Abstract
We have previously demonstrated that the proteasome serves as a central regulator of inflammation and macrophage function. Until recently, proteasomes have generally been considered to play a relatively passive role in regulation of cellular activity, i.e., any ubiquitinated protein was considered to be in discriminatively targeted for degradation by the proteasome. We have demonstrated, however, by using specific proteasome protease inhibitors and knockout mice lacking specific components of immunoproteasomes, that proteasomes (containing X, Y, and Z protease subunits) and immunoproteasomes (containing LMP7, LMP2, and LMP10 protease subunits) have well-defined functions in cytokine induction and inflammation based on their individual protease activities. We have also shown that LPS-TLR mediated signaling in the murine RAW 264.7 macrophage cell line results in the replacement of macrophage immunoproteasomal subunits. Such modifications serve as pivotal regulators of LPS-induced inflammation. Our findings support the relatively novel concept that defects in structure/function of proteasome protease subunits caused by genetic disorders, aging, diet, or drugs may well have the potential to contribute to modulation of proteasome activity. Of particular relevance, we have identified quercetin and resveratrol, significant constituents present in berries and of red wine respectively, as two novel proteasome inhibitors that have been previously implicated as disease-modifying natural products. We posit that natural proteasome inhibitors/activators can potentially be used as therapeutic response modifiers to prevent/treat diseases through pathways involving the ubiquitin-proteasome pathway (UP-pathway), which likely functions as a master regulator involved in control of overall inflammatory responses.
Keywords: proteasome, cytokines, resveratrol, quercetin, proteasome inhibitors, LMP knockout mice
1.1 Introduction
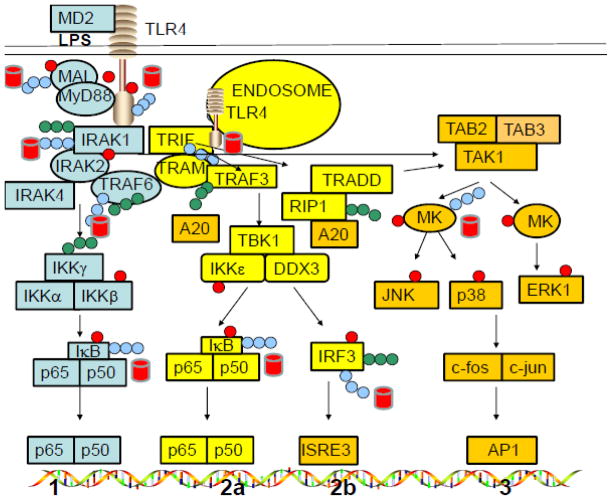
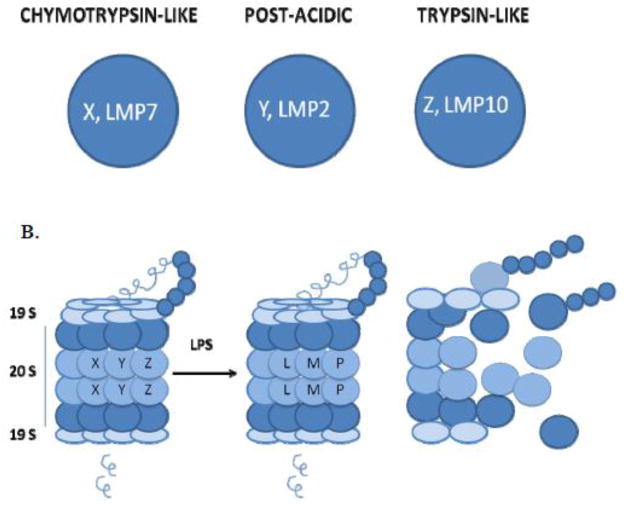
Lipopolysaccharides (LPS) serve as a prototype microbe-derived activator of macrophages via TLR4-dependent signaling, leading to generation of multiple pro- and anti-inflammatory mediators through either the MyD88-independent (TRIF/TRAM) or MyD88-dependent pathway (Fig. 1). When produced in excess in response to LPS, these proinflammatory mediators can promote the development of systemic inflammation that can progress to a life-threatening condition termed septic shock. Results from our recently completed studies have revealed that macrophage proteasomes play a pivotal role in regulation of LPS-induced signaling and regulation of gene expression and, via this mechanism, are key regulators of macrophage-dependent inflammatory responses [1–11]. The 26S proteasomes are multi-protein protease complexes which serve a key function in mammalian cells by degrading ubiquitinated proteins. ATP is required for this process. The 26S proteasome consists of a 19S regulatory complex and a 20S proteasome complex, the latter of which has been established to contain six different subunits (X, Y, Z, LMP2, LMP7, and LMP10), each of which possesses a specific protease activity. Subunits X and LMP7 possess chymotrypsin-like (CT) enzymatic activity that is responsible for protein sites after tyrosine or phenylalanine residues), Y and LMP2 possess post-acidic (PA) activity that is responsible for cleaving aspartic or glutamic acidic residues, while Z and LMP10 possess trypsin-like (T) activity that targets after arginine, lysine, or basic residues) [12–14] as indicated in Fig. 2A. One of the important contributions of proteasomes in host defenses was originally recognized because of their role in antigen presentation, but proteasomes have now been shown to contribute to regulation of a number of additional cellular functions that are important to innate cellular defenses [12–15].
Fig. 1. LPS-induced signaling pathways in macrophages.
Pathways 1 and 3 utilize MyD88/MAL as the adaptor molecules, while pathways 2a and 2b utilize the TRIF/TRAM adaptor molecules.
Fig. 2. A. The six proteases of the proteasome.
Subunits X, LMP7 have the chymotrypsin-like activity, Y, LMP2 have the post-acidic and Z, LMP10 have the trypsin-like activity. B. LPS treated RAW 264.7 cells manifest altered XYZ/LMP subunits and proteolytic activities of the proteasome. The ratio of chymotrypsin-like/post-acidic activity increases when the proteasomes contain LMP subunits upon LPS treatment. After use, the subunits of the proteasomes are ubiquitinated and degraded by the proteasome.
We initially undertook studies designed to assess the contribution of proteasomes to LPS-induced inflammatory responses in murine macrophages, particularly from the perspective of their potential to serve as therapeutic targets for the treatment of sepsis [6]. We have vigorously pursued this goal over the past several years, and the results of extensive published studies by us, and others, have served to highlight the role of the ubiquitin-proteasome (UP) pathway as a pivotal regulator of LPS-induced inflammation and cytokine induction [1–11]. Our overall objective in this review is to summarize our research findings that describe the function of the proteasome proteolytic subunits.
