Abstract
As the importance of ubiquitylation in certain disease states becomes increasingly apparent, the enzymes responsible for removal of ubiquitin (Ub) from target proteins, deubiquitylases (DUBs), are becoming attractive targets for drug discovery. For rapid identification of compounds that alter DUB function, in vitro assays must be able to provide statistically robust data over a wide dynamic range of both substrate and enzyme concentrations during high throughput screening (HTS). The most established reagents for HTS are Ubs with a quenched fluorophore conjugated to the C-terminus; however, a luciferase-based strategy for detecting DUB activity (DUB-Glo™, Promega) provides a wider dynamic range than traditional fluorogenic reagents. Unfortunately, this assay requires high enzyme concentrations and lacks specificity for DUBs over other isopeptidases (e.g. desumoylases), as it is based on an aminoluciferin (AML) derivative of a peptide derived from the C-terminus of Ub (Z-RLRGG-). Conjugation of aminoluciferin to a full-length Ub (Ub-AML) yields a substrate that has a wide dynamic range, yet displays detection limits for DUBs 100- to 1000-fold lower than observed with DUB-Glo™. Ub-AML was even a sensitive substrate for DUBs (e.g. JosD1 and USP14) that do not show appreciable activity with DUB-Glo™. Aminoluciferin derivatives of hSUMO2 and NEDD8 were also shown to be sensitive substrates for desumoylases and deneddylases, respectively. Ub/Ubl-AML substrates are amenable to HTS (Z′ =0.67) yielding robust signal, and providing an alternative drug discovery platform for Ub/Ubl isopeptidases.
Keywords: Ubiquitin, SUMO, luciferin, deubiquitylation, protease assay, desumoylation
Introduction
The complex and dynamic process of protein post-translational modification through the attachment and removal of ubiquitin (Ub) impacts many cellular processes, including proteasome-mediated degradation, DNA damage repair, and endocytic sorting of membrane bound receptors [1–3]. Protein ubiquitylation can vary in the number of sites in a single protein targeted for Ub conjugation, as well as the number of Ub molecules attached at any one site, with long chains of polyubiquitin (polyUb) being formed through the attachment of one Ub molecule to another. These modifications vary in type and extent from one target substrate to the next. Adding further combinatorial power to this system is the potential for assembly of polyUb chains with different functions through linkages at any of the seven lysines (K6, K11, K27, K29, K33, K48, and K63) present in Ub [4], as well as the N-terminus of the protein [5]. Heterogeneity can exist for polyUb, as well, with “branched” chains being composed of linkages through different lysines on the same Ub.
Considering this, it is not surprising that nearly 100 deubiuqitylase enzymes (DUBs) responsible for the removal of ubiquitin and/or polyUb chains from proteins appear to exist in the human genome [6]. The reader is referred to a number of excellent reviews that explore DUBs in more detail than will be done here[7–9]. Briefly, this class of enzymes is typically divided into five families based on structural homology and/or catalytic function. The ubiquitin C-terminal Hydrolase (UCH) family of DUBs is characterized by efficient cleavage of short peptides linked to the C-terminus of Ub via isopeptide or standard peptide bonds. Ubiquitin specific proteases (USPs), the largest family, are typically found to catalyze the catabolism of iso-peptide bond linkages between a Ub molecule and the target lysine. In addition to these two families, there exist the ovarian tumor ubiquitin (OTU) domain and Machado-Josephine domain (MJD) DUBs. These four DUB families are cysteine proteases, with a canonical catalytic triad consisting of Cys, Asp, and His residues. The fifth DUB family, Jab1/MNP metalloenzyme (JAMM), is unique in that it consists of metalloproteinases, requiring the co-ordination of a Zn+2 ion at the active site for catalytic activity. Investigation of substrate specificity for DUBs has mostly been carried out in two ways, with traditional biochemical techniques attempting to discern the individual contributions of both polyUb chain topology type and the target substrate to which it is covalently attached. More often, though, DUBs are associated with a disease state through their dysregulation or genetic deletion in mouse models. While this may make a DUB an attractive candidate for drug discovery, it is only through extensive biochemical/biophysical characterization that it becomes a viable one.
