Abstract
Background
Ultraconserved elements (UCEs) are non-coding genomic sequences completely identical among human, mouse, and rat species and harbor critical biological functions. We hypothesized that single nucleotide polymorphisms (SNPs) within UCEs are associated with clinical outcomes in colorectal cancer (CRC) patients.
Patients and Methods
Forty-eight SNPs within UCEs were genotyped in 662 patients with stage I–III CRC. The associations between genotypes and recurrence and survival were analyzed in stage II or III patients receiving fluoropyrimidine-based adjuvant chemotherapy using a training and validation design. The training set contained 115 stage II and 170 stage III patients, and the validation set contained 88 stage II and 112 stage III patients, respectively.
Results
Eight SNPs were associated with clinical outcomes stratified by disease stage. In particular, for stage II patients with at least one variant allele of rs7849, consistent association with increased recurrence risk was observed in the training set (HR: 2.39; 95%CI: 1.04–5.52), replication set (HR: 3.70; 95%CI: 1.42–9.64), and meta-analysis (HR: 2.89; 95%CI: 1.54–5.41). There were several other SNPs that were significant in training set, but not in the validation set. These include: rs2421099, rs16983007 and rs10211390 with recurrence, and rs6590611 with survival in stage II patients; and SNPs rs6124509 and rs11195893 with recurrence in stage III patients. In addition, we also observed significant cumulative effect of multiple risk genotypes and potential gene-gene interactions on recurrence risk.
Conclusions
This is the first study to evaluate the association between SNPs within UCEs and clinical outcome in CRC patients. Our results suggest that SNPs within UCEs may be valuable prognostic biomarkers for locally advanced CRC patients receiving 5FU-based chemotherapy.
Keywords: SNP, ultraconserved elements, colorectal cancer, recurrence
INTRODUCTION
Surgery is the primary treatment modality with curative intent in patients with localized colorectal cancer (CRC). However, approximately 50% of patients will develop recurrent or metastatic disease after radical resection1. Administration of 5-fluorouracil (FU) may be considered in patients with AJCC (version 6.0) high-risk stage II and III disease. Unfortunately, 40% to 50% of patients will not experience beneficial effects and suffer from treatment-related toxicities2. Recent studies have shown that single nucleotide polymorphisms (SNPs) can provide information for personalized chemotherapy3. It has been estimated that there are perhaps 50,000–250,000 SNPs that confer a biological effect, most of which are distributed in and around the 30,000 genes4. Therefore, it is advantageous to evaluate SNPs that are more likely to be functional and have a bearing in colorectal cancer recurrence.
Ultraconserved elements (UCEs) are 200 to 779 base pairs (bp), absolutely conserved non-coding sequences that show 100% sequence identity among orthologous genomic regions of the human, mouse, and rat species5. Recent studies suggest that UCEs are frequently located at fragile sites and genomic regions involved in cancers 6 and have important functions in vertebrate genomes, such as serving as long-range enhancers of flanking genes7 and regulating splicing 8, epigenetic modifications 9, and transcriptional coactivation 10. Alleles derived from SNPs within conserved regions are rarer than new alleles in nonconserved regions (P = 3×10−18) 11. The variants in these regions have been subjected to extreme evolutionary pressure and conserved in humans over long evolutionary periods, suggesting that the few common SNPs within UCEs may harbor critical biological functions.12 Thus, these SNPs could be excellent tools for studying cancer risk, treatment efficacy, and patient prognosis. However, there has been only one study published analyzing the association between genetic polymorphisms within UCEs and cancer risk revealing the potential impact of six SNPs on familial breast cancer risk13. However, another study found that UCEs have distinct expression signatures in CRC, and inhibiting overexpressed UCEs induced apoptosis,6 suggesting potential links between UCEs and prognosis and treatment response in CRC. Therefore, we hypothesize that SNPs within UCEs modulate clinical outcomes of patients with CRC. To test this hypothesis, we selected 48 potentially functional SNPs within UCEs and systematically evaluated their individual and joint associations with clinical outcomes of locally advanced CRC patients treated with adjuvant fluoropyrimidine-based chemotherapy.
MATERIALS AND METHODS
Study population and epidemiologic data
Six-hundred-and-sixty-two patients with histologically confirmed colorectal adenocarcinoma were enrolled at The University of Texas MD Anderson Cancer Center between March 1995 and May 2008. All patients were diagnosed with stage I–III disease (according to the American Joint Committee on Cancer TNM version 6.0 classification) and underwent radical surgery. There were no recruitment restrictions on age, gender, ethnicity, or cancer stage. Of the 662 patients, 435 had their disease diagnosed within 1 year prior to recruitment, and these patients were analyzed as the training set in this study. The remaining 227 patients had a longer history (>1 year) of CRC before referral to MD Anderson Cancer Center and were used as the validation set. Each patient signed an informed consent form and donated a 10- to 20-mL peripheral blood sample for the isolation of DNA.
Epidemiological data were collected using a structured questionnaire, including questions about demographic characteristics, smoking history, alcohol consumption, medical history, and family history of cancer. Clinical and follow-up data including date of diagnosis, performance status, clinical stage, tumor location, histological grade, primary surgery, pathological stage, chemotherapy, chemoradiation, radiation, and tumor recurrence/progression were abstracted from the patients’ medical records. Information on vital status was obtained from the medical records and Social Security Death Index (SSDI). The study was approved by Institutional Review Board of MD Anderson.
