligand and a distance probe.
Introduction
Structure determination of membrane proteins by magic-angle-spinning (MAS) solid-state NMR requires not only local structural constraints such as torsion angles and short-range distances 1–3, but also long distances (above 5 Å) to constrain the protein three-dimensional fold. These long-range distances are more difficult to measure than short-range ones because of the weakness of internuclear dipolar couplings for 13C and 15N nuclei and the complexity of measuring 1H-1H dipolar couplings in organic solids. In 13C and 15N 2D correlation spectra, cross peaks for distances above 5 Å require long mixing times to observe 4 and thus are limited by nuclear spin relaxation. Even when long-range cross peaks are present, they are usually dominated by high-intensity cross peaks from nuclei within an amino acid residue 5. Although techniques for measuring 1H distances to 13C or 15N 6 and 19F-19F distances 7,8 have been developed to increase the distance upper limit of NMR, these techniques require site-specific labels, thus do not easily yield many distances simultaneously.
Paramagnetic relaxation enhancement (PRE) is an effective way to measure long-range distances. The unpaired electrons in paramagnetic metal ions enhance longitudinal (T1) and transverse (T2) relaxation of nuclear spins according to the inverse sixth power of the electron-nucleus distance 9. Because PRE scales with the electron gyromagnetic ratio, which is two to three orders of magnitude larger than the gyromagnetic ratios of nuclear spins, distances up to ~20 Å can be measured using this effect 10. For paramagnetic ions with high numbers of unpaired electrons and long (nanosecond to microsecond) electron spin relaxation times, transverse PRE dominates, which broadens the lineshapes and quenches the signals in the NMR spectra. In comparison, paramagnetic ions with small numbers of unpaired electrons and short electron-spin relaxation times causes smaller transverse PRE, which allows longitudinal PRE to be detected in unbroadened spectra to quantify the electron-nucleus distances. In addition to PRE, paramagnetic ions cause distance-dependent dipolar hyperfine shifts, also called pseudocontact shifts (PCS) 11,12. This effect has been exploited in small molecules 13,14 as well as metalloproteins 15,16 under fast MAS.
Cu(II) is an especially favorable paramagnetic ion for structure determination by PRE, because it has relatively short electron spin relaxation times and small g-tensor anisotropy, thus causing moderate line broadening and small isotropic PCS, respectively. As a result, Cu(II)-induced T1 and T2 PRE can be measured readily, and has been used to determine the global fold of microcrystalline proteins 10,17 and Cu(II) binding sites in amyloid fibrils 18.
For membrane-bound proteins, paramagnetic ions have so far been mainly used to measure the depth of protein residues from the membrane surface where the metal ions bind 1. Both lanthanides such as Gd(III), Dy(III) and Pr(III) and non-lanthanides such as Mn(II) have been employed for this purpose, and pseudocontact shifts 19 and T2 PRE 20–22 were observed. In addition, Cu-chelated lipids have been employed to enhance the sensitivity of membrane protein NMR 23,24. However, Cu(II) has not been used to investigate the structure of membrane proteins, whether through its non-specific binding to the membrane surface or through site-specific binding to protein sidechains. Compared to globular and fibrous proteins, hydrated lipid membranes present additional complexity for Cu(II) PRE, since Cu(II) can bind lipid phosphates, buffer ions and hydroxide ions in water, in addition to specific protein sites. Therefore, distance determination of membrane proteins using Cu(II) PRE requires careful consideration of multiple binding equilibria.
The influenza M2 protein forms a tetrameric proton channel in the virus envelope that acidifies the virion to initiate virus uncoating 25–28. Proton selectivity is achieved by a single histidine in the transmembrane (TM) domain, His37 29, whereas channel gating is accomplished by Trp41 30, one helical turn away from His37. The channel is inhibited by the adamantane class of antiviral drugs 31,32 and by Cu(II) 33. Whole-cell electrophysiology experiments indicated that Cu(II) inhibition was biphasic: a low-affinity binding site exists outside the TM electric field with an equilibrium dissociation constant (KD) of ~100 μM, while a high-affinity binding site with a KD of ~2 μM exists in the TM electric field. High-affinity Cu(II) binding is sensitive to pH and applied voltage, and Cu(II) inhibits both inward and outward currents. Prior application of the hydrophobic drug BL-1743 prevented Cu(II) inhibition, indicating competitive binding 33. These biochemical data suggested His37 to be the high-affinity Cu(II) binding site; however, no structural data has been reported to determine the exact position of Cu(II). An atomic-level structural study of Cu(II) binding to M2TM will be useful for understanding the Cu(II) inhibition mechanism in comparison with the inhibition mechanism of hydrophobic drugs.
In this study, we investigate the Cu(II) binding site in the M2 TM domain (M2TM) using 13C PRE. We show that high-affinity Cu(II) binding to M2TM is achieved under appropriate pH and solution conditions where the competing binding equilibria are minimized. Based on spectral line broadening, 13C T1 PRE and the resulting distances, we have identified the high-affinity binding site and also propose the low-affinity site detected in the electrophysiology experiments. Our results indicate that the Cu(II) inhibition mechanism is distinct from the amantadine inhibition mechanism, and demonstrate that Cu(II) PRE is a useful and general probe of membrane protein structure and function.
Materials and Methods
Membrane sample preparation
All lipids, including 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), egg sphingomyelin (SM), and cholesterol (Chol), were obtained from Avanti Polar Lipids (Alabaster, AL). The transmembrane domain (residues 22–46) of the M2 protein of the Udorn strain of influenza A virus was synthesized by PrimmBiotech (Cambridge, MA) using Fmoc chemistry and purified to >95% purity. The amino acid sequence is SSDPLVVAASIIGILHLILWILDRL. Three site-specifically labeled peptides were used in this work, with 13C, 15N-labeled residues at Val28, Ser31, Ile32 and Leu36 (VSIL-M2TM), Gly34, His37 and Ile39 (GHI-M2TM) and Leu40 and Trp41 (LW-M2TM).
M2TM was reconstituted into a virus-envelope-mimetic membrane (termed VM+) consisting of POPC: POPE: SM: Chol at a molar ratio of 5: 5: 5: 4.5 34,35. The lipids were codissolved in chloroform and methanol, dried under nitrogen gas and lyophilized overnight. The homogeneous lipid powder was suspended in 1 mL of pH 4.5 water, vortexed, and subject to eight cycles of freeze-thawing to create uniform vesicles. No buffer was used in preparing these membrane samples because many common buffer ions such as phosphates and Tris form insoluble complexes with Cu(II) 36,37, which deplete free Cu(II) ions in solution. M2TM was solubilized in octyl-β-D-glucopyranoside (OG) in 1 mL of pH 4.5 water, then mixed with the 1 mL lipid vesicle solution to reach an OG concentration of 12 mg/mL. The solution was vortexed for 2 hours and dialyzed against 1 L pH 4.5 water at 4°C for 3 days with water changes every 12 hours to remove the detergent. The proteoliposome solution was centrifuged at 150,000 g and ~4°C for 4 hours to give a membrane pellet with ~45% hydration. The pellet was transferred into 4 mm MAS rotors for solid-state NMR experiments. The molar ratio of peptide to lipids (not including cholesterol) was 1: 15. A peptide-free POPC membrane was prepared without dialysis as a control for the PRE experiments. The tetrameric assembly and channel activity of the transmembrane M2 peptide in lipid bilayers have been confirmed by various biophysical experiments and functional assays 7,38,39.