The combination of disease relevance and the critical mass of available investigative means has spurred increased interest within the pharmaceutical industry to pursue DUBs as drug targets [10, 11]. The most precise methods for assessing DUB activity and specificity rely on the analysis of polyUb chain cleavage by SDS-PAGE or liquid chromatography-mass spectroscopy techniques (LC-MS), neither of which are compatible with high throughput screening (HTS). In order to rapidly identify potential medicinal compounds for either antagonism or agonism of catalytic function, biochemical assays must be developed that are amenable to both miniaturization and automation, as well as provide data that are sensitive and statistically robust with respect to variance.
Although a number of novel assay methodologies, including the Ub-CHOP reporter system [12, 13] and a FRET-based LanthaScreen™ DUB substrate (Life Technologies), have been developed to translate DUB activity to HTS, the most widely used screening rationale for HTS is perhaps the most simple: monomeric Ub linked at the C-terminus via an amide bond to a fluorophore such as amino-methylcoumarin (AMC) or rhodamine 110 (Rho110) resulting in quenching of the fluorophore[14, 15]. DUB amidohydrolase activity is monitored through the gain of signal associated with liberation of the fluorophore, with the modulation of this signal in a significant way being the criterion for identifying lead compounds. Although this assay configuration does not measure isopeptidase activity (the hallmark of most DUBs) nor even peptidolytic activity, due to its ease of use and convenience, it has been widely adopted as a surrogate for these activities. In addition, there are a number of limitations inherent in the use of fluorescence based Ub derivatives. In the case of Ub-AMC, the spectral requirements of the assay are especially problematic with respect to background, resulting in a relatively narrow dynamic range. The use of Ub-Rho110 provides a considerable improvement over Ub-AMC, due to the “red shift” in both excitation and emission spectra; however, the limits of fluorimetry still place constraints on the useful concentration ranges of both the enzyme and the substrate. The use of Ub-AMC or Ub-Rho110 could be precluded if situations arise that require either low enzyme concentrations and/or high substrate concentrations. This might occur during lead compound optimization, for example, when inhibitor potency dictates lower enzyme concentrations so as not to approximate those of the inhibitor.
The recent emergence of a luminescence strategy demonstrated to broadly detect activity of proteases that are responsible for the removal of Ub/Ubl from target proteins, DUB-Glo™, is an alternative to fluorimetry with little to no detectable background signal [16, 17]. The use of Photinus pyralis (firefly) luciferase in combination with the substrate aminoluciferin has recently been adapted to measure the activity of caspase proteases[18], based on the initial pioneering work measuring the activity of chymotrypsin[19]. Since light is only emitted when free aminoluciferin is in complex with luciferase, and the molecule itself has no intrinsic luminescence, this molecule is completely “dark” when conjugated to a peptide substrate [17–19]. Reactions are virtually background-free, allowing greater sensitivity at high substrate concentrations compared to fluorogenic substrates. Luciferase based protease assays are not without disadvantages, including the requirement of additional reagent components (e.g. luciferase), the inability to determine Henri-Michaelis-Menton kinetic constants, and the need to counter screen against luciferase activity. Nevertheless, luciferin/luciferase technology can provide a viable HTS platform through greatly enhanced sensitivity where other readouts cannot.