SNP selection and genotyping
Bejerano et al. discovered 481 UCEs using a bioinformatic comparison of the mouse, rat, and human genomes 14. All ultraconserved sequences used in this study is available in their report (http://www.cse.ucsc.edu/~jill/ultra.html)14 and from the UCbase & miRfunc database (http://microrna.osu.edu/UCbase4) 15. For each UCE, we first selected haplotype-tagging SNPs based on data from the International HapMap Project (http://www.hapmap.org) and obtained a list of 141 SNPs. After filtering the SNPs with the LD Select program (http://droog.gs.washington.edu/ldSelect.html) and the University of California, San Francisco Golden Path Gene Sorter program (http://genome.ucsc.edu), we retained 54 SNPs on the basis of a linkage disequilibrium r2 threshold of 0.8 and a minor allele frequency greater than 0.05 in Caucasian. The 54 SNPs were then submitted to Illumina (San Diego, CA) technical support, and those with low Illumina quality design scores (< 0.6) were excluded. Table 1 shows the 48 SNPs selected for genotyping in this study.
Table 1.
Polymorphisms in UCEs selected for this study
| Gene name (gene symbol) | SNP ID | Chromosome | UCE element | Position | Major/minor allele | MAF (%) |
|---|---|---|---|---|---|---|
| HS2ST1 | rs10747335 | 1 | uc.29 | Intron | C/A | 0.23 |
| PKN2 | rs10493807 | 1 | uc.31 | 5′ – FR* | G/A | 0.08 |
| C1orf110 | rs4412572 | 1 | uc.38 | 5′-FR | ||
| RGS4 | rs11580679 | 1 | uc.39 | 5′-FR | A/G | 0.38 |
| RGS4 | rs16847292 | 1 | uc.39 | 5′-FR | A/G | 0.07 |
| NUF2 | rs10917804 | 1 | uc.40 | 3′-FR | A/G | 0.08 |
| C1orf75 | rs7533689 | 1 | uc.41 | 5′-FR | G/A | 0.19 |
| LOC730134 | rs10211390 | 2 | uc.54 | 3′-FR | C/G | 0.36 |
| BCL11A | rs12473113 | 2 | uc.57 | 3′-FR | G/A | 0.09 |
| BCL11A | rs9784100 | 2 | uc.60 | 3′-FR | G/C | 0.39 |
| FLJ16124 | rs2954963 | 2 | uc.65 | 3′-FR | G/A | 0.27 |
| SFXN5 | rs2421099 | 2 | uc.66 | Intron | A/T | 0.14 |
| LOC730124 | rs786255 | 2 | uc.79 | 35′-FR | A/G | 0.39 |
| LOC728773 | rs1399685 | 2 | uc.81 | 3′-FR | T/A | 0.06 |
| LOC728304 | rs12619842 | 2 | uc.92 | 5′-FR | C/G | 0.17 |
| PDK1 | rs6710129 | 2 | uc.99 | Intron | A/G | 0.20 |
| ZBTB20 | rs16822925 | 3 | uc.119 | Intron | C/A | 0.14 |
| rs9838168 | 3 | uc.126 | G/A | 0.44 | ||
| RSRC1 | rs11713363 | 3 | uc.131 | Intron | A/G | 0.26 |
| C5orf36 | rs13154972 | 5 | uc.172 | 3′-FR | A/G | 0.17 |
| EBF1 | rs4921445 | 5 | uc.175 | Intron | A/G | 0.13 |
| C7orf30 | rs199657 | 7 | uc.208 | 5′-FR | G/C | 0.40 |
| LOC442660 | rs774265 | 7 | uc.213 | Intron | G/A | 0.15 |
| SHFM1 | rs6953983 | 7 | uc.220 | 5′-FR | G/A | 0.36 |
| EBF2 | rs9942838 | 8 | uc.235 | Intron | G/A | 0.50 |
| LOC347119 | rs7033100 | 9 | uc.264 | 3′-FR | G/A | 0.32 |
| LHX6 | rs1467737 | 9 | uc.278 | Intron | A/G | 0.47 |
| ZNF503 | rs12782308 | 10 | uc.287 | 5′-FR | G/C | 0.14 |
| PKD2L1 | rs2305386 | 10 | uc.294 | Intron | G/A | 0.05 |
| SCD | rs7849 | 10 | uc.298 | 3′ UTR | A/G | 0.17 |
| TECTB | rs11195893 | 10 | uc.310 | 3′-FR | G/A | 0.10 |
| RAB11FIP2 | rs12218935 | 10 | uc.311 | 3′-FR | A/G | 0.46 |
| MGMT | rs1711662 | 10 | uc.318 | Intron | G/A | 0.28 |
| DLG2 | rs3815988 | 11 | uc.331 | Intron | A/G | 0.35 |
| HNT | rs6590611 | 11 | uc.334 | Intron | C/A | 0.33 |
| LOC647277 | rs1395351 | 13 | uc.351 | 3′-FR | A/G | 0.29 |
| ARHGEF7 | rs9560010 | 13 | uc.357 | Intron | G/A | 0.17 |
| AKAP6 | rs8007042 | 14 | uc.367 | 5′-FR | C/A | 0.12 |
| AKAP6 | rs1956211 | 14 | uc.369 | Intron | A/G | 0.22 |
| MAP2K5 | rs8037887 | 15 | uc.391 | Intron | A/C | 0.15 |
| IKZF3 | rs13313561 | 17 | uc.410 | Intron | G/C | 0.05 |
| TCF4 | rs12455881 | 18 | uc.436 | Intron | G/A | 0.16 |
| ZNF407 | rs4243289 | 18 | uc.437 | Intron | G/C | 0.44 |
| RBL1 | rs6124509 | 20 | uc.455 | 3′-FR | A/G | 0.14 |
| FAM48B1 | rs16983007 | X | uc.465 | 5′-FR | G/A | 0.08 |
| LOC729188 | rs1029496 | X | uc.466 | 3′-FR | A/G | 0.14 |
| PDK3 | rs10482283 | X | uc.469 | 5′-FR | G/A | 0.19 |
| GRIA3 | rs1293524 | X | uc.481 | 5′-FR | A/T | 0.11 |
FR: flanking region
DNA was isolated from the peripheral blood samples using a QIAampDNA extraction kit (Qiagen, Valencia, CA). SNP genotyping was conducted using an Illumina VeraCode GoldenGate Assay kit. The BeadXpress Reader was used for microbead code identification and fluorescent signal detection. Genotype clustering and calling were performed using Illumina GenomeStudio software. Ten duplicate DNA samples showed 100% concordance. The mean call rate for the SNP array was 99.9%. One SNP, rs4412572 in C1orf110, failed in all samples due to low signals and was therefore discarded from further analysis.