Cu(II)-containing membrane samples
Cu(II) ions were incorporated into the membrane sample by direct titration of copper chloride solution into the MAS rotor. The M2TM monomer concentration in our samples at a hydration level of ~45 wt% is about 0.1 M, thus requiring a minimum amount of 0.025 M Cu(II) to occupy all tetrameric channels. Cu(II) also complexes strongly with hydroxide ions in water to form insoluble copper hydroxide, with a solubility product constant Ksp ≡ [Cu2+]·[OH−]2 of 4.8·10−20 M3. To minimize hydroxide binding and maintain a free Cu(II) concentration of 0.025 M, a low bulk pH of 5.1 is necessary (Fig. S1). In addition, Cu(II) also binds to lipid phosphates. Our titration tests showed that a moderate excess of 4 equivalents of Cu(II) per M2TM tetramer at a monomer: lipid molar ratio of 1:15 saturate the lipid phosphate binding site while yielding sufficient channel binding, as indicated by the 13C PRE. At this Cu(II): tetramer: lipid molar ratio of 4: 1: 60, the sample pH was controlled at 4.8 to avoid Cu(II) precipitation by hydroxide ions. An appropriate amount of Cu(II) chloride solution was titrated into the membrane to reach the desired pH and concentration. After titration, the sample was subject to freeze-thawing again, followed by incubation overnight at 37 °C in the closed rotor to allow Cu(II) equilibration within the membrane. The low pH of the final sample was confirmed by the His37 15N spectra shown below.
Solid-state NMR experiments
Solid-state NMR experiments were carried out on a Bruker DSX-400 MHz (9.4 Tesla) spectrometer using 4 mm triple-resonance MAS probes. Typical radiofrequency pulse lengths were 3.5 – 5.0 μs for 13C, 6.0 μs for 15N, 3.0 – 4.0 μs for 1H and 5.0 μs for 31P. 13C, 15N, and 31P chemical shifts were respectively referenced to the α-Gly 13CO signal at 176.465 ppm on the TMS scale, the 15N signal of N-acetyl-valine at 122.0 ppm on the liquid ammonia scale, and the hydroxyapatite signal at +2.73 on the phosphoric acid scale. Sample temperature was regulated using the built-in heater in the MAS probes and a Kinetics Thermal System XR air-jet sample cooler.
13C T1 relaxation times were measured using Torchia’s z-filter sequence 40 under 8 kHz MAS. 31P spectra were measured by direct polarization under 5–6 kHz MAS at 298 K. 1D double-quantum (DQ) filtered 13C spectra were measured to detect the 13C signals of the labeled peptide without lipid background signals. The SPC5 recoupling sequence 41 was used to excite and reconvert 13C-13C DQ coherence under 5–7 kHz MAS. 2D 13C-13C and 15N-13C correlation spectra were measured at 243 K for resonance assignment of the peptide signals. 2D 13C-13C correlation spectra were measured using a 40 ms spin diffusion mixing time under 6–9 kHz MAS. 2D 15N-13C correlation spectra were measured under 7 kHz MAS using a REDOR-based pulse sequence 42 with a mixing time of 0.8 ms, which mainly gave one-bond 15N-13Cα cross peaks.
Results
Distance extraction using Cu(II) PRE
We first summarize quantitative aspects of distance extraction from Cu(II)-induced PRE. The longitudinal relaxation enhancement, Γ1, is the difference between the relaxation rates in the absence and presence of the paramagnetic ion,
| (1) |
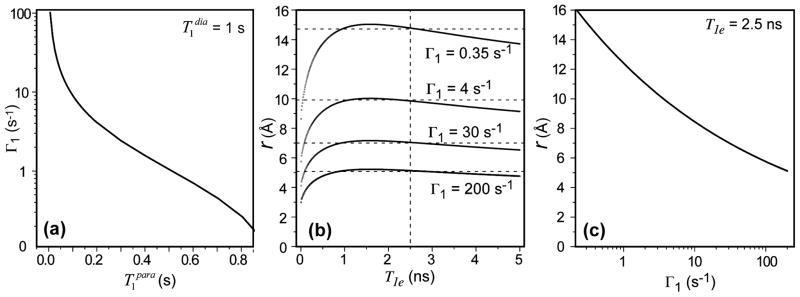
The transverse PRE, Γ2, can be similarly defined. If we assume a diamagnetic 13C of 1.0 s, which is typical for the backbone carbons of membrane-bound proteins at moderate MAS frequencies, then we obtain the dependence of 13C Γ1 on as shown in Fig. 1a. To obtain a Γ1 larger than 10 s−1, which corresponds to a Cu(II) - 13C distance of less than 8 Å (eq. 4), must be shorter than 0.1 s.
Figure 1.
Distance extraction from Cu(II)-induced 13C T1 PRE. (a) Dependence of Γ1 on , assuming a diamagnetic T1 of 1.0 s. (b) 13C-Cu(II) distances as a function of T1e for different Γ1. The distances are insensitive to T1e for T1e > 1 ns. (c) Distances as a function of Γ1 for T1e = 2.5 ns.
The paramagnetic relaxation enhancement of nuclear spins under a metal-centered approximation for the unpaired electron depends on the electron-nucleus distances r according to the Solomon-Bloembergen equations 9,12,43:
| (2) |
Here, μ0 = 4π × 10−7 Tm/A is the vacuum permeability, γn is the nuclear-spin gyromagnetic ratio and equals 2π × 10.705 MHz/T for 13C, ge is the electron spin g-factor, μB = 9.274 × 10−24 J/T is the Bohr magneton, S is the electron-spin quantum number of the paramagnetic ion, and ωn and ωe are the Larmor frequencies of the nucleus and electron at the magnetic field of interest. The longitudinal and transverse correlation times, τc1 and τc2, depend on the electronic relaxation time T1e and T2e, the rotational correlation time (τr) of the molecule, and the exchange correlation time (τM):
| (3) |
To a good approximation, T1e = T2e. For solids without chemical exchange, τr and τM are long, thus τc1 and τc2 are approximately equal to the electronic relaxation times. The T1e of Cu(II) has been estimated to be in the 1 – 5 ns range 44. Since Γ1 is roughly linear with T1e but to the inverse sixth power of distance, it is much more sensitive to the distance than to T1e. Fig. 1b shows the dependence of r on T1e for Γ1 values of 0.35 – 200 s−1. For a given Γ1, the distance is largely invariant with T1e for T1e > 1 ns. For example, for Γ1 = 4 s−1, a distance of 9.4 ± 0.4 Å is obtained for T1e of 1–5 ns. For Γ1 = 0.35 s−1, a longer distance of 15 Å with a small uncertainty of ± 0.70 Å is obtained. Therefore, we use an intermediate T1e value of 2.5 ns in the following distance analysis.