1.2 Current paradigm of agonist-induced signaling in murine macrophages
The current paradigm for TLR4 signaling (Fig. 1) has evolved considerably since our initial work first provided evidence that the UP-pathway and, more specifically, chymotrypsin (CT)-like and trypsin (T)-like sites in the proteasome, were involved in LPS-induced signaling in murine macrophages [2–3, 6–7], as described later in 1.3.
LPS is known to interact with the TLR4/MD2 complex at the macrophage surface, leading to recruitment of two pairs of adapter proteins (TIRAP (Mal)/MyD88 and TRAM/TRIF) to the TLR4 intracellular “Toll-IL-1R resistance” (TIR) domain [16–18]. MyD88 and TRIF facilitate docking of downstream kinases and other proteins that trigger two major signal transduction pathways termed the MyD88-dependent (pathway 1) and the TRIF-dependent (pathways 2a and 2b). IRAK4 and IRAK1 dock onto MyD88 and are activated through phosphorylation [phosphate groups are shown in red circles in Figure 1] [9]. These events lead to ubiquitination of IRAK1 (ubiquitinated via K48- or K63-linked ubiquitins [blue and green chains, respectively, in Figure 1]), which results in their dissociation from the TLR4 complex, and subsequent association of IRAK1 with TRAF6 and associated molecules. TRAF6, an E3 ubiquitin ligase, becomes auto-ubiquitinated with K63 chains, which do not signal degradation by the proteasome. Ubiquitinated TRAF6 then ubiquitinates IκB kinase (IKKγ) that, in turn, phosphorylates IκB, that is complexed to NF-κB in the cytosol [19–24]. Once phosphorylated, the IκB component of the IκB-NF-κB complex is ubiquitinated with K48 chains, leading to its degradation by the proteasome. IκB serves as the primary inhibitory component of the NF-κB complex, so its degradation leads to the activation of NF-κB, meaning that it can now translocate to the nucleus where it serves as a key transcription factor (Pathway 1).
Similar to what happens with Pathway 1, LPS-stimulated macrophages triggers the activation of TRAM which is known to recruit TRIF to endosomally associated TLR4 and to activate both NF-κB (relatively late, in comparison to the MyD88-dependent pathway; Pathway 2a and IRF-3 (Pathway 2b). Proteins that are ubiquitinated at K48 as expected, are substrates and generally become targets for degradation by the proteasome (red barrels) [19–26]. The MAPK pathway (Pathway 3) is induced through activation of both MyD88/Mal and TRIF/TRAM pathways. A20 deubiquitinates K63-ubiquitinated proteins, whenever its ubiquitinated substrates are present. Another bacterial agent, CpG DNA, activates cells via MyD88 only (without the requirement for TIRAP [Mal]), whereas poly I:C activates cells via the TRIF/TRAM pathway exclusively.
Since we first proposed that signaling proteins might serve as proteasome substrates [3], a relatively large number of TLR signaling and regulatory proteins have been established to be ubiquitinated and degraded by the proteasome. The UP-pathway that functions to regulate TLR4 signaling is known to consist of E1, E2, and E3 ligases (e.g., TRAF6) that covalently ubiquitinate proteins. However, the actual ubiquitination process is relatively complex; attachment of ubiquitin to proteins via lysine at the 63rd position of ubiquitin (K63 ubiquitination) results in formation of molecular scaffolds that facilitate signaling events. In contrast, ubiquitination at the K48 position (via lysine at position 48 of ubiquitin) selectively targets proteins for degradation by the proteasome [19–20]. LPS signaling through TLR4 promotes ubiquitination of signaling proteins with both K48- and/or K63-linked ubiquitin chains leading to selective signaling and/or degradation events of targeted proteins. The proteasome degrades K48-linked proteins, while the deubiquitinating enzyme DUB, A20, removes K63-ubiquitin chains from the molecular scaffolds. These proteins are subsequently K48-ubiquitinated, and thereby targeted for proteasome degradation.
In the following subsections we will summarize our findings that center on mechanisms by which the UP pathway might serve as a potential therapeutic target for the treatment of septic shock and other inflammatory disorders. This information discussed below has already provided new insights into possibilities for improved understanding of regulation of inflammatory mediators in general, as well as development of therapeutic interventions for treatment of other inflammation-related diseases [2, 6].
1.3 The pivotal role of proteasome proteases in modulating LPS-induced signal transduction and cytokine induction
After initially demonstrating that LPS physically binds to specific proteasome subunits, we undertook a series of studies to assess the potential physiological relevance of these interactions [2]. To this end, we first carried out studies to determine the extent to which LPS would modulate the proteasome’s proteolytic activity. We showed that, when LPS was added to partially purified proteasomes in vitro, it activated the chymotrypsin-like activity of both rabbit muscle and macrophage proteasomes [2, 3]. Given this observation we next undertook studies to determine the extent to which well-defined proteasome inhibitors might have the capacity to block LPS-induced inflammatory response in macrophages. To address this question, we pretreated the macrophage-like murine RAW 264.7 cell line in vitro, with the well-characterized proteasome inhibitor, lactacystin (low dose 5 μM, that primarily affects the CT-like activity), and observed a dose-dependent inhibition of LPS-induced cytokine secretion [2–3]. We also found that the level of expression of many LPS-inducible genes was markedly inhibited by pretreatment of primary mouse macrophages with lactacystin, as assessed by real-time RT-PCR. Importantly, expression of TNF-α gene was a notable exception to this finding; lactacystin was not particularly effective at inhibiting its expression following LPS stimulation of mouse macrophages. Thus, the effects of lactacystin appear to be at least partially selective within the TLR4 signaling pathway.
We have also demonstrated for the first time, the important role of the proteasome in regulating the phosphorylation of mitogen activated protein kinases (MAPK), since there are three specific MAPK’s, (ERK-1, 2, JNK-1, 2 and p38 MAPK) that are activated in LPS-stimulated macrophages. The addition of lactacystin to macrophages had no detectable effect on the phosphorylation of ERK-1, 2, however, when macrophages were pretreated with lactacystin and then treated with LPS, the levels of this phosphorylation event were greatly reduced. In contrast, lactacystin pretreatment alone and lactacystin pretreatment followed by LPS stimulation resulted in increased levels of phosphorylation of JNK and p38 kinases, thus supporting the concept that the proteasome differentially regulates the phosphorylation of the three macrophage associated MAPK’s [2]. Although, lactacystin was found to be effective in inhibiting LPS-induced phosphorylation of ERK 1, 2, it failed to detectably inhibit IRAK-1 associated kinase activity [2]. Collectively, these results also provide additional support for the concept that the proteasome is intimately involved in the activation of many LPS-inducible genes, and that these effects may well be mediated through the activation of certain upstream IRAK’s, MAPK’s and transcription factors.