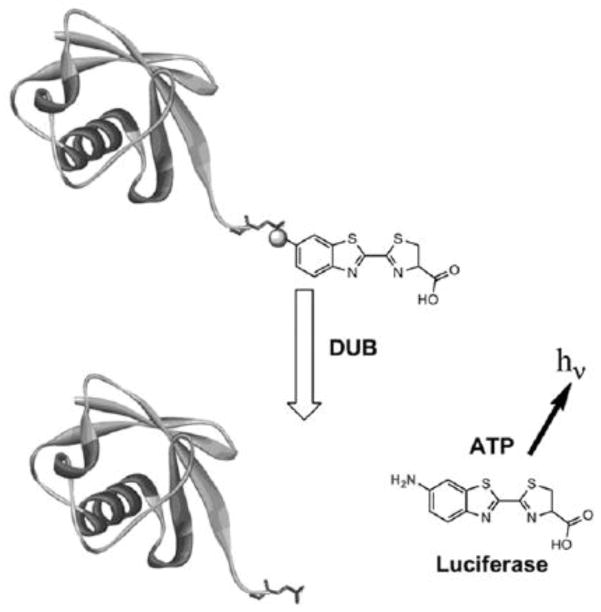
DUB-Glo™ employs a peptide derived from the C-terminus of Ub conjugated to aminoluciferin, (Z-RLRGG-AML) and has been demonstrated to be compatible with measuring the activity of DUBs, desumoylases, and deNEDDylases. Unfortunately, the use of this peptide requires relatively high enzyme concentrations compared to full-length Ub, consistent with the fact that this peptide is widely reported to be a poor substrate for DUBs. Essentially, the increased dynamic range of this assay is countered by the need for high enzyme concentrations, potentially complicating downstream compound optimization as mentioned above. This might be explained by the fact that Z-RLRGG-AML does not (by definition) possess extended binding sites thought to convey substrate specificity for DUBs [20]. These structural features likely play a dominant role in favoring the cleavage of Ub by these enzymes over other Ub-like proteins, such as small ubiquitin-like modifier (SUMO) [21] or neuronal precursor cell expressed developmentally down-regulated 8 (NEDD8) [22]. The lack of these structural features might also explain the utility of this substrate with a broad range of Ub/Ubl proteases. We hypothesized, then, that conjugation of aminoluciferin to a full-length Ub (Ub-AML, Scheme I), as opposed to a peptide derived from its C-terminus (Z-RLRGG-AML), would result in a reagent displaying the specificity and catalytic efficiency of full length ubiquitin substrates (e.g. Ub-Rho110), as well as providing the robustness and dynamic range typically observed for luciferin/luciferase assay platforms, such as DUB-Glo™.
Scheme 1.
Cleavage of Ub-AML through DUB action at the C-terminus liberates aminoluciferin, a substrate for Photinus pyralis luciferase (firefly). This light producing reaction provides large signal amplification with near non-existent background.
In order to investigate this hypothesis, comparative studies with Ub-AML, DUB-Glo™, Ub-AMC, and Ub-Rho110 were carried out to characterize this reagent as a substrate for a wide range of DUBs. The additional ubiquitin-like (Ubl) molecule aminoluciferin derivatives, human SUMO2 (hSUMO2-AML) and NEDD8 (NEDD8-AML), were also generated and tested as analogous reagents for HTS in desumoylation and deneddylation assays. Finally, Ub-AML was used with a representative USP to simulate the process of assay development and determine statistical robustness of this reagent in an HTS campaign for this class of enzymes.
Materials and Methods
2.1 Expression and purification of Ub/Ubl-MESNa
DNA constructs containing human Ub (CAA44911), SUMO2 (NM_006937.3), and NEDD8 (NP_006147) cloned as C-terminal intein fusions at Gly75 (pYTB, New England Biolabs) were generous gifts of Dr. Keith Wilkinson (Emory University). Fusion proteins were expressed (overnight, 18°C) in BL21 (DE3pLysS) strain under the IPTG inducible T7 promoter, and purified essentially as described [15, 23]. Briefly, cell pellets were lysed (sonication) in cold 50mM HEPES, 50mM sodium acetate, pH 6.5, 75mM NaCl (HSS) containing 1mM phenylmethylsulfonylfluoride (PMSF) and 5mM benzamidine, prior to clarification by high speed centrifugation. Soluble Ub/Ubl-intein fusions were captured (4°C) by chitin bead affinity chromatography (New England Biolabs), and subjected to “on-column” cleavage (overnight, 4°C) in the presence of 100mM mecaptoethansulfonic acid sodium salt (MESNa) at pH 6.5. Eluted protein was concentrated and stored at −80°C at pH 6.5. The precise concentration of an aliquot of purified Ub-MESNa was determined by amino acid analysis, which was then used to generate a calibration curve by RP-HPLC. All subsequent Ub/Ubl-MESNa and Ub/Ubl-AMLs were subjected (at least three injection amounts) to RP-HPLC and liquid chromatography-mass spectroscopy (LC-MS), to determine molar concentrations, purity, and integrity. It was assumed that the contribution of the aminoluciferin molecule contributed negligibly to these measurements.