Statistical analysis
The χ2-test was used to assess the differences in the distributions of categorical variables, and the Student’s t test was used to evaluate continuous variables. Cox’s proportional hazards model was used to estimate Hazard ratios (HRs) and their 95% confidence intervals (CIs) for the multivariate survival analyses, while adjusting for age, gender, ethnicity, smoking status and histologic grade. For each SNP, we tested three different genetic models, specifically, a dominant model, a recessive model and an additive model. The model with most significant P value was considered the best-fitting model. Fixed and random effects meta-analyses were used to calculate the pooled HRs. The Cochrane Q statistics test was used to assess heterogeneity between different data sets. When the Q test is significant, a random-effects model is used to accommodate the diversity in the magnitude of treatment effects. Otherwise, the pooled HR was estimated using the random effects model. The associations between genotype and survival time were plotted using the Kaplan-Meier method and analyzed using the log-rank test. We also evaluated the combined effects of the SNPs by the number of genotypes identified from the main effects analysis of single SNPs. Higher-order gene-gene interactions were evaluated using survival-tree analysis, as implemented in the STREE program (http://masal.med.yale.edu/stree/), which uses recursive partitioning to identify subgroups of individuals with similar risk. All statistical analyses were performed using STATA software (version 10, STATA Corporation, College Station, TX). All P-values were two-sided, and a P-value < 0.05 was considered statistically significant.
RESULTS
Patient characteristics
The demographic and clinical characteristics of patients are presented in Table 2. Of the 435 patients in the training set, 352 (80.9%) were Caucasian and 263 (60.5%) were male. There were 61 patients with stage I disease, 171 patients with stage II disease, and 203 patients with stage III disease. All patients underwent primary surgery with curative intent. Of the 435 patients, we obtained genotype data from a total of 285 patients (66%) patients who had received fluoropyrimidine-based chemotherapy: 115 patients with stage II disease and 170 patients with stage III disease. During the median follow-up time of 45.1 months, there were 65 deaths and 93 recurrences. Gender, race, smoking pack-years, tumor location, and histology grade were not significantly associated with clinical outcomes. AJCC stage was significantly associated with both recurrence (P = 0.005) and survival (P = 0.02), and age correlated with survival (P = 0.01).
Table 2.
Demographic and clinical variables for CRC patients
| Variables | Training Set (N=435)
|
Replication Set (N=227)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Recurrence | No recurrence | P Value | Dead | Alive | P value | Recurrence | No recurrence | P value | Dead | Alive | P value | |
| Age, mean (SD) | 59.2 (13.7) | 58.4 (12.8) | 0.6 | 62.3(13.3) | 57.9 (12.8) | 0.01 | 57.06(12.30) | 53.00(16.28) | 0.23 | 56.77(12.26) | 56.86(13.00) | 0.95 |
| Smoking pack-years (SD) | 37.4(36.9) | 30.8(39.3) | 0.33 | 37.2 (36.4) | 31.4(39.2) | 0.47 | 27.30(20.74) | 22.70(15.12) | 0.63 | 25.30(21.68) | 30.20(21.05) | 0.29 |
| Gender, N (%) | ||||||||||||
| Male | 57(61.3) | 206(60.2) | 41(63.1) | 222(60.0) | 124(58.8) | 7(46.7) | 72(58.1) | 60(58.3) | ||||
| Female | 36(38.7) | 136(39.8) | 0.85 | 24(36.9) | 148(40.0) | 0.64 | 87(41.2) | 8(53.3) | 0.36 | 52(41.9) | 43(41.7) | 0.98 |
| Race, N (%) | ||||||||||||
| Caucasian | 73(78.5) | 279(81.8) | 51(78.5) | 301(81.6) | 179(84.8) | 12(80.0) | 105(84.7) | 87(84.5) | ||||
| African-American | 9(9.7) | 33(9.7) | 7(10.8) | 35(9.5) | 11(5.2) | 1(6.7) | 7(5.6) | 5(4.9) | ||||
| Others | 11(11.8) | 29(8.5) | 0.61 | 7(10.8) | 33(8.9) | 0.84 | 21(10.0) | 2(13.3) | 0.88 | 12(9.7) | 11(10.7) | 0.94 |
| Tumor location, N (%) | ||||||||||||
| Proximal | 21(23.3) | 100(29.4) | 14(22.6) | 107(29.1) | 64(32.0) | 3(20.0) | 40(35.1) | 27(26.5) | ||||
| Distal | 20(22.2) | 80(23.5) | 17(27.4) | 83(22.6) | 70(35.0) | 8(53.3) | 40(35.1) | 39(38.2) | ||||
| Rectal | 49(54.4) | 160(47.1) | 0.41 | 31(50.0) | 178(48.4) | 0.51 | 66(33.0) | 4(26.7) | 0.35 | 34(29.8) | 36(35.3) | 0.38 |
| Stage, N (%) | ||||||||||||
| Stage I | 4(4.3) | 57(16.7) | 2(3.1) | 59(15.9) | 27(12.8) | 0(0.0) | 15(12.1) | 12(11.7) | ||||
| Stage II | 36(38.7) | 135(39.5) | 26(40.0) | 145(39.2) | 78(37.0) | 9(60.0) | 43(34.7) | 45(43.7) | ||||
| Stage III | 53(57.0) | 150(43.9) | 0.005 | 37(56.9) | 166(44.9) | 0.02 | 106(50.2) | 6(40.0) | 0.13 | 66(53.2) | 46(44.7) | 0.36 |
| Histology grade, N (%) | ||||||||||||
| Well-differentiated | 4(4.4) | 11(3.3) | 3(4.8) | 12(3.3) | 8(3.9) | 0(0.0) | 5(4.2) | 3(3.0) | ||||
| Moderate-differentiated | 73(80.2) | 283(84.2) | 49(77.8) | 307(84.3) | 159(77.9) | 11(78.6) | 88(74.6) | 83(82.2) | ||||
| Poorly-differentiated | 14(15.4) | 42(12.5) | 0.65 | 11(17.5) | 45(12.4) | 0.43 | 37(18.1) | 3(21.4) | 0.73 | 25(21.2) | 15(14.9) | 0.40 |
Among the 227 patients in the validation set, 200 patients (88%) had received fluoropyrimidine-based chemotherapy; 88 patients with stage II disease and 112 patients with stage III disease. Overall, there were 124 deaths and 211 recurrences during the median follow-up time of 57.5 months. These patients were diagnosed outside of MD Anderson Cancer Center at least a year before presenting to MD Anderson Cancer Center for treatment due to potential tumor recurrence or progression; therefore, the recurrence and death rates were higher than newly diagnosed patients (Table 2).