1.3.1 Affymetrix gene-chip array analyses of macrophages treated with LPS and/or proteasome inhibitors and Ingenuity pathway analysis
We next addressed questions regarding the underlying mechanisms and consequences of lactacystin treated macrophages in terms of the potential to be stimulated by LPS. Pathway analysis using gene-chip array data revealed that addition of lactacystin to macrophages in culture markedly affects key inflammatory signaling pathways induced by LPS. Using Affymetrix microarray analysis, we confirmed and extended our original findings with lactacystin (in which we analyzed a relatively limited panel of LPS-inducible genes by RT-PCR and Southern blot analysis); most LPS-induced early genes in murine macrophages were found to be highly sensitive to inhibition by lactacystin [2, 3]. We tested lactacystin at a concentration of 5 μM for the microarray studies because it resulted in no detectable cellular toxicity over the 5 h experimental time, as assessed by the MTT (3-(4,5)-dimethylthiozol-2,5-diphenyltetrazolium bromide) assay. Interestingly, however, and as pointed out earlier at this concentration, the chymotryptic activity of the proteasome was preferentially inhibited.
In these studies, we pretreated, in vitro-cultured LPS-responsive C3HeB/FeJ primary peritoneal macrophages (1 × 106) without or with lactacystin for 1 h, followed by stimulation with medium only or with LPS (10 ng/ml) for 4 h. This experimental protocol, therefore, allowed us to evaluate the effects of treating macrophages with LPS alone, lactacystin pretreatment alone, and LPS plus lactacystin pretreatment, with untreated macrophages as controls. RNA extraction and its conversion to cDNA was carried out as described in the Affymetrix expression analysis technical manual. Our major findings from this series of experiments can be summarized as follows:
Of 120 genes whose level of expression was upregulated in C3HeB/FeJ peritoneal macrophages following treatment with LPS alone (normalization ratio of 5.0 – 404), levels of expression of 102 of these genes were significantly reduced by pre-treatment of macrophages with lactacystin (5 μM). The remaining LPS-induced genes were not significantly suppressed by lactacystin treatment, in part, because lactacystin alone compensatorily upregulated their expression. When this unanticipated effect of lactacystin was taken into account, the results of analysis provided evidence that ~ 90% of the LPS-inducible genes could be characterized as being lactacystin-sensitive.
Genes in 14 distinct, well-defined signaling pathways were found to be affected by LPS, and levels of expression of most of these genes were inhibited by lactacystin pretreatment. The antigen presentation pathway (12 genes, MHC class I and II), NF-κB pathway (42 genes), death receptor signaling (23 genes), IL-6 signaling (32 genes), IFN-γ signaling (8 genes), and p38 signaling pathway (23 genes) were among the key signaling pathways induced in macrophages in response to LPS that were inhibited by lactacystin [3].
As expected, LPS-induced upregulation of expression of a large number of key pro-inflammatory genes, e.g., IL-1β, IL-1α IL-6, TNF-α, IL-12, iNOS, VCAM1, ICAM1, prostaglandin-endoperoxide synthase 2, endothelin 1, STAT5A, complement component 3, MAIL, TRAF1, cyclin D2, IL-23α, MAFF, CXCL9, Adrenomedullin, Adenosine A2a receptor, BCL3 were >80% inhibited when macrophages were pretreated with lactacystin [3]. One critical observation, which was somewhat unexpected was that 5 μM lactacystin blocked LPS-induced TNF-α gene expression by only 24%, yet this same concentration of lactacystin reduced level of expression of IL-1β, IL-6, and IL-12 mRNA expression by >90%. This observation supported the notion that TNF-α is a cytokine which is induced by a pathway that may well be different than that required for IL-1 β, IL-6, and IL-2 and thus dependent upon the protease site(s) of the proteasome.
Taken collectively, the results of these DNA microarray analyses provided us with novel information that suggested that approximately 90% of LPS-induced genes (with a normalization ratio of ≥5.0) in macrophages are lactacystin-sensitive (i.e. they are dependent upon proteolytic activity of the proteasome). Further, our successful completion of these studies provided strong evidence that the combined use of the gene-chip array, coupled with Ingenuity Pathways analysis, can serve as informative approach for both confirming the participation of the proteasome in known signaling pathways, and for identifying additional novel pathways that are either positively or negatively affected by lactacystin and, potentially, other novel proteasome inhibitors.
1.3.2 Lactacystin suppresses peptidoglycan- and CpG DNA-induced inflammatory gene expression and also dysregulates phosphorylation of MAPK in macrophages
Our studies, summarized above, demonstrated that lactacystin pretreatment of macrophages results in the inhibition of most LPS-induced cytokines, as well as the phosphorylation of MAPK, and that LPS induced upregulation of TNF-α is relatively resistant to suppression by lactactystin pretreatment [2]. It is well established that other microbial stimuli, such as peptidoglycan and bacterial unmethylated CpG DNA, can also trigger TLR-dependent stimulation of proinflammatory cytokine production by macrophages that may also contribute to the development of systemic inflammation and shock. Therefore, we hypothesized that lactacystin pretreatment of macrophages would also result in inhibition of pro-inflammatory gene-expression induced by these agonists as well. We tested this hypothesis by examining the effect of lactacystin in CpG DNA- and peptidoglycan-induced signaling pathways in RAW 264.7 macrophages. Lactacystin pretreatment once again inhibited, in a dose-dependent manner, both CpG DNA- and peptidoglycan-induced expression of TNF-α, IL-1β and iNOS genes without detectable alterations in β-actin expression or any detectable manifestation of cellular cytotoxicity [4]. In addition, lactacystin pretreatment also inhibited both CpG DNA- and peptidoglycan-induced phosphorylation of ERK’s. Collectively, these data strongly support the conclusion that the proteasome also plays an important role in regulating not only TLR4-dependent signaling, but also TLR9 (CpG DNA) and TLR2 (peptidoglycan)-induced signal transduction in macrophages [4, 5]. Of importance, although we have noted that lactacystin pretreatment at low doses was not a particularly potent inhibitor of LPS-induced TNF-α gene expression in macrophages, it nevertheless still serves as a potent inhibitor of TNF-α gene expression when CpG DNA was used to stimulate RAW 264.7 cells in primary mouse macrophages. This observation implies that activation of different TLRs leading to TNF-α gene expression may involve signaling pathways that show different levels of sensitivity to proteasome inhibition [4, 5].