2.2 Synthesis of Ub-AMC and Ub-Rho110
Ub-AMC and Ub-Rho110 were prepared through C-terminal conjugation of each fluorophore to Ub-MESNa, essentially as previously described for Ub-Rho110 [15]. Ub conjugates were purified (>95%) by HPLC, lyophilized, and stored at −80°C until use.
2.3 Generation of Ub/Ubl-AML substrates
2.3.1 Synthesis of glycyl-D-amino-luciferin
The synthesis of glycyl-D-amino-luciferin was essentially as previously described [24]with the intermediate 2-Cyano-6-amino-N-Boc-glycine-benzothiazole being generated first, followed by generation of a chemically blocked form of glycine-D-aminoluciferin.
2.3.2 Synthesis of 2-Cyano-6-amino-N-Boc-glycine-benzothiazole
Commercially available Boc-Gly-OH (3.85 mmol) was dissolved in 18 mL CH2Cl2. Diisopropylcarbodiimide (3.9 mmol) and DMAP (0.39 mmol) were added at 0° to the reaction, with stirring (0°C) for 20 min. 2-Cyano-6-aminobenzothiazole (2.57 mmol) was added to the mixture and the reaction was stirred for an additional 3h (RT). The reaction was diluted with CH2Cl2 and washed with brine. The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated. The crude product was purified by preparative RP-HPLC (Axia Gemini C18, 30 × 100 mm, Phenomenex, CA) using a linear gradient of 10–90% AcN over 10 min with a flow rate of 20 mL/min. After lyophilization, 2-Cyano-6-amino-N-Boc-glycine-benzothiazole was obtained in 75% yield as a white solid and characterized by LC-MS (Exact Mass: 332.0943).
2.3.3 Synthesis of Boc-glycine-D-aminoluciferin
2-Cyano-6-amino-N-Boc-glycine-benzothiazole (0.11mmol) was dissolved in 0.7mL of dimethylformamide (DMF); H-D-cysteine-HCl (0.12mmol, dissolved in 0.1mL of degassed water) was added drop-wise to the organic solution, while protected from light. The mixture was adjusted to pH 8.0 by addition of saturated K2CO3 in degassed water, and the reaction was stirred for 1h protected from light (RT). Analytical LC-MS showed complete conversion to the product. The crude reaction mixture was purified by preparative RP-HPLC as described above. After lyophilization, Boc-glycyl-D-aminoluciferin was obtained as a white solid in 80% yield and characterized by LC-MS (Exact Mass: 436.0875).
2.3.3 Deprotection of Glycyl-D-aminoluciferin
Boc-glycyl-D-aminoluciferin (0.2 M) was dissolved in a 1:1 mixture of TFA:CH2Cl2 with 5% Et3SiH and allowed to stir for 1.5h. The mixture was concentrated under vacuum, and the residue was directly purified by preparative RP-HPLC using a linear gradient of 0–45% AcN over 15min with a flow rate of 20 mL/min. After lyophilization, pure glycyl-D-aminoluciferin was obtained as a light yellow solid in 90% yield with mass determinations (LC-MS) consistent with those predicted (Exact Mass: 336.0351).
2.3.4 Preparation and Characterization of Ub/Ubl-AML substrates
Purified Ub/Ubl-MESNa was conjugated to glycyl-D-amino-luciferin using a modified protocol for Ub-Rho110 that improved solubility of the luciferin molecule and optimized conjugation efficiency. Ub-MES and glycyl-D-aminoluciferin were mixed in a molar ratio of 1:100 in 2:1 DMF/DMSO, plus 2% triethylamine. The final concentration of Ub-MES was 0.5–1mM. The reaction was monitored by LC-MS. After stirring for 16h (40°C, protected from light), the conjugation was complete; prolonged reaction time did not result in higher product yields. The crude reaction mixture was purified by preparative RP-HPLC column using a linear gradient of 20–50% AcN over 20 min at a flow rate of 5 mL/min (Viva C18, 10 × 150 mm, Restek, PA). After lyophilization, pure Ub (1–76)-aminoluciferin was obtained in 8–12% yield as a white solid and characterized by LC-MS (See Figure 1). The procedure for the generation of additional Ubl-AML substrates was essentially identical to one described above for Ub-AML. Ub/Ubl-AML purity was typically >95% for each batch (Figure 1). Lyophilized Ub/Ubl-AML substrates were stored at −20°C under desiccating conditions. They were reconstituted in DMSO and the actual concentration determined by RP-HPLC immediately prior to use.