Individual SNPs and clinical outcomes
We assessed the association of each individual SNP with disease recurrence and death using a multivariate Cox model, adjusting for age, gender, ethnicity, smoking status, and histologic grade. Eight genetic loci were found to be associated with clinical outcomes of patients treated with fluoropyrimidine-based chemotherapy stratified by stage (Table 3). We next evaluated the associations between genotype and clinical outcome to fluoropyrimidine-based chemotherapy in patients with stage II and III disease. We did not analyze stage I diseases which are typically treated with surgery only with excellent prognosis and there were very few recurrence or death events in our training set. For stage II patients receiving fluoropyrimidine-based chemotherapy (N=115), the homozygous variant and heterozygous genotypes of rs7849 showed an increased risk of recurrence (HR: 2.39, 95%CI: 1.04–5.52; P=0.04) and a decrease in the median recurrence-free time (log-rank P = 0.03) compared with the wild-type genotype. This association was confirmed in the replication set (HR: 3.70, 95%CI: 1.42–9.64; P=0.007) and meta-analysis (HR: 2.89; 95%CI: 1.54–5.41; P=0.001). For other SNPs that were significant in training set, patients carrying a homozygous variant genotype of rs10211390 had a significantly increased risk of recurrence (HR: 2.79; 95%CI: 1.16–6.71; P=0.02) and a shorter median recurrence-free time (log-rank P = 0.03) compared with those with wild-type and heterozygous genotypes. A significant increase in the risk of recurrence was also bestowed on patients with the homozygous variant and heterozygous genotypes for rs2421099 (HR: 2.44; 95%CI: 1.08 –5.51; P=0.03) and rs16983007 (HR: 2.81, 95% CI: 1.02–7.70; P=0.04). Moreover, patients carrying at least one variant allele of rs16983007 had a significantly shorter recurrence-free survival time than those with the wild-type genotype (log-rank P = 0.03). The variant alleles for rs6590611 were associated with an increased risk of dying in a dose-dependent manner (per-allele HR: 2.92; 95%CI: 1.22–7.02).
Table 3.
SNPs associated with clinical outcome in patients receiving fluoropyrimidine -based adjuvant chemotherapy
| SNP | Model | Training Set
|
Replication Set
|
Pooled Analysis
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR(95%CI) | P value* | Log rank p | Genotype Distribution MM/MV/VV# | HR(95%CI) | P value* | Genotype Distribution MM/MV/VV | HR(95%CI) | P value* | Genotype Distribution MM/MV/VV | Cochran’s Q-test P value† | ||
| Recurrence: | ||||||||||||
| Stage II | ||||||||||||
| rs10211390 | recessive | 2.79(1.16–6.71) | 0.02 | 0.03 | 39/52/24 | 1.93(0.73–5.13) | 0.18 | 26/18/8 | 2.37(1.23–4.54) | 0.01 | 65/70/32 | 0.58 |
| rs2421099 | dominant | 2.44(1.08–5.51) | 0.03 | 0.16 | 86/25/4 | 0.50(0.24–1.08) | 0.08 | 38/14/0 | 1.04(0.60–1.80) | 0.90 | 124/39/4 | 0.005 |
| rs7849 | dominant | 2.39(1.04–5.52) | 0.04 | 0.03 | 80/28/7 | 3.70(1.42–9.64) | 0.007 | 36/11/5 | 2.89(1.54–5.41) | 0.001 | 116/39/12 | 0.50 |
| rs16983007 | dominant | 2.81(1.02–7.70) | 0.04 | 0.03 | 102/3/10 | 1.23(0.28–5.34) | 0.78 | 44/6/2 | 2.16(0.94–4.97) | 0.07 | 146/9/12 | 0.36 |
| Stage III | ||||||||||||
| rs10211390 | recessive | 2.70(1.12–6.50) | 0.03 | 0.007 | 63/84/22 | 1.60(0.77–3.35) | 0.21 | 45/47/14 | 1.98(1.13–3.49) | 0.02 | 108/131/36 | 0.37 |
| rs6124509 | dominant | 0.38(016–0.91) | 0.03 | 0.04 | 118/47/4 | 1.00(0.59–1.70) | 1.00 | 71/32/3 | 0.77(0.49–1.21) | 0.26 | 189/79/7 | 0.06 |
| rs11195893 | dominant | 0.26(0.08–0.91) | 0.04 | 0.61 | 139/30/1 | 0.83(0.42–1.61) | 0.58 | 90/15/1 | 0.63(0.35–1.14) | 0.13 | 229/45/2 | 0.10 |
| Survival: | ||||||||||||
| Stage II | ||||||||||||
| rs6590611 | additive | 2.92(1.22–7.02) | 0.02 | 0.05 | 54/52/9 | 1.46(0.63–3.35) | 0.38 | 28/22/2 | 2.03(1.11–3.72) | 0.02 | 82/74/11 | 0.26 |
| Stage III | ||||||||||||
| rs9942838 | recessive | 3.23(1.17–8.92) | 0.02 | 0.17 | 44/88/38 | 1.27(0.57–2.83) | 0.55 | 27/47/32 | 1.82(0.97–3.41) | 0.06 | 71/135/70 | 0.16 |
Adjusted by age, gender, ethnicity, smoking status and histologic grade
Cochran’s Q statistic to test for heterogeneity between studies
M: major allele, V: variant allele
For patients with stage III disease receiving fluoropyrimidine-based chemotherapy (N=170), a significantly decreased risk of recurrence was shown for the homozygous variant and heterozygous genotypes of rs6124509 (HR: 0.38, 95%CI: 0.16–0.91; P=0.03) and rs11195893 (HR: 0.26, 95%CI: 0.08–0.91; P=0.04), whereas the homozygous variant genotype for rs10211390 was associated with an increased risk of recurrence (HR: 2.70; 95%CI: 1.12–6.50; P=0.03). In addition, patients carrying the homozygous variant genotype of rs10211390 had shorter recurrence-free interval than those with the wild-type and heterozygous genotype (log-rank P = 0.007). Patients carrying the homozygous variant genotype of rs9942838 were also at an increased risk of death (HR: 3.23; 95%CI: 1.17–8.92; P=0.02). These SNPs in stage III patients were not validated in the replication set.