1.3.3 The proteasome protease subunits function to regulate LPS-induced MyD88/TIRAP and TRIF/TRAM signaling pathways
Again, using lactacystin as a relatively selective proteasome inhibitor, we undertook a series of studies designed specifically to identify sites in LPS-induced signaling pathways that are directly, or indirectly, regulated by the proteasome [1–10]. For these studies, RAW 264.7 murine macrophages were pretreated with either vehicle or lactacystin, followed by stimulation with LPS (TLR4; MyD88/TIRAP and TRIF/TRAM pathways), CpG-DNA (TLR9; MyD88 pathway only), poly I:C (TLR3; TRIF pathway only), or peptidoglycan (TLR2; MyD88/TIRAP pathway). Macrophage activation by these varied TLR agonists induces gene expression of iNOS as well as a wide variety of inflammatory cytokines, activation of MAP kinases and several signaling pathways and, in most cases, as expected lactacystin treatment had a significant impact on all of these TLR agonist induced activities.
More recently, we have explored the relationship between proteasomal activity and macrophage activation. Our results provided strong evidence to support the conclusion that the differential properties of distinct proteolytic subunits of the proteasome play a critical role in dictating the characteristics of the LPS-induced responses in mouse macrophages [3,7,8]. To determine the relative contribution of each individual proteasomal protease in regulating LPS-induced cytokine production, we used selective inhibitors of the individual LMP subunits, and in addition performed experiments using macrophages obtained from LMP-null mice, the results of which will be summarized below.
1.3.4 Use of selective proteasome inhibitors to determine function of proteasomal subunits
Although textbooks of cell biology generally characterize proteasomes as rather indiscriminate proteolytic complexes (i.e., any ubiquitinated protein will be indiscriminately degraded by the proteasome), the situation is now recognized to be significantly more complex. We recently demonstrated, for example, that pretreatment of macrophages with selective proteasome inhibitors (NC-005, NC-001, and lactacystin) to selectively target various protease sites of the proteasome, that murine constitutive proteasomes (containing X, Y, and Z protease subunits) and immunoproteasomes (in which LMP subunits have partially replaced X, Y, and Z protease subunits) results in modification of several LPS-induced cellular functions, based on their individual protease activities [7,8].
In this respect, evidence documenting the differential capacity of NC-005, NC-001, and lactacystin to inhibit distinct protease activities of the proteasome is summarized in Table 1. It should be noted that NC-001 is a comparatively potent inhibitor of PA activity, but is relatively ineffective at inhibiting CT and T enzymatic activity. In contrast, lactacystin is considerably more effective at inhibiting CT than either T or PA activity. NC-005 is not a particularly potent inhibitor of PA activity, but does inhibit both CT and T, though it is considerably more active against CT than against T. We have also been able to demonstrate that inhibition of PA activity alone with NC-001 is insufficient to suppress TNF-α gene expression in LPS-stimulated RAW 264.7 macrophages. Similarly, concentrations of lactacystin or NC-005 that are sufficiently low enough to inhibit only CT activity, failed to suppress TNF-α gene expression and TNF-α production, but do suppress iNOS gene expression and NO production in LPS-stimulated RAW 264.7 macrophages [7]. In contrast, inhibition of both CT- and T-like protease activities, achieved with higher concentrations of NC-005, markedly reduces LPS-induced TNF-α production in the absence of measurable effects on cell viability (90–100%) as assessed by the MTT assay [7]. These results suggest that CT-like (subunits X, LMP7) and T-like (subunits Z, LMP10) proteolytic activities must be simultaneously suppressed in order to inhibit LPS-induced TNF-α production through the proteasome [7]. As a reminder, we discussed earlier the fact that inhibition of CT-like activity alone with low concentrations of lactacystin is sufficient to inhibit NO production by LPS stimulated murine macrophages [7].
Table 1.
The Ki values for (50%) inhibition of CT-like (LMP7), T-like (LMP10) and PA (LMP2) activities of different natural and synthetic proteasome inhibitors. These compounds are most useful for blocking the proteasome’s protease activities that are underlined. The three protease sites are in close proximity, therefore proteasome inhibitors tend to affect other sites simultaneously. The advantage of using natural inhibitors over synthetic inhibitors is that these are non toxic and no cell death was observed at the concentrations using the MTT assay [6, 7].
| Inhibitors/Cell types | CT activity (Ki (μM) | T activity (Ki μM) | PA activity (Ki μM) |
|---|---|---|---|
| Lactacystin RAW 264.7 | 0.6 | 20 | >20 |
| NC-005 RAW 264.7 | 0.1 | 3 | >20 |
| NC-001 RAW 264.7 | ≫20 | ≫20 | 5.0 |
| Resveratrol RAW 264.7 | 4.0 | 9.0 | <2.7 |
| Quercetin RAW 264.7 | 25 | 15 | 50 |
| Resveratrol THP-1 | <2.7 | 10 | 10 |
| Quercetin THP-1 | 20 | 20 | 15.8 |
1.3.5. Use of macrophages and splenocytes from LMP knockout mice to determine function of proteasomal subunits
To begin to dissect where in the signaling pathway that the effects of lactacystin pretreatment manifest, we capitalized upon the fact that IFN’s are known to have the capacity to amplify the macrophages’ response to TLR agonists. For example, the TRIF/TRAM pathway is recognized to be responsible for the transcription of IFN-β protein after translation then binds to receptors on cells and activates via autocrine, paracrine mechanisms, Janus kinases (JAK kinases) which leads to phosphorylation of STAT1 and STAT2. P-STAT1 is known to be required for the induction of iNOS. Thus IFN’s induced via the TRIF/TRAM (amplification) pathway play an amplifying role in the induction of certain cytokines and NO induction in agonist-treated cells. Thus IFN’s are induced by the TRIF/TRAM pathway and also mediate their effects via the JAK/STAT pathway. We hypothesized that these agonist-induced pathways are dependent on the proteasome proteases.