Inputting the various constants and using a magnetic field of 9.4 Tesla, we obtain the distance-dependent 13C T1 PRE:
| (4) |
Fig. 1c plots r as a function of Γ1, which allows us to estimate the limiting measurable 13C-Cu(II) distances. Assuming a minimum of 50 ms before the onset of severe line broadening, a Γ1 of ~20 s−1 is obtained, corresponding to a minimum 13C-Cu(II) distance of about 7.5 Å. At the other extreme, if the closest that can be resolved from a of 1.0 s is 0.9 s, then the Γ1 of 0.11 s–1 corresponds to a maximum 13C-Cu(II) distance of 18 Å. This is a conservative upper limit because at higher MAS frequencies, the 13C T1 will lengthen due to suppression of spin diffusion, which will allow smaller Γ1 values to be resolved to detect longer distances. For distances shorter than 7.5 Å, transverse PRE is expected to become dominant to broaden the spectra. Isotropic PCS may also become observable; however this effect is generally weak for Cu(II) due to its small g-tensor anisotropy and thus less useful for structure determination.
Cu(II) binding to the lipid membrane
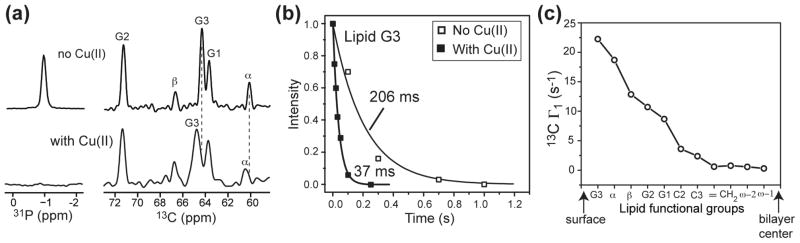
Mutagenesis and electrophysiological experiments suggested the high-affinity Cu(II) binding site to be His37, with a possible equilibrium reaction of Cu2+ + 4His Ä [Cu(His)4]2+. However, in hydrated lipid membranes, two other Cu(II) binding equilibria are also present. Cu(II) can bind to lipid phosphate groups with 2: 1 or 1: 1 stoichiometry 45,46. No binding constant has been reported, but the dissociation constants for a number of other divalent cations such as Mn2+ and Co2+ were found to be in the 1–4 M range 45. Cu(II) also binds hydroxide ions in solution, but this binding is minimized by using acidic pH for our samples. Below we first assess the extent of Cu(II) binding to the membrane surface and its effects on the lipid signals. We measured the 31P and 13C spectra of protein-free POPC bilayers and the lipid 13C T1 in the absence and presence of Cu(II). A low Cu(II): lipid mole ratio of 1: 15, which was identical to the molar ratio of most protein-containing samples, was used. Fig. 2a shows that the 31P MAS signal is completely suppressed after addition of Cu(II), confirming ion binding to the phosphate. In comparison, the headgroup Cα and glycerol G3 carbons, which are the closest carbons to the phosphate, retain the 13C signals but exhibit line broadening and small frequency changes of 0.3 – 0.4 ppm. These frequency changes are not due to Fermi contact shift, since there is no covalent bonding or orbital overlap between the unpaired electron of Cu(II) and lipid carbons. They are also unlikely to be due to lipid conformational changes. First, even at higher ion concentrations than used here, multivalent metal ions only affect the polar headgroup α and β segments, as detected through 2H quadrupolar couplings 47,48. No 2H quadrupolar coupling change was observed for glycerol backbone groups, in contrast to the G3 chemical shift change detected here. Second, our previous PRE studies using the divalent cation Mn(II), which has similar charge-charge interactions with the lipid phosphates as Cu(II), did not show any 13C chemical shift changes 20,22. Therefore, the G3 and Cα chemical shift changes in the Cu(II)-bound membrane most likely result from pseudocontact shifts. Fig. 2b shows a representative pair of 13C T1 relaxation curves: the T1 of glycerol G3 decreased from 206 ms to 37 ms upon Cu(II) binding. Plotting the 13C Γ1 for all lipid groups as a function of their depths from the membrane surface, we obtained the expected monotonic trend of decreasing Γ1 with increasing distance from the membrane surface (Fig. 2c). The Γ1 values range from 22 to 0.3 s−1 (Table S1), corresponding to effective Cu(II) - 13C distances of 7.4 – 15 Å. These values correlate well qualitatively with the depth of the lipid carbons to the membrane surface, which are known from diffraction data 49. Quantitatively, the glycerol G3 and headgroup Cα carbons have longer PRE distances (~7.5 Å) than their depths or distances to the phosphate. This discrepancy can be understood by the fact that Cu(II) ions laterally diffuse on the surface of the neutral lipid membrane 45,46, with residence times at an individual lipid headgroup much shorter than 10−6 s 47,48, thus reducing the PRE effect through 〈r〉−6 and a shorter correlation time 50. For acyl-chain carbons, this distance-averaging effect is attenuated because the significant depths of these carbons make the instantaneous distances to the surface Cu(II) ions more comparable. Overall, Table S1 shows that for the bottom half of the acyl chain carbons, the relaxation enhancement due to the surface-bound Cu(II) is 0.1 – 1 s−1. Therefore, if protein residues well embedded in the middle of the membrane experience 13C T1 PREs that are much larger than this range, then a Cu(II) binding site in the protein must exist at much closer proximity than the surface-bound Cu(II).
Figure 2.
Effects of Cu(II) binding on the NMR spectra of POPC bilayers. (a) 31P and 13C spectra without (top) and with (bottom) Cu(II) at the 1: 15 Cu(II): lipid molar ratio. The 31P signal is quenched by T2 PRE and the glycerol and headgroup 13C signals show line broadening and small pseudocontact shifts for G3 and Cα. (b) 13C T1 relaxation curves of G3. (c) 13C T1 PRE for lipid functional groups from the bilayer surface to bilayer center, showing monotonically decreasing Γ1.
Cu(II)-induced 13C PRE of membrane-bound M2TM
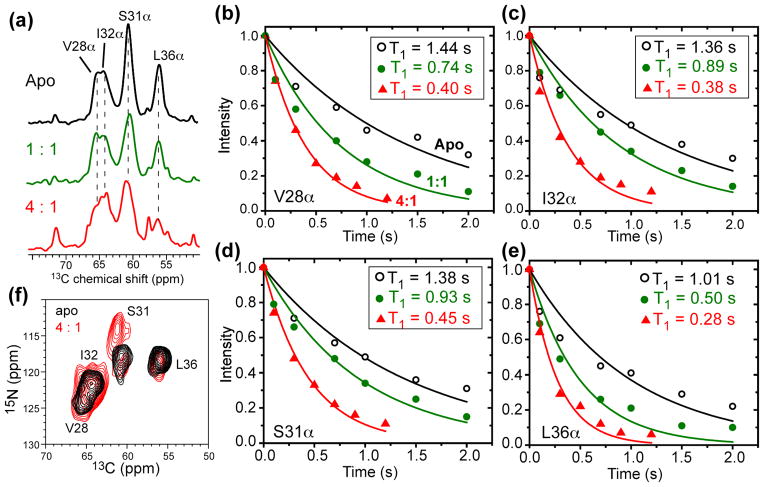
To determine the specific binding site of Cu(II) in M2, we measured the 13C T1 PRE of nine transmembrane residues, including Val28, Ser31, Ile32, Gly34, Leu36, His37, Ile39, Leu40 and Trp41. Fig. 3a shows the aliphatic region of the 13C spectra of Val28, Ser31, Ile32, Leu36-labeled M2TM in the absence and presence of Cu(II). Among the four labeled residues, Leu36 Cα and Cβ exhibited the strongest line broadening, as shown in both 1D and 2D 13C spectra (Fig. S2), indicating that Leu36 is the closest to Cu(II) among the four labeled residues. Fig. 3b–e shows the 13C T1 relaxation curves of Cu(II)-free and Cu(II)-bound samples. In addition to the 4:1 Cu(II): tetramer sample, we also examined the spectra of a 1:1 Cu(II): tetramer sample to observe the progression of PRE effects. Sizeable T1 relaxation enhancement was detected for all residues. At the 4:1 ratio, 13C Γ1 ranges from 1.5 to 2.6 s−1 (Table 1), which corresponds to distances of 11.6 – 10.6 Å to the Cu(II) center.