Cumulative effects of unfavorable genotypes on clinical outcome
We defined those genotypes that were associated with increased risks of disease recurrence or death as unfavorable genotypes. We next asked whether combining the unfavorable genotypes would have an additive effect on the clinical outcomes of patients treated with fluoropyrimidine-based chemotherapy. We performed a joint-effect analysis using four SNPs that were significantly associated with recurrence risk in patients with stage II disease. There was a significant dose-response trend of increased risk of CRC recurrence with increasing number of unfavorable genotypes. Compared with the low-risk group (0–1 unfavorable genotypes), the medium-risk (2 unfavorable genotypes) and high-risk (3–4 unfavorable genotypes) groups had a 4.36 times (95% CI: 1.66–11.47) and 9.67 times (95% CI: 2.99–31.25) higher risk of recurrence, respectively (P for trend = 1.78×10−5). The median recurrence-free survival times were >143.3, 30.2, and 16.8 months for patients in the low-, medium-, and high-risk groups, respectively (Figure 1a; log-rank P = 8.14×10−6).
Figure 1.
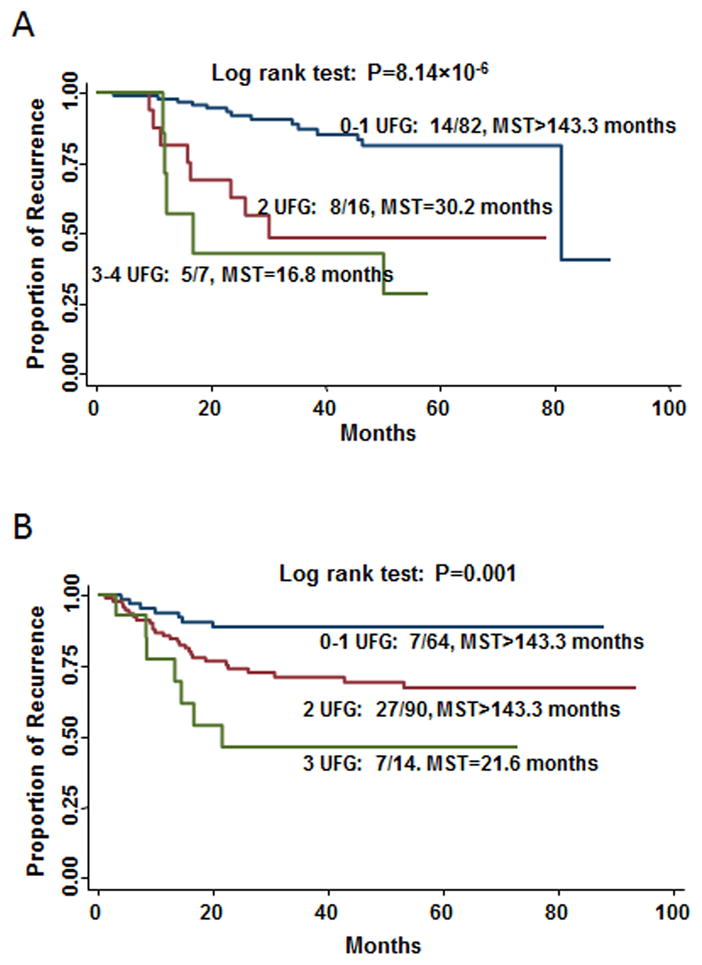
Kaplan-Meier curves for recurrence-free survival by the number of unfavorable genotypes (UFG) for patients with a) stage-II or b) stage-III disease. MST: median recurrence-free survival.
We also evaluated the combined effects of the three SNPs significantly associated with disease recurrence in patients with stage III disease. Compared with the reference group (those with 0–1 unfavorable genotypes), the HRs for individuals with two and three unfavorable genotypes were 3.21 (95% CI: 1.34–7.74) and 6.98 (95% CI: 2.12 22.97), respectively (P for trend = 0.001). Cumulative effect analysis also showed a significant dose-dependent effect on median recurrence-free survival times (Figure 1b; log-rank P =0.001).