One of our major objectives has been to determine the contribution of selective LMP proteasome protease subunits to TLR-agonist-induced production of cytokines, activation of signaling mediators, or to nitric oxide (NO) production in primary murine macrophages and splenocytes. Our findings with LMP knockout mice summarized below have provided novel insights into the pathways of regulation of inflammatory responses by the macrophage proteasome. As reported above, LPS induced TNF-α production was essentially normal in peritoneal macrophages derived from LMP2, LMP7, LMP10 and LMP7/LMP10 double knockout mice [8] allowing the conclusion that these subunits are not responsible for induction of this cytokine. Therefore, CT-like and T-like proteasome activities in macrophages of knockout mice lacking both LMP7 and LMP10 must be attributable solely to subunits X and Z. Consequently, the capacity of LMP7/LMP10 double knockout mice to still generate normal levels of production of TNF-α in response to LPS stimulation suggests that constitutive proteasomes (i.e. lacking LMP7 and LMP10, but containing X and Z) are fully capable of regulating signaling mechanisms responsible for TNF-α LPS induced macrophage production of NO, iNOS, IRF3, IFN-γ, IL-1β, IL-6, P-STAT1,3; in contrast, are markedly reduced in LMP7/LMP10 double knockout mice [8], as shown in Table 2, allowing the conclusion that LMP subunits are essential for their production while, X, Y, and Z subunits are responsible for induction of TNF-α in LPS stimulated murine macrophages. Treating these macrophages with interferons and LPS, however, reverses this defect, leading to robust NO induction [8]. Collectively, these experiments clearly demonstrate that NO production is highly dependent on LMP containing immunoproteasomes, while this is not the case for LPS-induced TNF-α responses. Importantly, TNF-α responses occur relatively rapidly after LPS exposure (regulated by X,Y, and Z containing constitutive proteasomes within 4h of stimulation), whereas NO production occurs much later (within 24 hours after stimulation, allowing sufficient time for LMP containing immunoproteasomes to be assembled) in normal macrophages.
Table 2.
Levels of gene expression and protein expression of cytokines and signaling mediators with LPS as the agonist in thioglycollate-elicited macrophages and PMA/ionomycin as an agonist in splenocytes from C57BL/6 LMP7/LMP10 double knockout mice [8, 27].
| C57BL/6 LMP7/LMP10 knockouts | mRNA not significantly affected | mRNA Affected | Proteins Affected |
|---|---|---|---|
| Macrophages | TNF-α | IL-1β, IL-6, IL-12, iNOS | IFN-γ, IFN-β, iNOS, P-STAT-1, P-STAT-3 |
| Splenocytes | IL-2, IL-13, TNF-α, IL-2Rα, | IFN-γ, IL-4, IL-10, IL-2Rβ, GATA-3, t-bet |
Next we also wanted to investigate if the proteasome proteases in splenocytes in the wild type and double knockout mice (C57BL/6) are also responsible for the agonist-induced cytokines, their receptors and transcription factors. Since LPS does not directly activate CD4+ T cells, we used phorbol-12-myristate-13-acetate (PMA)/ionomycin as an agonist, thus bypassing the receptor. LMP7/LMP10-null splenocytes exhibited reduced levels of gene expression of IL-10, IL-4, IFN-γ, IL-2Rβ t-bet and GATA-3 (transcription factors) in response to agonists, whereas, induction of IL-13, IL-2, TNF-α and IL-2Rα was essentially normal in these knockout mice as compared to wild-type mice, and were found to be LMP-independent [27] and may therefore be X, Y, Z- dependent. Lactacystin (which predominantly inhibits the chymotrypsin-like activity of the proteasome) pretreatment of LMP7/LMP10-null splenocytes, followed by PMA/ionomycin, reduces gene expression of IL-2, IFN-γ IL-4, IL-13, TNF-α(cytokines), IL-2Rα and IL-2Rβ (receptors) t-bet and GATA-3 (transcription factors). In contrast, levels of expression of IL-10 (anti-inflammatory cytokine) were increased upon pretreatment of cells with lactacystin in splenocytes from null as compared to controls [27]. Thus, as with macrophages, production of subsets of inflammatory cytokines by stimulated T cells, is influenced by proteasome subunit composition, or differential protease activities of cellular proteasomes.
Collectively, these results underscore the importance of LMP subunits in the proteasome of macrophages and splenocytes [27]. We have established that different proteasome proteases regulate levels of cytokine gene expression and protein expression differentially.
1.4 Signaling proteins accumulate in macrophages upon treatment with LPS and lactacystin that are normally degraded by the proteasome
Lactacystin, (low levels, 2.5 μM) binds to chymotrypsin-like binding sites in the proteasome and it blocks the degradation of ubiquitinated proteins [7]. We have identified several signaling proteins such as IRAK-1, TRAF6, IRF3, P-STAT1, P-IκB-α that accumulate in RAW 264.7 cells after they have been pretreated with low dose lactacystin, followed by stimulation with LPS, suggesting that these signaling proteins are degraded via the CT-like activity of the proteasome.
1.5 Proposed model for the role of proteasomes in signaling
Interactions between LPS and the TLR4/MD2 complex are known to lead to the recruitment of two pairs of adapter molecules to TLR4, specifically TIRAP (Mal)/MyD88 and TRAM/TRIF [18–22]. TLR4 becomes ubiquitinated by a series of enzymes, E1, E2, and E3 ligase, and is degraded rapidly within 10–30 min, (our unpublished data) by proteasomes that contain predominantly the constitutively expressed X, Y, and Z subunits. These proteasomes have CT/PA ratios of ~1.0 in RAW 264.7 cells. LPS stimulation leads to a cascade of events that phosphorylate IκB, which then becomes ubiquitinated with the K48-linked ubiquitin, and thereby targets IκB for proteasome degradation. This allows the activation of NF-κB, which translocates to the nucleus and induces gene expression of TNF-α and other proinflammatory cytokines. Both CT-like and T-like activities associated with subunits X, and Z, are responsible for LPS-induced TNF-α, while the CT-like activity appears to be required for LPS-induced gene expression of other proinflammatory cytokines [7, 8]. Through this pathway, LPS stimulates the synthesis and secretion of TNF-α and induces new immunoproteasome synthesis in cells that then become characterized predominantly by the presence of LMP7, LMP2, and LMP10 proteolytic subunits (CT/PA ratio ~4.5, RAW 264.7 cells), that are accordingly, characterized by increased CT-like and T-like activity, with correspondingly decreased PA activity (relative to X, Y, and Z subunits) [7]. Differences in the relative specificity of these new protease activities lead to differential cleavage of the already generated signaling proteins. The TRIF/TRAM pathway is activated upon endocytosis of LPS [36, 37], and this latter event ultimately leads to generation of NO, and other mediators whose production is dependent on the TRIF/TRAM pathway and through production of IFN-β. RAW 264.7 cells can also be induced to produce NO by LPS and IFN-γ thus bypassing the TRIF/TRAM pathway although these cells remain potent inducers of TNF-α in response to LPS. In conclusion; activation and regulation of both LPS-induced pathways are critically dependent on the presence of specific proteasome proteases in RAW 264.7 cells [7]. After use, the proteasome subunits are ubiquitinated at specific lysines and degraded by the proteasome itself (Fenselau and Qureshi, unpublished data).