Figure 1. Physical characterization of Ub-AML by LC-MS.
The modifiied protein was analyzed by RP-HPLC (Restek Viva C18 column, 3 micron particle size, 2.1 × 150mm). The column was equilibrated in 30% AcN/0.05% TFA at 0.2 mL/min and developed with a linear gradient to 50% AcN/0.05% TFA over 11 min. The absorbance at 214 nm (upper panel, black line) of the effluent from the column was recorded prior to injection into the ionization chamber of an ESI/ToF mass spectrometer. The total ion chromatogram is shown as the dark gray line in the upper panel. The mass spectrum (lower left panel) was extracted for the peak eluting at ~7 min and deconvoluted using the program Max Entropy to derive the parent mass spectrum shown in the lower right panel.
2.4 Expression and purification of Ub/Ubl proteases
The majority of Ub/Ubl proteases used in this study were expressed in a BL21 strain of E. coli as either His-tagged proteins, or fusions to SUMO or SUMOstar (LifeSensors, Inc). These proteases were purified by a combination of immobilized metal affinity chromatography (IMAC) and size exclusion chromatography (SEC). They include: full-length Associated Molecule with the SH3-domain of STAM (AMSHFL) and the corresponding catalytic domain (AMSHcore), ADD05037; Ataxin3, NP_004984.2; Ataxin3-like, NP_001129467.1; Deneddylase-1 (DEN1), NP_001159812.1; Josephin-1 (JosD1), NP_055691.1; Josephin-2 (JosD2), NP_612207.1; Otubain-2, NP_075601.1; Sentrin/SUMO Specific Protease 1 (SENP1), NP_055369.1, SENP2, NP_067640.2; SENP6, NP_001093879.1; Ubiquitin C-terminal Hydrolase L3 (UCHL3), NP_005993.1; UCHL5, NP_057068.1; catalytic domain of Ubiquitin-Specific Protease 2 (USP2core), NP_004196.4; USP8, NP_001122082.1; and USP14, NP_005142.1. Full-length forms of USP7 (NP_003461.2), and USP20 (NP_001008563.2) were generated in a baculovirus expression system (BEVS), through the use of the Bac-to-Bac system (Life Technologies), with recombinant viral characterization (titer) and protein production carried out in Spodoptera fugiperda (Life Technologies) cells. Proteins were expressed intracellularly, and purified from soluble insect extracts by IMAC. After purification, all proteases were buffer exchanged into either 50mM Tris or 20mM HEPES (pH 7.5) containing 0.15M NaCl, 2mM DTT, and 10% glycerol. After buffer exchange and concentration, proteins were aliquoted and stored at −80°C until use. Stock protein concentrations were determined by Bio Rad Protein Assay (Bio Rad) relative to bovine serum albumin as a standard. In some cases, protein concentration was also determined by absorbance, using a calculated molar extinction coefficient at 280nm (Vector NTI).
2.5 Assessment of Ub/Ubl protease activity with Ub/Ubl-AML
All proteases were serially diluted into 50mM HEPES, pH 7.5, 10mM DTT, 0.1% Prionex® (Sigma-Aldrich). In order to assess the reactivity of the peptide substrate Z-RLRGG-AML to these enzymes, the substrate was diluted into reconstituted DUB-Glo™ reagent (Promega, Inc.), according to the manufactures’ instructions. This included pre-incubation for 30 min (RT, protected from light) in order to allow for the consumption of any contaminating free D-aminoluciferin. For comparative studies, Ub/Ubl-AML substrates were substituted for Z-RLRGG-AML and prepared in a similar manner. Reactions were assembled in either 96- or low volume 384-well white plates, and luminescence was monitored (EnVision Multi-label Plate Reader, Perkin Elmer) approximately 30min post-DUB addition, unless otherwise noted. Reactions were typically carried out in triplicate, with signal-to-background (S/B) calculated by first subtracting mean relative luminescence (RLU) in the absence of enzyme (RLUbackground) from RLU in the presence of enzyme (RLUsignal), then dividing by RLU in the absence of the enzyme so that S/B = (RLUsignal − RLUbackground)/RLUbackground. RLU values used to determine (S/B) were taken from progress curve time points observed to be at “steady-state” of light generation, ensuring appropriate relative comparisons across the plate. In most cases, RLUbackground was insignificant relative to the signal in the presence of the enzyme.