SNP-SNP interactions and clinical outcomes
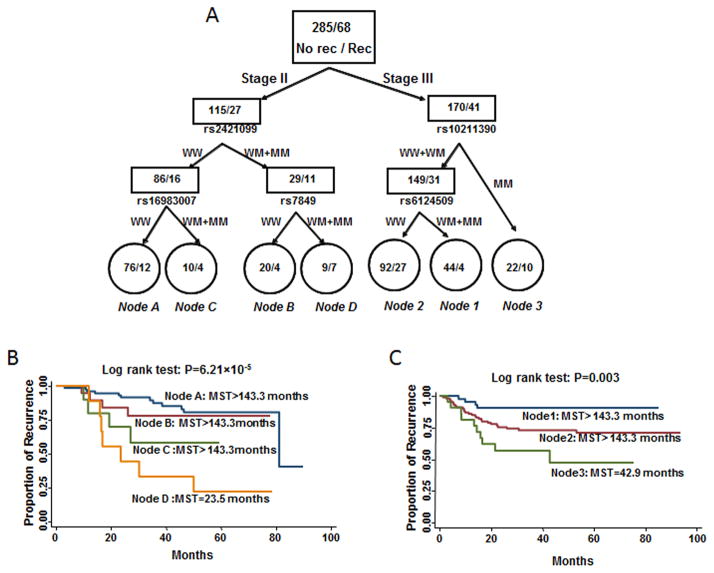
We next used survival tree analysis to further evaluate the potential interactions among the SNPs significantly associated with recurrence in patients with stage II and III disease (Figure 2a). For those with stage II disease, the tree structure resulted in four terminal nodes, ranging from low to high recurrence risk. The initial split was rs2421099, suggesting its value as a prognostic marker for patients receiving adjuvant chemotherapy. When using terminal node A as the reference group (wild-type genotypes of rs2421099 and rs16983007), the HR was 1.46 (95%CI: 0.46–4.68) for terminal node B (heterozygous and homozygous variant genotypes of rs2421099 and wild-type genotypes of rs7849), 3.18 (95%CI: 0.90–11.19) for terminal node C (wild-type genotype of rs2421099 and heterozygous and homozygous variant genotypes of rs16983007), and 6.27 (95%CI: 2.25–17.41) for terminal node D (heterozygous and homozygous variant genotypes of rs2421099 and rs7849), respectively (P for tend=0.0004). The increase in recurrence risk resulted in a decrease in the median recurrence-free survival times for subgroups corresponding to terminal nodes A–D (Figure 2b; log-rank P = 6.21×10−5).
Figure 2.
Potential SNP-SNP interactions. a) Tree structure identifying subgroups of patients with different genetic backgrounds. Kaplan–Meier curves for recurrence-free survival based on survival-tree analysis in patients with b) stage-II or c) stage-III disease; MST: median recurrence-free survival time.
We performed a similar analysis for patients with stage III disease. The analysis resulted in three terminal nodes, with rs10221390 as the initial split. When using terminal node 1 (subjects carrying wild-type and heterozygous genotypes of rs10211390 and heterozygous and homozygous variant genotypes of rs6124509) as the reference group, the HRs for terminal node 2 (subjects carrying wild-type and heterozygous genotypes of rs10211390 and wild-type genotype of rs6124509) and terminal node 3 (subjects carrying homozygous variant genotypes of rs10211390) were 3.75 (95% CI: 1.23–11.50) and 7.96 (95% CI: 2.07–30.65), respectively (P for tend=0.001). The corresponding decreased median recurrence-free survival times was highly significant (Figure 2c; log-rank P =0.003).
DISCUSSION
We have completed a comprehensive study to identify polymorphisms within UCEs that influence clinical outcomes of locally advanced CRC patients treated with adjuvant fluoropyrimidine-based chemotherapy. We identified eight genetic loci that are most likely to have an impact on the sensitivity to fluoropyrimidine agents. These SNPs can be used as prognostic biomarkers to assist stratify patients for fluoropyrimidine-based chemotherapy. For patients with stage II disease, rs7849 was consistently associated with disease recurrence in the training, validation, and meta-analysis. Moreover, we showed that the genotype-drug interaction was much more pronounced when multiple gene variants were considered in combination.
Chemotherapy is currently considered in patients with AJCC high-risk stage II and stage III disease. Thus, it is very important to define individual risk to determine who may or may not benefit from adjuvant chemotherapy. In this study, rs10211390 allowed us to identify the patients with stage II and III disease who had an increased recurrence risk after fluoropyrimidine-based chemotherapy. Rs10211390 affects the non-exonic element uc.54. This category of non-exonic elements has been shown to act as a long-range enhancer to control flanking gene expression 16,17. Such long-range enhancers can act at distances greater than 2630 kb from their target genes 16. An in vivo analysis confirmed that 45% of human conserved non-coding sequences, including uc.54, function as tissue-specific enhancers of gene expression 7,18. The nearest gene downstream of rs10211390 is FANCL (Fanconi anemia, complementation group L), one of 13 known Fanconi anemia genes that are 705-kb from rs10211390. FANCL was recently identified as the putative catalytic E3 ubiquitin ligase subunit of the Fanconi anemia core complex, which monoubiquitinates FANCD2 to allow proper repair of exogenous DNA damage 19,20. Moreover, cross-links between the Fanconi anemia core complex and BRCA2 appear to be involved with multiple DNA repair mechanisms 21. Thus, the FANC protein network has an important role in promoting chromosomal instability and tumor development and determining the sensitivity of cancer cells to chemotherapy 22,23. Recently, Fei et al reported that a splice variant of FANCL resulted in decreased FANCL expression, which provided lung cancer cells with a growth advantage 22. Nevertheless, the biological mechanisms underlying the associations of rs10211390 with cancer and the function of uc.54 are still unclear and need further research.