The signaling pathways in C57BL/6 mouse macrophages in response to LPS are somewhat more complex. In contrast to RAW 264.7 cells, both resident and thioglycollate-ellicited macrophages from C57BL/6 mice contain “mixed proteasomes” containing both X, Y, Z, and LMP7, LMP2, and LMP10 proteasome protease subunits. The macrophages obtained via the peritoneal lavage from the C57BL/6 mouse can be readily induced to generate cytokines, such as TNF-α and also NO efficiently in response to LPS. LPS-induced TNF-α by these cells is dependent on X, Y and Z subunits, while IL-1β, IL-6, IFN-β and IL-12 are dependent on the presence of LMP subunits of the proteasome [8]. We have shown that levels of signaling proteins and enzymes, such as P-IRF3, iNOS, IFN-β, IFN-γ P-STAT1, and P-STAT3 (S727); and levels of gene expression of cytokines, such as IL-1βIL-6, IL-12; and NO production are reduced in LPS-stimulated macrophages from LMP7/LMP10 double knockout mice, thus underscoring the importance of LMP subunits to production of these effector molecules. The TRIF/TRAM pathway also functions to activate NF-κB [8] (pathway 2a). Macrophages from LMP knockout mice show intrinsic defects in the TRIF/TRAM pathway [8], which can be overridden by addition of IFN-γ. Thus, the availability of mixed proteasomes appears to be important for LPS-induced synthesis of NO in macrophages. In contrast, LPS-induced TNF-α and the MyD88 pathway are still able to function normally in macrophages from LMP knockout mice, albeit at a lower level [8]. Once again, we would conclude that the type of proteasome proteolytic subunits dictates the specific cellular response to LPS and associated signaling events.
1.6. Screening of small molecules for use as potential proteasome inhibitors. Several natural compounds and licensed drugs act at the proteasome level
In preliminary experiments, we have found that pretreatment of mice with lactacystin [2] or mevinolin (structurally related to lactacystin) are both effective in protecting galactosamine-sensitized mice and cecal-ligation and puncture mice from LPS-induced mortality, and both are known to function as inhibitors of proteasome activities [6]. Mevinolin (closed ring form of lovastatin, normally lovastatin is 80% open ring and 20% closed ring form) is well-established as a cholesterol-lowering compound, and has been shown to function as a competitive inhibitor for the β-hydroxy-β-methylglutaryl coenzyme A reductase, the rate-limiting enzyme in the biosynthesis of cholesterol. Results of recent investigations by others have shown that mevinolin can act as a proteasome inhibitor [38] in MDA-MB-157 tumor cells, and serve to either increase or decrease inflammation, depending on relative dose. Both lactacystin and mevinolin are β-lactones. Therefore, we queried the extent to which other FDA-approved drugs or natural compounds with complex lactone structures might also manifest inhibitory activity at the proteasome level. We have now screened several known drugs and natural compounds that would be predicted to function as anti-inflammatory compounds in vitro [39–41]. We have evaluated their relative effect on the three protease activities (CT, T and PA) of the purified 20S proteasome of rabbit muscle and also by using the Pro-Glo proteasome protease enzymatic assays in macrophages [39–41]. We also tested the inhibition of expression of LPS-induced cytokines of several drugs in several experimental models [39–42] and degradation of P-IκB to determine the extent to which these compounds may function at the level of the proteasome when added before and after LPS treatment of primary cultured mouse macrophages in vitro and in vivo [39–41]. We identified mevinolin, quercetin (present in berries and other natural fruits and vegetables), resveratrol (compound found in red wine) and as the positive control, lactacystin, as compounds that function as potent anti-inflammatory agents and cause inhibition of LPS-induced gene expression levels of iNOS and TNF-α IL-6, IL-1β; activation of NF-κB, and NO in macrophages. Importantly, using this approach, we identified quercetin and resveratrol for the first time, as potent proteasome inhibitors, by the data summarized in Table 1. The advantage of using natural inhibitors over synthetic inhibitors is that these have already been established to be non-toxic and no detectable cell death has been observed at any concentration tested using the MTT assay.
1.7 Quercetin’s and Resveratrol’s (proteasome inhibitors) activities are similar to that of lactacystin, but these agents are considerably less toxic
Lactacystin has been extensively employed as an effective anti-inflammatory compound, but it is relatively toxic to cells primarily because it functions as an irreversible proteasome inhibitor. To assess the differential effects of lactacystin, quercetin and resveratrol on cellular toxicity, we treated RAW 264.7 cells with each of these agents at a variety of concentrations and monitored cell-death at different time points using the MTT assay as described previously [7, unpublished]. We found that, for up to 8 h, none of the compounds induced significant cell death. However, after 24 h, lactacystin, even at doses as low as 6.5 μM, was toxic to those cells, whereas quercetin and resveratrol did not detectably kill the cells even at 100 μM. The results from these experiments provided a strong rationale for our studies to identify novel compounds that have anti-inflammatory effects similar to lactacystin, but are relatively non-toxic. The data on Ki values shown in Table 1 have been obtained in RAW 264.7 and THP-1 (human monocytes) cells. There are differences in the Ki values of inhibitors with RAW 264.7 and THP-1 cells, because these cells contain different proteasome subunits. Our experiments clearly suggested that quercetin and resveratrol both function as potent proteasome inhibitors and can be used to inhibit inflammation, without the deleterious effects.