Figure 3.
Effects of Cu(II) binding on the chemical shifts and 13C T1 relaxation of VSIL-M2TM in the VM+ membrane. (a) 13C spectra in the absence (black) and presence of Cu(II) at 1:1 (green) and 4:1 (red) Cu(II): tetramer ratios. Leu36 showed the strongest line broadening (also see Fig. S2). (b–e) Representative 13C T1 relaxation curves for the apo, 1:1, and 4:1 samples. (b) Val28 Cα. (c) Ile32 Cα. (d) Ser31 Cα. (e) Leu36 Cα. (f) 2D 15N-13C correlation spectra of apo and 4:1 Cu(II)-bound VSIL-M2TM samples. Ser31 exhibits a clear 15N chemical shift change.
Table 1.
13C T1 and T2 PREs of membrane-bound M2TM. A Cu(II): tetramer ratio of 4:1 was used for the bound samples.
| Residue | Site | Ipara/Idia | (s) | (s) | Γ1 (s−1) | r (Å) |
|---|---|---|---|---|---|---|
| V28 | Cα | 0.92 | 1.4 ± 0.2 | 0.40 ± 0.01 | 1.80±0.12 | 11.2±0.1 |
| S31 | Cα | 0.70 | 1.4 ± 0.2 | 0.45 ± 0.02 | 1.51±0.14 | 11.6±0.2 |
| I32 | Cα | 1.0 | 1.4 ± 0.2 | 0.38 ± 0.03 | 1.92±0.23 | 11.1±0.2 |
| Cγ2 | 0.77 | 2.4 ± 0.2 | 0.53 ± 0.01 | 1.47±0.05 | 11.6±0.1 | |
| Cδ1 | 0.74 | 2.4 ± 0.3 | 0.46 ± 0.02 | 1.76±0.11 | 11.3±0.1 | |
| G34 | Cα | 0.22 | 2.6 ± 0.2 | 0.25 ± 0.01 | 3.62±0.16 | 10.0±0.1 |
| L36 | Cα | 0.52 | 1.0 ± 0.1 | 0.28 ± 0.02 | 2.57±0.27 | 10.6±0.2 |
| Cβ | 0.61 | 0.7 ± 0.1 | 0.30 ± 0.02 | 1.90±0.30 | 11.1±0.3 | |
| H37 | Cα | 0.18 | 1.9 ± 0.3 | - | - | < 7.5 |
| Cγ | 0.13 | - | - | - | < 7.5 | |
| Cδ2 | 0.16 | - | - | - | < 7.5 | |
| Nδ1 | 0.11 | - | - | - | < 7.5 | |
| I39 | Cα | 0.58 | 0.77 ± 0.09 | 0.20 ± 0.03 | 3.70±0.77 | 10.0±0.3 |
| Cγ2 | 0.60 | 1.2 ± 0.1 | 0.36 ± 0.04 | 1.94±0.32 | 11.1±0.3 | |
| Cδ1 | 0.73 | 1.2 ± 0.1 | 0.34 ± 0.03 | 2.11±0.27 | 11.0±0.2 | |
| L40 | Cα | 0.23 | 2.4 ± 0.1 | 0.11 ± 0.02 | 8.67±1.65 | 8.7±0.3 |
| Cβ | 0.45 | 1.5 ± 0.3 | 0.28 ± 0.01 | 2.90±0.19 | 10.4±0.1 | |
| W41 | Cα | 0.33 | 2.5 ± 0.3 | 0.19 ± 0.03 | 4.86±0.83 | 9.5±0.3 |
| Cδ1 | 0.40 | 3.3 ± 0.1 | 0.23 ± 0.02 | 4.04±0.38 | 9.8±0.2 | |
| Cη2 | 0.70 | 2.4 ± 0.2 | 0.25 ± 0.03 | 3.58±0.48 | 10.0±0.2 | |
| Cε3 | 0.63 | 2.4 ± 0.1 | 0.23 ± 0.02 | 3.93±0.38 | 9.9±0.2 | |
| Cζ3, Cγ | 0.50 | 3.4 ± 0.3 | 0.26 ± 0.01 | 3.55±0.15 | 10.0±0.1 |
In addition to enhancing 13C longitudinal and transverse relaxation, Cu(II) binding also perturbed the 15N chemical shift of Ser31 from 117.9 ppm in the apo state to 114.1 ppm in the Cu(II) bound state (Fig. 3f). This large shift cannot be due to PCS but must result from Cu(II)-induced conformational change of this residue. We previously showed that Ser31 is the site of maximal chemical shift perturbation by amantadine, where the 15N chemical shift increased from 115 ppm in the apo state to 121 ppm in the drug-bound state in DLPC bilayers 51. Similar 15N chemical shift increase was also observed when M2TM was bound to virus-mimetic membranes 34. The fact that Cu(II) binding changed the Ser31 15N chemical shift in the opposite direction indicates that Cu(II) affects M2TM conformation differently from amantadine.
Fig. 4 shows the 13C and 15N spectra of GHI-M2TM in the absence and presence of Cu(II). The 13C spectra were measured with a 13C double-quantum filter to suppress the lipid natural abundance 13C signals. Among the three residues, the His37 signals were nearly completely quenched by Cu(II) (<18% residual intensity), while Ile39 retained the most intensities. The aromatic signals of His37 were largely undetectable, as shown by both 1D and 2D 13C correlation spectra (Fig. 4b, c). These results strongly indicate that Cu(II) binds to His37. For the detectable Gly34 and Ile39 residues, most sites exhibited clear T1 relaxation enhancement (Fig. S3a, b). In addition, the resolved lipid 13C signals such as glycerol backbone G2 showed comparable T1 PRE as the protein-free membrane (Fig. S3c), indicating that Cu(II) binds to the membrane surface similarly whether the protein is present or not. In addition to causing PRE, Cu(II) also perturbed the conformation of Gly34: the apo peptide exhibited two Gly34 CO peaks at 175.6 and 173.2 ppm while the Cu(II)-bound peptide showed a CO peak at 172.2 ppm. Previous chemical shift analysis 52 and orientation measurements 53 indicated that the downfield CO peak at 175 ppm corresponded to M2TM with a kink at Gly34, while a straight TM helix exhibited the more upfield Gly34 CO chemical shift.
Figure 4.
13C and 15N MAS spectra of GHI-M2TM in the VM+ membrane without Cu(II) (black) and with Cu(II) at the 4:1 Cu(II): tetramer ratio (red). (a) 1D 13C double-quantum filtered spectra detecting only peptide signals. The His37 aliphatic and carbonyl signals are mostly suppressed by Cu(II). (b) Aromatic region of the 1D 13C MAS spectra with and without Cu(II). Except for the lipid signal at 130 ppm (denoted by a star), all imidazole signals are suppressed by Cu(II). (c) Regions of the 2D 13C-13C correlation spectra of GHI-M2TM. Cu(II) binding suppressed the His37 signals and perturbed the Gly34 CO chemical shift. (d) 1D 15N MAS spectra. The His37 Nε2 signal is removed while a small fraction (~10%) of the Nδ1 intensity remains. The two spectra are scaled such that the Nδ1 intensity is matched to show the preferential suppression of the Nε2 peak. No 250-ppm 15N signal is observed, verifying the acidic pH of the samples. The Cu(II)-Nε2 chelated imidazole structure is shown in the box.