Administration of adjuvant chemotherapy for all patients with stage II disease remains controversial. Our results suggest that individual outcomes after fluoropyrimidine treatment can be determined based on the genotypes of rs7849 and rs6590611. Notably, rs7849 was consistently associated with an increased recurrence risk in both the validation and combined sets. The rs7849 SNP is located in uc.298, one of 12 paralogous UCE sets. The fact that paralogous sets have minimally changed in the past 300 million years suggests that they have crucial functions. The nearest gene upstream of rs7849 is stearoyl-CoA desaturase 1, a critical mediator of fatty acid synthesis. The rare allele of rs7849 has been shown to have an effect on body mass index, waist circumference, and insulin sensitivity, suggesting its potential physiologic significance. Recently, Luyimbaz et al. linked this cell fat metabolism gene to the mTOR oncogenic cell signaling pathway 24. The mTOR pathway functions through its effectors to mediate protein synthesis and cell cycle progression and is involved in multiple anticancer drug resistance. Rs6590611 affects uc.334, which was included in the HNT intron, a cell adhesion molecule family member. The intronic polymorphisms of HNT were identified as possible susceptibility loci for IgA nephritis and Alzheimer’s disease25,26. However, no study has reported the genetic effects of HNT polymorphisms on CRC treatment response. Interestingly, HNT expression was associated with recurrence for patients with stage I–II disease after surgery 27. Our results further suggest that these patients may be good candidates for chemotherapy but may not benefit from the fluoropyrimidine regimen.
Though pooled analysis of fluoropyrimidine-based adjuvant therapy trials showed a beneficial treatment effect in patients with stage III disease 28, we found that patients with minor alleles of rs9942838 had poorer survival. The rs9942838 genotype is located in the intron of early B-cell factor 2 (EBF2). The EBF family is a group of DNA-binding transcription factors with a basic helix-loop-helix domain 29. Several studies have shown that EBF inactivation due to genomic deletion, epigenetic silencing, or somatic point mutations exists in several types of cancer, including leukemia, glioblastoma, and pancreatic cancer, supporting the emerging roles of the EBF family in tumor suppression 30,31,32. Another study showed that silencing EBF2 led to a reduced resistance to apoptosis in chemo-naive tumor-derived cell populations from patients diagnosed with sporadic osteosarcoma33. However, EBF2 has not been investigated within the context of a fluoropyrimidine regimen.
To enhance the identification of patients with CRC who would benefit from the fluoropyrimidine regimen, we completed a combined effects analysis of unfavorable genotypes within the identified prognostic loci. A clear and significant trend was evident for increased risk with increasing number of unfavorable genotypes. These results suggest that the cumulative influence of multiple genetic variants within the UCEs can further enhance the separation of patients based on clinical outcome. Complex interactions between the SNPs could determine the functional outcome more than the independent main effects of any one susceptibility gene. We also performed an exploratory analysis of the SNP-SNP interactions and identified subgroups of patients with dramatically different RFS times after fluoropyrimidine treatment. However, statistical modeling of an interaction doesn’t amount to a true biological interaction, and these results should be interpreted with caution.
This study has several strengths. First, we reviewed and analyzed all the variations within 481 UCEs and reported systematic SNPs within the UCEs. Second, this is the first study to date specifically designed to identify the genetic effects of SNPs within UCEs in locally advanced colorectal cancer patients and treatment outcome following adjuvant therapy. In addition, we have comprehensive epidemiologic and clinical data for all locally advanced CRC patients, with a prolonged period of surveillance. The main limitation of this study is that the replication set was not newly diagnosed patients but consisted of those patients came to MD Anderson mainly due to potential recurrence or progression. Therefore, there was an over-representation of recurrence and progression in the replication set patients. The reason for our current means of splitting training and replication sets was to keep the training set as clean as possible to identify promising and more reliable candidate SNPs for further validations by us and other investigators in the field. The relative small and heterozygous population in the replication set may result in false negatives. Another weakness was that although the overall patient cohort has a large number, we stratified analysis by stage and treatment to limit the confounding of stage and treatment on recurrence or survival, which caused smaller numbers in the analyses and limited our power to detect additional significant associations. Only one SNP, rs7849, was validated in our replication set. Other SNPs were not significant in the replication set, although several of them showed consistent trend and remained significant in pooled analysis (e.g., rs10211390 for recurrence in both stage II and stage III patients and rs6590611 for survival in stage II patients). In addition, given the multiple comparison issue, there is a possibility that the significant results may be due to chance. Future validations with comparable patient populations to our training set and larger sample sizes are needed to confirm our results and validate more significant SNPs.
In conclusion, we have identified genetic variations within UCEs as prognostic markers in locally advanced CRC patients receiving fluoropyrimidine based adjuvant chemotherapy. The validation and incorporation of the identified SNPs and interactions with the clinical variables may allow clinicians to stratify patients for optimal adjuvant chemotherapy to achieve a step forward in personalized cancer care.
Acknowledgments
Funding sources:
This work was supported by a Multidisciplinary Research Program (MRP) grant on colorectal cancer from The University of Texas MD Anderson Cancer Center and by grant NIH-NCI CA-16672 from the National Cancer Institute.
Footnotes
Financial disclosures: The authors have no financial disclosures related to the content of this article.