1.8 Proteasome inhibitors, in combination with antibiotics, provide protection in a CLP model of polymicrobial septic shock
Since septic shock occurs primarily due to production of excessive levels of inflammation, we sought to assess the potential clinical significance of these proteasome inhibitors in vivo in a cecal ligation and puncture (CLP) model of polymicrobial sepsis. Co-administration of either mevinolin or quercetin with standard antibiotic therapy (Primaxin) afforded significantly greater protection against septic shock in a mouse CLP model than either treatment alone. In untreated mice, model resulted in 95% 3-day mortality, and this was reduced to 70% with mevinolin alone, 58% with Primaxin alone, but was reduced drastically to 35% using a combination of mevinolin and Primaxin. The combination of quercetin and Primaxin showed similar results. These studies strongly support the conclusion that therapeutic targeting of cellular proteasomes to dampen mediator production [39], in conjunction with standard antimicrobial therapy, may be of considerable survival benefit in the treatment of infection by mitigating the cytokine storm normally elicited in sepsis [6].
1.9 Summary of Findings
In this review, we have summarized our recent findings that center on the mechanisms involving the proteasome as a potential therapeutic target in the treatment of inflammation. LPS treated RAW macrophages manifest altered relative levels of XYZ/LMP protease subunits and associated proteolytic activities of the cellular proteasome. Our collective evidence would strongly support the conclusion that the proteasome proteases serve as pivotal regulators of LPS-induced inflammation in macrophages. These functions include modulation of: levels of gene expression, generation of specific transcription factors, degradation of proteins involved in cell metabolism, relative rates of degradation of proteins involved in multiple signaling pathways, relative levels of cytokines secreted, NO production, and cell death/growth. Therapeutic targeting of cellular proteasomes, in conjunction with standard antimicrobial therapy, may be of considerable survival benefit in treatment of septic shock by mitigating the ‘cytokine storm’ normally observed in sepsis. Defects in structure/function of proteasome subunits caused by genetic disorders (such as shown in knockout mice) [8], aging [39], diet (quercetin, resveratrol) [39–42, Qureshi, unpublished data], or drugs [lovastatin] [6] contribute to changes in proteasome’s protease activity. Natural proteasome inhibitors/activators can potentially be used as therapeutic response modifiers to prevent/treat diseases based on inflammation.
Highlight points.
The proteasome serves as a pivotal regulator of agonist-induced inflammation and cytokine induction.
Agonist treatment of RAW 264.7 cells induces changes in relative proteolytic activity of the proteasomes.
Proteasomes and immunoproteasomes have well-defined functions affecting multiple signaling pathways.
Several proteins are ubiquitinated and modulated during cellular treatment with LPS.
Quercetin and Resveratrol are novel natural proteasome inhibitors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Qureshi N, Vogel SN, Van Way C, III, Papasian CJ, Qureshi AA, Morrison DC. The proteasome. A central regulator of Inflammation and macrophage function. Immunol Res. 2005;31:243–260. doi: 10.1385/IR:31:3:243. [DOI] [PubMed] [Google Scholar]
- 2.Qureshi N, Perera P-Y, Splitter G, Morrison DC, Vogel SN. The Proteasome as a LPS-binding protein in macrophages. Toxic lipopolysaccharide activates the proteasome complex. J Immunol. 2003;171:1515–1525. doi: 10.4049/jimmunol.171.3.1515. [DOI] [PubMed] [Google Scholar]
- 3.Shen J, Reis J, Morrison DC, Papasian C, Raghavaikaimal S, Kolbert C, Qureshi AA, Vogel SN, Qureshi N. Key inflammatory signaling pathways are regulated by the proteasome. Shock. 2006;25:472–484. doi: 10.1097/01.shk.0000209554.46704.64. [DOI] [PubMed] [Google Scholar]
- 4.Shen J, Gao JJ, Zhang G, Tan X, Morrison DC, Papasian CJ, Vogel SN, Qureshi N. Proteasome inhibitor, lactacystin blocks CpG DNA- and peptidoglycan induced inflammatory genes, cytokines and mitogen-activated protein kinases in macrophages. Shock. 2006;25:594–599. doi: 10.1097/01.shk.0000209555.46704.2d. [DOI] [PubMed] [Google Scholar]
- 5.Gao JJ, Shen J, Kolbert C, Raghavakaimal S, Papasian CJ, Qureshi N, Vogel SN, Morrison DC, Qureshi N. The proteasome regulates bacterial CpG DNA-induced signaling pathways in murine macrophages. Shock. 2010;34:390–401. doi: 10.1097/SHK.0b013e3181d884ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reis J, Tan X, Yang R, Rockwell CE, Papasian CJ, Vogel SN, Morrison DC, Qureshi AA, Qureshi N. A combination of proteasome inhibitors and antibiotics prevents lethality in a septic shock model. Innate Immun. 2008;14:319–329. doi: 10.1177/1753425908096855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reis J, Guan X-G, Kisselev AF, Papasian CJ, Qureshi AA, Morrison DC, Van Way CW, Vogel SN, Qureshi N. LPS-induced formation of immunoproteasomes: TNF-α and nitric oxide production are regulated by altered composition of proteasome-active sites. Cell Biochemistry and Biophysics. 2011;60:77–88. doi: 10.1007/s12013-011-9182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reis J, Hassan F, Guan X-Q, Shen J, Monaco JJ, Papasian CJ, Qureshi AA, Van Way CW, Morrison DC, Vogel SN, Qureshi N. The immunoproteasomes regulate LPS-induced TRIF/TRAM signaling pathway in murine macrophages. Cell Biochemistry and Biophysics. 2011;60:119–126. doi: 10.1007/s12013-011-9183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins D, Qureshi N, Vogel SN. A TLR-Responsive Kinase, Protein Kinase R (Pkr), Is Inactivated in Endotoxin Tolerance Through Differential K63/K48 Ubiquitination. Mbio. 2010;1:e00239–10. doi: 10.1128/mBio.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockwell CE, Qureshi N. Differential effects of lactacystin on cytokine production in activated Jurkat cells and murine splenocytes. Cytokine. 2010;51:12–17. doi: 10.1016/j.cyto.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rockwell CE, Morrison DC, Qureshi N. Lipid A-mediated tolerance and cancer therapy. Adv Exp Med Biol. 2009;667:81–99. doi: 10.1007/978-1-4419-1603-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaczynska M, Goldberg AL, Tanaka K, Hendil KB, Rock KL. Proteasome subunits X and Y alter peptidase activities in opposite ways to the interferon-γ-induced subunits LMP2 and LMP7. J Biol Chem. 1996;271:17275–17280. doi: 10.1074/jbc.271.29.17275. [DOI] [PubMed] [Google Scholar]