In principle, Cu(II) can bind to either Nε2 or Nδ1 of His37, which faces the C-terminus and N-terminus of the channel, respectively, due to the trans-trans rotamer of the His37 sidechain 54,55. To determine which nitrogen coordinates Cu(II), we measured the 15N MAS spectra of His37-labeled M2TM. Previous 2D 15N-13C correlation spectra showed that the two nitrogens of the imidazolium ring differed in their isotropic chemical shifts by 4.1 ppm: Nε2 resonates at 176.3 ppm while Nδ1 resonates at 180.4 ppm 54, due to the higher proton affinity of Nε2 throughout a large pH range. Fig. 4d shows that Cu(II) binding completely suppressed the upfield Nε2 peak while retaining ~10% of the Nδ1 signal, suggesting that Cu(II) binds preferentially to Nε2.
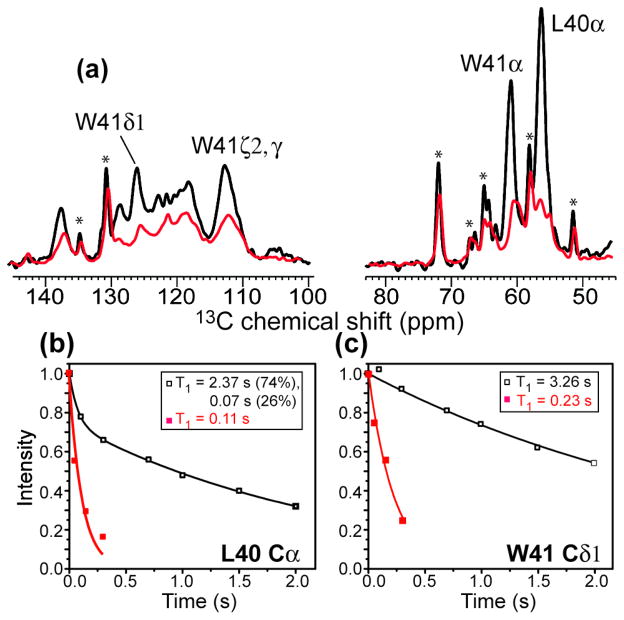
If Nε2 binding is correct, then residues immediately C-terminal to His37 should manifest larger PRE than residues immediately N-terminal to His37. To test this hypothesis, we measured the 13C PRE of LW-M2TM. Indeed, both the backbone and sidechain 13C signals of Leu40 and Trp41 exhibited significant line broadening (Fig. 5a), with Cα peaks decreasing to 20–30% of the apo intensities. 13C T1 relaxation rates also increased substantially. Leu40 Cα gave a Γ1 of 8.67 s−1, corresponding to a distance of ~8.7 Å to Cu(II) (Table 1). This Γ1 was much larger than the Gly34 Cα Γ1 of 3.46 s−1. Since Leu40 and Gly34 are both three residues away from His37, their different PRE lends further support to the conclusion that Cu(II) binds the imidazolium rings from the C-terminal side, between His37 and Trp41.
Figure 5.
13C T1 PRE of Leu40 and Trp41 in M2TM. (a) 1D 13C spectra of LW-M2TM without Cu(II) (black) and with Cu(II) at the 4:1 Cu(II): tetramer ratio (red). Key peptide signals are assigned. Lipid 13C signals are indicated by stars. (b–c) 13C T1 relaxation curves for Leu40 Cα (b) and Trp41 Cδ1 (c). Strong T1 relaxation enhancements are observed, supporting Cu(II) binding to Nε2 between His37 and Trp41. A biexponential function was used to fit the Leu40 Cα decay data due to partial overlap with the lipid Cγ peak.
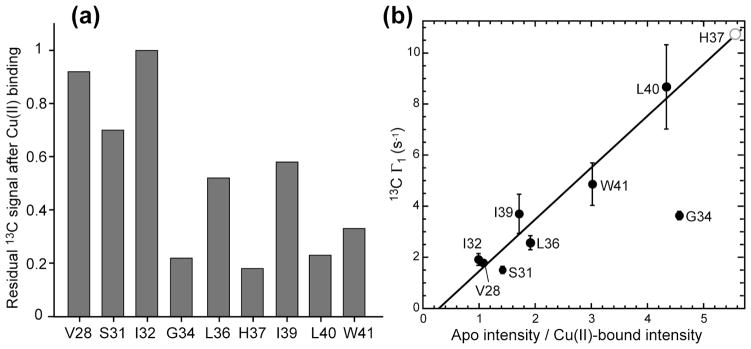
Table 1 lists the 13C T1 PREs and the resulting distances for all labeled residues. The 13C intensity reduction, measured as the ratio Ipara/Idia between the Cu(II) bound sample and the diamagnetic sample, is also listed to reflect the qualitative T2 relaxation enhancement. For the intensity-suppressed His37, a conservative distance upper limit of 7.5 Å was used, although the actual distances of Cu(II) to the imidazolium sidechain atoms are almost certainly much shorter. The central segment of the TM helix, from Gly34 to Leu40, manifests longitudinal PRE in the range of 3–9 s−1 (Table 1), which is an order of magnitude larger than the PRE of lipid chain carbons at comparable depths from the membrane surface (Table S1). This confirms the presence of a high-affinity Cu(II) binding site within this segment, which is chiefly responsible for the PRE effects of these residues. Fig. 6a plots the residual Cα intensities after Cu(II) binding. The minimum intensity occurs at His37, indicating the Cu(II) binding site. Gly34 Cα also showed low intensity. However, this value does not accurately reflect a very short distance to the Cu(II) center, because the CH2 Cα group of Gly experiences more severe 1H signal quenching by Cu(II) than the CH Cα group of other amino acids, thus disproportionally reducing the Cα intensity through 1H-13C cross polarization. Fig. 6b compares the 13C T1 PRE of Cα sites with their intensity reduction factors. The non-Gly residues show a significant correlation between Γ1 and intensity reduction, consistent with the common r−6 dependence of Γ1 and Γ2. Although the His37 Cα Γ1 could not be measured, based on its intensity reduction factor, a Γ1 of about 10 s−1 is expected.
Figure 6.
Correlation between 13C T1 and T2 PRE. (a) Cu(II) induced reduction of the Cα intensities of M2TM, reflecting T2 PRE. (b) Correlation between the 13C T1 PRE and the 13C intensity reduction factor, which is the inverse of the residual intensity plotted in (a). Residues that show large T1 PRE also have large intensity reductions. Gly34 Cα shows anomalously low intensities (strong T2 PRE) because the CH2 Cα group is more sensitive to 1H quenching by Cu(II) than the CH Cα groups of all other amino acid residues. The His37 Cα intensity reduction suggests a 13C Γ1 of ~10 s−1 (open circle).