References
- 1.Staib L, Link KH, Blatz A, Beger HG. Surgery of colorectal cancer: surgical morbidity and five- and ten-year results in 2400 patients--monoinstitutional experience. World J Surg. 2002;26(1):59–66. doi: 10.1007/s00268-001-0182-5. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11898035. [DOI] [PubMed] [Google Scholar]
- 2.Pohl A, Lurje G, Manegold PC, Lenz HJ. Pharmacogenomics and -genetics in colorectal cancer. Adv Drug Deliv Rev. 2009;61(5):375–80. doi: 10.1016/j.addr.2008.10.002. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19100300. [DOI] [PubMed] [Google Scholar]
- 3.Sengupta N, Gill KA, MacFie TS, Lai CS, Suraweera N, McDonald S, et al. Management of colorectal cancer: a role for genetics in prevention and treatment? Pathol Res Pract. 2008;204(7):469–77. doi: 10.1016/j.prp.2008.03.003. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18534770. [DOI] [PubMed] [Google Scholar]
- 4.Risch N. The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev. 2001;10(7):733–41. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11440958. [PubMed] [Google Scholar]
- 5.Baira E, Greshock J, Coukos G, Zhang L. Ultraconserved elements: genomics, function and disease. RNA Biol. 2008;5(3):132–4. doi: 10.4161/rna.5.3.6673. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18708752. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–29. doi: 10.1016/j.ccr.2007.07.027. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17785203. [DOI] [PubMed] [Google Scholar]
- 7.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444(7118):499–502. doi: 10.1038/nature05295. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17086198. [DOI] [PubMed] [Google Scholar]
- 8.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21(6):708–18. doi: 10.1101/gad.1525507. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17369403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16630819. [DOI] [PubMed] [Google Scholar]
- 10.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20(11):1470–84. doi: 10.1101/gad.1416106. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16705037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drake JA, Bird C, Nemesh J, Thomas DJ, Newton-Cheh C, Reymond A, et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet. 2006;38(2):223–7. doi: 10.1038/ng1710. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16380714. [DOI] [PubMed] [Google Scholar]
- 12.Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, et al. Human genome ultraconserved elements are ultraselected. Science. 2007;317(5840):915. doi: 10.1126/science.1142430. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17702936. [DOI] [PubMed] [Google Scholar]
- 13.Yang R, Frank B, Hemminki K, Bartram CR, Wappenschmidt B, Sutter C, et al. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis. 2008;29(2):351–5. doi: 10.1093/carcin/bgm290. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18174240. [DOI] [PubMed] [Google Scholar]
- 14.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304(5675):1321–5. doi: 10.1126/science.1098119. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15131266. [DOI] [PubMed] [Google Scholar]
- 15.Taccioli C, Fabbri E, Visone R, Volinia S, Calin GA, Fong LY, et al. UCbase & miRfunc: a database of ultraconserved sequences and microRNA function. Nucleic Acids Res. 2009;37(Database issue):D41–8. doi: 10.1093/nar/gkn702. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18945703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302(5644):413. doi: 10.1126/science.1088328. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14563999. [DOI] [PubMed] [Google Scholar]
- 17.Licastro D, Gennarino VA, Petrera F, Sanges R, Banfi S, Stupka E. Promiscuity of enhancer, coding and non-coding transcription functions in ultraconserved elements. BMC Genomics. 11:151. doi: 10.1186/1471-2164-11-151. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20202189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40(2):158–60. doi: 10.1038/ng.2007.55. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18176564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole AR, Lewis LP, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol. 17(3):294–8. doi: 10.1038/nsmb.1759. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20154706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35(2):165–70. doi: 10.1038/ng1241. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12973351. [DOI] [PubMed] [Google Scholar]
- 21.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–48. doi: 10.1038/nrg2159. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17768402. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Zhao D, Park HK, Wang H, Dyer RB, Liu W, et al. FAVL elevation in human tumors disrupts Fanconi anemia pathway signaling and promotes genomic instability and tumor growth. J Clin Invest. 120(5):1524–34. doi: 10.1172/JCI40908. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20407210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyakhovich A, Surralles J. Disruption of the Fanconi anemia/BRCA pathway in sporadic cancer. Cancer Lett. 2006;232(1):99–106. doi: 10.1016/j.canlet.2005.07.038. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16246487. [DOI] [PubMed] [Google Scholar]
- 24.Luyimbazi D, Akcakanat A, McAuliffe PF, Zhang L, Singh G, Gonzalez-Angulo AM, et al. Rapamycin regulates stearoyl CoA desaturase 1 expression in breast cancer. Mol Cancer Ther. 9(10):2770–84. doi: 10.1158/1535-7163.MCT-09-0980. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20876744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo KT, Lau YK, Wong KS, Zhao Y, Chan CM. Parallel genotyping of 10,204 single nucleotide polymorphisms to screen for susceptible genes for IgA nephropathy. Ann Acad Med Singapore. 2009;38(10):894–9. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19890582. [PubMed] [Google Scholar]
- 26.Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P, et al. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet. 2007;81(1):17–31. doi: 10.1086/518720. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17564960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalady MF, Dejulius K, Church JM, Lavery IC, Fazio VW, Ishwaran H. Gene signature is associated with early stage rectal cancer recurrence. J Am Coll Surg. 211(2):187–95. doi: 10.1016/j.jamcollsurg.2010.03.035. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20670856. [DOI] [PubMed] [Google Scholar]
- 28.Gill S, Loprinzi CL, Sargent DJ, Thome SD, Alberts SR, Haller DG, et al. Pooled analysis of fluorouracil-based adjuvant therapy for stage II and III colon cancer: who benefits and by how much? J Clin Oncol. 2004;22(10):1797–806. doi: 10.1200/JCO.2004.09.059. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15067028. [DOI] [PubMed] [Google Scholar]
- 29.Liao D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol Cancer Res. 2009;7(12):1893–901. doi: 10.1158/1541-7786.MCR-09-0229. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19996307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–64. doi: 10.1038/nature05690. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17344859. [DOI] [PubMed] [Google Scholar]
- 31.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–12. doi: 10.1126/science.1164382. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18772396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–6. doi: 10.1126/science.1164368. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18772397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patino-Garcia A, Zalacain M, Folio C, Zandueta C, Sierrasesumaga L, San Julian M, et al. Profiling of chemonaive osteosarcoma and paired-normal cells identifies EBF2 as a mediator of osteoprotegerin inhibition to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Clin Cancer Res. 2009;15(16):5082–91. doi: 10.1158/1078-0432.CCR-09-0300. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19671856. [DOI] [PubMed] [Google Scholar]