- 13.Elenich LA, Nandi D, Kent AE, McCuskey TS, Cruz M, Lyer MN, Woodward EC, Conn CW, Ochoa AL, Ginsburg DB, Monaco JJ. The complete primary structure of mouse 20S proteasomes. Immunogenetics. 1999;49:835–842. doi: 10.1007/s002510050562. [DOI] [PubMed] [Google Scholar]
- 14.Dahlman B, Hendil KB, Kristensen P, Uerkvitz W, Sobek A, Kopp F. Subunit arrangement in the human proteasome. In: Hilt W, Wolf DH, editors. Proteasomes; The World of Regulatory Proteolysis. Landes Bioscience; Georgetown, Texas: 2000. pp. 37–47. [Google Scholar]
- 15.Kloetzel PM, Ossendorp F. Proteasome and peptidase function in MHC class I- mediated antigen presentation. Curr Opin Immunol. 2004;16:76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freundenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 17.Janeway CA, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2000;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 18.Vogel SN, Fitzgerald KA, Fenton MJ. TLRs: Differential adapter utilization by toll-like receptors mediates TLR-specific patterns of gene expression. Molecular Interventions. 2003;3:466–477. doi: 10.1124/mi.3.8.466. [DOI] [PubMed] [Google Scholar]
- 19.O’Neill LA, Bryant JCE, Doyle SI. Therapeutic targeting of Toll-like receptors for infectious and inflammatory diseases and cancer. Pharmacological reviews. 2009;61:177–197. doi: 10.1124/pr.109.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wertz IE, Dixit VM. Signaling to NF-κB: regulation by ubiquitination. Cold Spring Harb Perpect Biol. 2010;2:a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 22.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 23.Abott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schauvilege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: a role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006;580:4697–4702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 25.Aisling D, Carpenter S, Brikos C, Gray P, Strelow A, Wesche H, Morrice N, O’Neill LAJ. IRAK1 and IRAK4 Promote Phosphorylation, Ubiquitination, and Degradation of MyD88 Adaptor-like (Mal) J Biol Chem. 2010;285:18276–18282. doi: 10.1074/jbc.M109.098137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgs R, Gabhann JN, Larbi NB, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rockwell CE, Monaco JJ, Qureshi N. A Critical Role for the Inducible Proteasomal Subunits LMP7 and MECL1 in Cytokine Production by Activated Murine Splenocytes. Pharmacology. 2012;89 doi: 10.1159/000336335. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hjerpe, Aillet RF, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO. 2009:rep. 1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasilescu J, Smith JC, Ethier M, Figeys D. Proteomic analysis of ubiquitinated proteins from human MCF-7 breast cancer cells by immunoaffinity purification and mass spectrometry. J Prot Res. 2005:2192–2200. doi: 10.1021/pr050265i. [DOI] [PubMed] [Google Scholar]
- 31.Wang D, Cotter RJ. Approach for determining protein ubiquitination sites by MALDI-TOF mass spectrometry. Anal Chem. 2005;77:1458–1466. doi: 10.1021/ac048834d. [DOI] [PubMed] [Google Scholar]
- 32.Peng J. Evaluation of proteomic strategies for analyzing ubiquitinated proteins. BMB reports. 2008;41:177–183. doi: 10.5483/bmbrep.2008.41.3.177. [DOI] [PubMed] [Google Scholar]
- 33.Sliter DA, Aguiar M, Gygi SP, Wojcikiewicz RJ. Activated inositol 1,4,5-trisphosphate receptors are modified by homogeneous Lys-48 and Lys-63 linked ubiquitin chains, but only Lys-48 linked chains are required for degradation. J Biol Chem. 2011;286:1074–1082. doi: 10.1074/jbc.M110.188383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards N, Wu X, Tseng C-W. An unsupervised, Model-Free, Machine-Learning Combiner for Peptide Identifications from Tandem Mass Spectra. Clinical Proteomics. 2009;5:23–36. [Google Scholar]
- 35.Seyfried NT, Su P, Duong DM, Cheng D, Hanfelt J, Peng J. Systematic approach for validating the ubiquitinated proteome. Anal Chem. 2008;80:4161–4169. doi: 10.1021/ac702516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutuzova GD, Albrecht RM, Erickson CM, Qureshi N. Diphosphoryl lipid A from Rhodobacter sphaeroides blocks the binding and internalization of lipopolysaccharide in RAW 264.7 cells. J Immunol. 2001;167:482–489. doi: 10.4049/jimmunol.167.1.482. [DOI] [PubMed] [Google Scholar]
- 37.Kagan JC, Su T, Hornig T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nature Immunology. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao S, Porter DC, Chen X, Herliczek T, Lowe M, Keyomarsi K. Lovastatin-mediated G1 arrest is through inhibition of the proteasome, independent of hydroxymethyl glutaryl-CoA reductase. PNAS. 1999;96:7797–7802. doi: 10.1073/pnas.96.14.7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qureshi AA, Tan X, Reis JC, Badr MZ, Papasian CJ, Morrison DC, Qureshi N. Inhibition of nitric oxide in LPS-stimulated macrophages of young and senescent mice by δ-tocotrienol and quercetin. Lipids Health and Dis. 2011;10:239. doi: 10.1186/1476-511X-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qureshi AA, Reis J, Qureshi N, Papasian CJ, Morrison DC, Schaefer DM. δ-Tocotrienol and quercetin reduce serum levels of nitric oxide and lipid parameters in female chickens. Lipids Health and Dis. 2011 Mar;10:39. doi: 10.1186/1476-511X-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qureshi AA, Tan X, Reis J, Badr MZ, Papasian CJ, Morrison DC, Qureshi N. Suppression of nitric oxide production and pro-inflammatory cytokines by novel proteasome inhibitors in various experimental models. Lipids Health and Dis. 2011;10:177. doi: 10.1186/1476-511X-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qureshi AA, Guan XQ, Reis JC, Papasian CJ, Jabre S, Morrison DC, Qureshi N. Inhibition of nitric oxide and inflammatory cytokines in LPS-stimulated murine macrophages by Resveratrol, a potent proteasome inhibitor. Lipids in Health and Dis. doi: 10.1186/1476-511X-11-76. (In press, 2012) [DOI] [PMC free article] [PubMed] [Google Scholar]