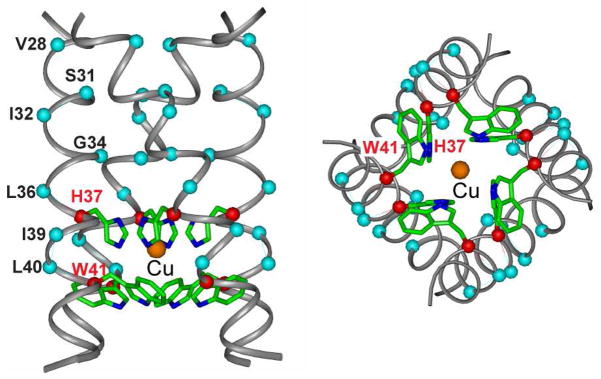
Fig. 7 shows the structural model of Cu(II) binding to His37 in M2TM. We used the 1.65 Å crystal structure (PDB: 3LBW) to model Cu(II) distances to various residues, but the same conclusion was reached when using the recent solid-state NMR structure (PDB: 2KQT) of M2TM 56. We placed Cu(II) at the center of the square formed by the four equivalent His37 Nε2 atoms. Thus positioned, the Cu(II) ion is ~3.0 Å from the nitrogens at the four corners, which is within the literature range of Cu(II) distances to histidine nitrogens 57. This Nε2 binding site yields distances in good agreement with the PRE-measured values for the central segment of the TM helix (Table S2). Compared to the alternative Nδ1 binding model, the Nε2 binding model has longer distances to the N-terminal residues and shorter ones to the C-terminal residues. The largest distance difference between the two models occurs at Gly34 Cα, which is 8.7 Å from the Nε2-bound Cu(II) but only 6.3 Å from the Nδ1 binding site. The latter would correspond to a Γ1 of about 60 s−1, which is not detected. Moreover, such a short distance would imply complete quenching of the Gly34 signals, which is inconsistent with the experimental data (Fig. 4a). Another key difference between the two binding sites is the relative distance between Leu36 and Leu40. Cu(II) binding at the upper Nδ1 site would predict a shorter distance to Leu36 than to Leu40, which is opposite the trend seen in the spectral intensities (less residual intensities for Leu40) and the T1 PRE (higher Γ1 for Leu40). Taken together, the quantitative 13C Γ1 and the spectral line broadening consistently support His37 Nε2 as the Cu(II) binding site.
Figure 7.
Proposed model of the Cu(II) binding site in M2TM. Cu(II) (orange), bound to the center of the square formed by the Nε2 atoms of the His37 tetrad, is trapped in an aromatic cage formed by the four His37 imidazolium rings from the top and the four Trp41 indole rings from the bottom, thus explaining the high affinity of the binding and the mechanism of inhibition of proton conduction.
For residues close to the termini of the TM peptide, the measured PRE distances are shorter than the predicted distances to the His37 Nε2 binding site. The discrepancy is especially obvious for Val28, whose predicted distances are 18 and 16 Å for the Nε2 or Nδ1 binding sites, while the measured distance is 11 Å (Table S2). This discrepancy results from the fact that Val28 is much closer to the membrane surface (10–11 Å) than to His37 (Table S2), thus should experience significant PRE effects from the surface-bound Cu(II).
Discussion
High affinity binding site of Cu(II) at His37 Nε2: a His-Trp “cage” binding model
The observed 13C and 15N spectral broadening (T2 PRE) and 13C T1 PRE indicate that Cu(II) binds to His37 Nε2, not far from the gating residue Trp41 (Fig. 7). Metal chelation to imidazole Nε2 in favor of Nδ1 has been often observed in proteins 18,57,58. In the case of M2, Cu(II) entrapment in the aromatic cage formed by the four imidazolium rings and four indole rings enhances the Cu(II) affinity compared to binding to Nδ1 on the N-terminal side, where Cu(II) can dissociate more readily. By binding between the His37 and Trp41 tetrads, Cu(II) can inhibit the proton current in multiple ways, including perturbing His37-water proton exchange 59, preventing His37 conformational dynamics 54, and interfering with His-Trp cation-π interaction 60,61. The Nε2 coordination of Cu(II) implicitly disagrees with a structural model where Nε2 of a charged imidazolium hydrogen-bonds with Nδ1 of a neutral imidazole 62. At the low pH used for the NMR samples, both Nε2 and Nδ1 are initially protonated, thus Cu(II) binding to Nε2 must at first displace a proton, which is consistent with the slower onset of Cu(II) inhibition at low pH than at high pH.
When Cu(II) solution was injected into internally high-pH oocytes that are bathed in externally low pH solution, it did not inhibit wild-type M2 channels but inhibited the W41A mutant channel. This was interpreted as the inability of Cu(II) to pass the bulky Trp indole rings from the C-terminal side, an effect that was removed by mutation to the smaller Ala. At the symmetric low pH condition used in our SSNMR samples, Cu(II) entry to the His37 binding site is more likely bidirectional, since the Trp41 gate should be open at acidic pH, as indicated by NMR distance measurements 8 and crystal structures 55,63. In principle, it would be interesting to measure Cu(II) binding at higher pH where the Trp41 gate is closed and the Cu(II) affinity for His37 is higher. However, to avoid Cu(II) binding to the more abundant hydroxide ions present at higher pH, a much lower concentration of Cu(II) must be used, which would in turn require a much lower concentration of protein than feasible by solid-state NMR in the absence of sensitivity-enhancement techniques.
Low affinity binding site for Cu(II)
The current 13C PRE data strongly suggest that the low affinity Cu(II) binding site seen in the electrophysiological data 33 is the lipid phosphate on the membrane surface. Even at a low Cu(II) concentration, the surface-bound Cu(II) caused detectable PRE to Val28, which is relatively far from His37 but closer to the membrane surface. Inspection of the 31P spectra of the lipids and 2D 15N-13C spectra of VSIL-M2TM (Fig. S4) upon Cu(II) titration from 0, 1: 1 to 4: 1 Cu(II): tetramer ratios gave useful insight into the Cu(II) binding process. The 31P spectra showed a small residual intensity at the 1:1 ratio but no intensity at the 4:1 ratio. The Val28 peak became broadened already at the 1:1 ratio and broadened further at the 4:1 ratio. In comparison, the Ser31 peak did not shift from the apo position at the 1:1 ratio but moved significantly in the 15N dimension at the 4:1 ratio. These results indicate that the first Cu(II) ions bind to the membrane surface, causing weak PREs to the most surface-proximal residues. When the Cu(II) concentration increases, the membrane surface binding sites become saturated, fully suppressing the 31P intensity. Additional Cu(II) ions then enter the channels to bind His37, and in so doing cause significant conformational changes at Ser31 and Gly34. Because of the immediate accessibility of the membrane surface to water and the necessity for ion diffusion into the channel pore to reach the His37 binding site, Cu(II) binding to the membrane surface is faster than to His37, which explains the fast initial kinetics seen in the proton conductivity measurements. However, the channel binding site with the slower kinetics has higher affinity, due to Cu(II) chelation with the imidazolium Nε2 as well as entrapment by the His-Trp cage.
The lipid phosphate binding model is consistent with biochemical evidence that the low-affinity site is non-specific, has fast kinetics, and does not depend on various sidechains in the protein that could bind metal ions, including Cys17, Cys19, Asp24 and Ser31 33. Membrane-surface bound Cu(II) may hinder proton conduction by influencing the conformation of the cytoplasmic domain of the full-length protein, which may in turn subtly perturb the conformation and packing of the TM helix.
Cu(II) versus amantadine-induced conformational changes of M2TM
Electrophysiological data indicate both similarities and differences between Cu(II) inhibition and amantadine inhibition of the M2 proton channel. While both compounds inhibit the channel bidirectionally and better at high pH, the Cu(II) binding affinity is about 10-fold weaker than amantadine 33. Cu(II) binding is also competitive with hydrophobic drugs: prior application of BL-1743 to M2 prevented Cu(II) inhibition. Since BL-1743 is believed to act similarly as amantadine, we can understand this competitive inhibition by the fact that amantadine and BL-1743 binding to the N-terminal pore 56,63, at Ser31, occludes the channel, thus preventing Cu(II) entry to His37. An additional effect may be that amantadine-induced dehydration of the channel 64 and the resulting perturbation of the His37 protonation equilibria 53,59 may interfere with Cu(II) binding.
Consistent with the functional data, the observed 13C and 15N chemical shifts indicate several differences between the Cu(II)-bound and amantadine-bound M2TM conformations. Ser31 and Gly34 exhibit Cu(II)-bound chemical shifts that are distinct from the amantadine-bound chemical shifts 51,65. The Cu(II)-bound chemical shifts at these sites are more consistent with a straighter TM helix at low pH 52. These differences are clearly related to the distinct binding sites at His37 and Ser31. Therefore, the structural and mechanistic information obtained here suggests that it should be possible to design new drugs 66 to target the His37 binding site as a route to inhibit the naturally occurring amantadine-resistant M2 mutants such as S31N and V27A.
Cu(II) PRE for studying membrane proteins
The distances extracted from 13C T1 PRE (Table 1) have relatively small uncertainties of 0.1–0.3 Å, as propagated from the random uncertainties in the measured T1 values. However, it is useful to evaluate the extent of systematic uncertainties, such as dynamics of the protein, uncertainty in the occupancy of the His37 binding site, the presence of 13C spin diffusion, and the effects of multiple Cu(II) ions on the 13C PRE. In general, protein anisotropic motion and incomplete Cu(II) occupancy at its binding site will attenuate the PRE, whereas multi-spin effects due to multiple Cu(II) ions should increase the PRE. Thus, these error sources should partially cancel. More careful considerations indicate that each of these potential sources of uncertainties is in fact minimal under the conditions of our experiments. Due to the use of the cholesterol- and SM-rich VM+ membrane for M2TM reconstitution, no large-amplitude motion of the tetramer backbone is present 35, in contrast to the situation in DMPC and POPC membranes 67. The occupancy of the His37 binding site is close to 100%, otherwise significant residual imidazole intensities would be detected. While 13C spin diffusion is present at the MAS frequency of 8 kHz for the T1 experiments, the use of site-specifically labeled samples should present significant barrier to inter-residue spin diffusion, thus making 13C T1 values relatively site-specific, as verified by the measured values (Table 1). Finally, at the Cu(II): tetramer: lipid molar ratio of 4: 1: 60, the surface density of Cu(II) is low, with about 3 Cu(II) per 60 lipids. With a typical lipid headgroup area of 60 Å2, the average distance between two Cu(II) ions on the membrane surface is about 35 Å. This distance is much larger than the thickness of one leaflet of the bilayer, thus membrane-embedded residues should experience at most only one surface-bound Cu(II) in addition to the His37-bound Cu(II). The surface-bound Cu(II) is also dynamic with respect to the protein, which further weakens its PRE effect. Finally, the PRE of surface-bound Cu(II) can in principle be internally calibrated using the lipid 13C signals if the approximate depths of the residues are known (Table S2). Therefore the surface Cu(II) PRE effects can be taken into account more quantitatively, if necessary, to analyze the PRE due to the protein-bound Cu(II).
This study demonstrates that functionally important paramagnetic ions in membrane proteins can be used as a structural probe to both determine the metal binding site and provide long distance constraints. For M2, Cu(II) binding to histidine is sufficiently tight due to the binding site geometry that no dissociation is detected, and additional binding equilibria to hydroxide ions and lipid phosphates can be minimized or controlled using appropriate pH and Cu(II) concentration. For such functional binding sites in proteins, metal chelating agents cannot be used, thus alternative paramagnetic NMR approaches where metal chelates are covalently attached to protein sidechains 10 or incorporated into lipids or solution 68 are not applicable. This study shows that membrane proteins containing high-affinity paramagnetic metal ions should be generally amenable to structural investigation.
Supplementary Material
Acknowledgments
The authors would like to thank Professors Yoshitaka Ishii, Klaus Schmidt-Rohr and Claudio Luchinat for insightful discussions. This work is supported by NIH grant GM088204 to M. H.
Footnotes
Supporting Information Available
Additional tables and NMR spectra are provided. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Hong M. Acc Chem Res. 2006;39:176–183. doi: 10.1021/ar040037e. [DOI] [PubMed] [Google Scholar]
- 2.Hong M, Zhang Y, Hu F. Annu Rev Phys Chem. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luca S, Heise H, Baldus M. Acc Chem Res. 2003;36:858–65. doi: 10.1021/ar020232y. [DOI] [PubMed] [Google Scholar]
- 4.Loquet A, Giller K, Becker S, Lange A. J Am Chem Soc. 2010;132:15164–15166. doi: 10.1021/ja107460j. [DOI] [PubMed] [Google Scholar]
- 5.De Paëpe G, Lewandowski JR, Loquet A, Böckmann A, Griffin RG. J Chem Phys. 2008;129:245101. doi: 10.1063/1.3036928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt-Rohr K, Hong M. J Am Chem Soc. 2003;125:5648–5649. doi: 10.1021/ja0344415. [DOI] [PubMed] [Google Scholar]
- 7.Luo W, Hong M. J Am Chem Soc. 2006;128:7242–7251. doi: 10.1021/ja0603406. [DOI] [PubMed] [Google Scholar]
- 8.Luo W, Mani R, Hong M. J Phys Chem. 2007;111:10825–10832. doi: 10.1021/jp073823k. [DOI] [PubMed] [Google Scholar]
- 9.Solomon I. Phys Rev. 1955;99:559–565. [Google Scholar]
- 10.Nadaud PS, Helmus JJ, Kall SL, Jaroniec CP. J Am Chem Soc. 2009;131:8108–8120. doi: 10.1021/ja900224z. [DOI] [PubMed] [Google Scholar]
- 11.Nayeem A, Yesinowski JP. J Chem Phys. 1988;89:4600–4608. [Google Scholar]
- 12.Bertini I, Turano P, Vila AJ. Chem Rev. 1993;93:2833–2932. [Google Scholar]
- 13.Lee H, Polenova T, Beer RH, McDermott AE. J Am Chem Soc. 1999;121:6884–6894. [Google Scholar]
- 14.Wickramasinghe NP, Shaibat MA, Ishii Y. J Phys Chem B. 2007;111:9693–9696. doi: 10.1021/jp0727289. [DOI] [PubMed] [Google Scholar]
- 15.Balayssac S, Bertini I, Bhaumik A, Lelli M, Luchinat C. Proc Natl Acad Sci U S A. 2008;105:17284–17289. doi: 10.1073/pnas.0708460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pintacuda G, Giraud N, Pierattelli R, Böckmann A, Bertini I, Emsley L. Angew Chem Int Ed Engl. 2007;46:1079–1082. doi: 10.1002/anie.200603093. [DOI] [PubMed] [Google Scholar]
- 17.Nadaud PS, Helmus JJ, Höfer N, Jaroniec CP. J Am Chem Soc. 2007;129:7502–7503. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- 18.Parthasarathy S, Long F, Miller Y, Xiao Y, McElheny D, Thurber K, Ma B, Nussinov R, Ishii Y. J Am Chem Soc. 2011;133:3390–3400. doi: 10.1021/ja1072178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aime S, Botta M, Mainero V, Terreno E. Magn Reson Med. 2002;47:10–13. doi: 10.1002/mrm.10048. [DOI] [PubMed] [Google Scholar]
- 20.Buffy JJ, Hong T, Yamaguchi S, Waring A, Lehrer RI, Hong M. Biophys J. 2003;85:2363–2373. doi: 10.1016/s0006-3495(03)74660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grobner G, Glaubitz C, Watts A. J Magn Reson. 1999;141:335–9. doi: 10.1006/jmre.1999.1894. [DOI] [PubMed] [Google Scholar]
- 22.Su Y, Mani R, Hong M. J Am Chem Soc. 2008;130:8856–8864. doi: 10.1021/ja802383t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang M, Berthold DA, Rienstra CM. J Phys Chem Lett. 2011;2:1836–1841. doi: 10.1021/jz200768r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto K, Xu J, Kawulka KE, Vederas JC, Ramamoorthy A. J Am Chem Soc. 2010;132:6929–6931. doi: 10.1021/ja102103n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cady SD, Luo WB, Hu F, Hong M. Biochemistry. 2009;48:7356–7364. doi: 10.1021/bi9008837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb RA, Holsinger KJ, Pinto LH. In: Cellular Receptors of Animal Viruses. Wemmer E, editor. Cold Spring Harbor Lab Press; Plainview, NY: 1994. pp. 303–321. [Google Scholar]
- 27.Pinto LH, Lamb RA. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Qiu JX, Soto CS, DeGrado WF. Curr Opin Struct Biol. 2011;21:68–80. doi: 10.1016/j.sbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Lamb RA, Pinto LH. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Y, Zaitseva F, Lamb RA, Pinto LH. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 31.Wang C, Takeuchi K, Pinto LH, Lamb RA. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balannik V, Wang J, Ohigashi Y, Jing X, Magavern E, Lamb RA, DeGrado WF, Pinto LH. Biochemistry. 2009;48:11872–11882. doi: 10.1021/bi9014488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gandhi CS, Shuck K, Lear JD, Dieckmann GR, DeGrado WF, Lamb RA, Pinto LH. J Biol Chem. 1999;274:5474–5482. doi: 10.1074/jbc.274.9.5474. [DOI] [PubMed] [Google Scholar]
- 34.Cady SD, Wang T, Hong M. J Am Chem Soc. 2011;133:11572–11579. doi: 10.1021/ja202051n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo W, Cady SD, Hong M. Biochemistry. 2009;48:6361–6368. doi: 10.1021/bi900716s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masi D, Mealli CMS, Sabatini A, Vacca A, Zanobini F. Helv Chim Acta. 1984;67:1818–1826. [Google Scholar]
- 37.Hegetschweiler K, Saltman P. Inorg Chem. 1986;25:107–109. [Google Scholar]
- 38.Howard KP, Lear JD, DeGrado WF. Proc Natl Acad Sci USA. 2002;99:8568–72. doi: 10.1073/pnas.132266099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Schön A, Magavern E, Jing X, Lear JD, Freire E, Lamb RA, DeGrado WF, Pinto LH. Proc Natl Acad Sci USA. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torchia DA, Szabo A. J Magn Reson. 1982;49:107–121. [Google Scholar]
- 41.Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. J Chem Phys. 1999;110:7983–7992. [Google Scholar]
- 42.Hong M, Griffin RG. J Am Chem Soc. 1998;120:7113–7114. [Google Scholar]
- 43.Bloembergen N. J Chem Phys. 1957;27:572–573. [Google Scholar]
- 44.Bertini I, Luchinat C, Parigi G. Solution NMR of paramagnetic molecules: applications to metallobiomolecules and models. Amsterdam; New York: Elsevier; 2001. [Google Scholar]
- 45.McLaughlin A, Grathwohl C, McLaughlin S. Biochim Biophys Acta. 1978;513:338–357. doi: 10.1016/0005-2736(78)90203-1. [DOI] [PubMed] [Google Scholar]
- 46.Puskin JS. J Membr Biol. 1977;35:39–55. doi: 10.1007/BF01869939. [DOI] [PubMed] [Google Scholar]
- 47.Altenbach C, Seelig J. Biochemistry. 1984;23:3913–3920. doi: 10.1021/bi00312a019. [DOI] [PubMed] [Google Scholar]
- 48.Seelig J, Macdonald PM, Scherer PG. Biochemistry. 1987;26:7535–7541. doi: 10.1021/bi00398a001. [DOI] [PubMed] [Google Scholar]
- 49.Wiener MC, White SH. Biophys J. 1992;61:434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeMarco ML, Woods RJ, Prestegard JH, Tian F. J Am Chem Soc. 2010;132:1334–1338. doi: 10.1021/ja907518x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cady SD, Mishanina TV, Hong M. J Mol Biol. 2009;385:1127–1141. doi: 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu F, Luo W, Cady SD, Hong M. Biochim Biophys Acta. 2011;1808:415–423. doi: 10.1016/j.bbamem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, Fu R, Cross TA. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu F, Luo W, Hong M. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Acharya A, Carnevale V, Fiorin G, Levine BG, Polishchuk A, Balannick V, Samish I, Lamb RA, Pinto LH, DeGrado WF, Klein ML. Proc Natl Acad Sci U S A. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brinen LS, Willett WS, Craik CS, Fletterick RJ. Biochemistry. 1996;35:5999–6009. doi: 10.1021/bi9530200. [DOI] [PubMed] [Google Scholar]
- 58.Klabunde T, Eicken C, Sacchettini JC, Krebs B. Nat Struct Biol. 1998;5:1084–1090. doi: 10.1038/4193. [DOI] [PubMed] [Google Scholar]
- 59.Hu F, Schmidt-Rohr K, Hong M. J Am Chem Soc. 2012;134:3703–3713. doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okada A, Miura T, Takeuchi H. Biochemistry. 2001;40:6053–6060. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- 61.Nishimura K, Kim S, Zhang L, Cross TA. Biochemistry. 2002;41:13170–13177. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- 62.Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath D, Zhou HX, Cross TA. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo W, Hong M. J Am Chem Soc. 2010;132:2378–2384. doi: 10.1021/ja9096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cady SD, Hong M. Proc Natl Acad Sci USA. 2008;105:1483–1488. doi: 10.1073/pnas.0711500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, Ma C, Fiorin G, Carnevale V, Wang T, Hu F, Lamb RA, Pinto LH, Hong M, Klein ML, DeGrado WF. J Am Chem Soc. 2011;133:12834–12841. doi: 10.1021/ja204969m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cady SD, Goodman C, Tatko C, DeGrado WF, Hong M. J Am Chem Soc. 2007;129:5719–5729. doi: 10.1021/ja070305e. [DOI] [PubMed] [Google Scholar]
- 68.Linser R, Fink U, Reif B. J Am Chem Soc. 2009;131:13703–13708. doi: 10.1021/ja903892j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.