2.6 Assessment of Ub/Ubl protease activity with Ub-Rho110 and Ub-AMC
Ub-AMC and Ub-Rho110 (250nM) were diluted into reaction buffer consisting of 50mM Tris, pH 8.0, 0.15M NaCl, 5mM DTT, and 0.05% CHAPS. Reactions were assembled in 96-well black plates, and initiated by the addition of serially diluted DUBs. Fluorescence intensity was monitored (30 min at intervals of 30 sec) immediately after DUB addition on an (EnVision Multi-label Plate Reader, Perkin Elmer) using the Excitation/Emission filter pair of 340/460nm for Ub-AMC or 485/531nm for Ub-Rho110.
3. Results and Discussion
3.1 Ub-AML is a robust substrate with femtomolar limits of detection for DUBs
Analysis of aminoluciferin conjugates by RP-HPLC determined each substrate was >95% pure. For each Ub/Ubl-AML substrate, determined masses (Da) were found to be consistent with those predicted for conjugation products, and were as follows: Ub-AML, 8826.1 (calculated mass, 8826.2, Figure 1); hSUMO2-LUC, 10,722.9. (calculated, 10,723.0); and NEDD8-LUC, 8821.5 (calculated 8821.3).
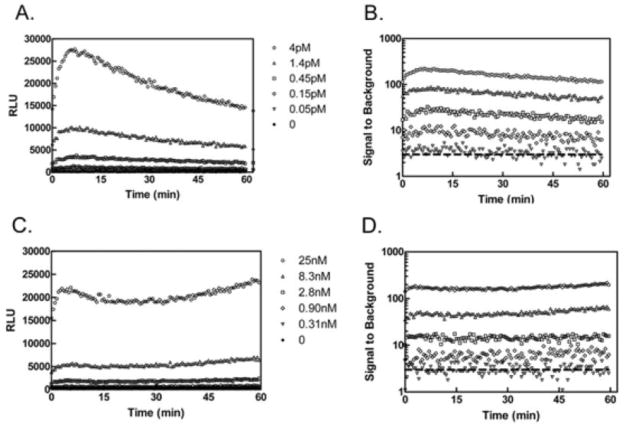
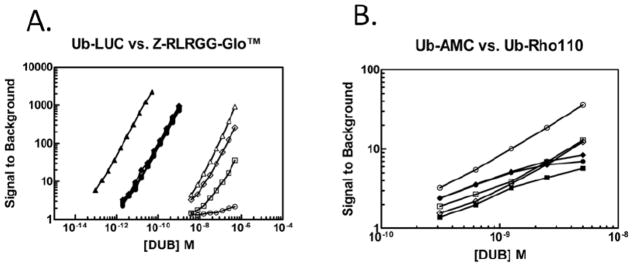
The DUB-Glo™ system was used for the functional characterization of Ub-AML, allowing independent comparison of Ub-AML to Z-RLRGG-AML (DUBGlo™) under identical assay conditions. The use of Ub-AML is expected to be compatible with other luciferase/luciferin based systems following user optimization. Figure 2 illustrates representative RLU traces for Ub-AML and Z-RLRGG-AML (Panels A and C, respectively), and calculated (S/B) values for both substrates (Panels B and D, respectively). The addition of UCHL3 to both Z-RLRGG-AML and Ub-AML was associated with increased luminescence over time. An initial “spike” of observed light in the first 30m was followed by relatively prolonged and steady light emission for more than an hour. This RLU signal trace is typically indicative of “coupled” luminescence based protease assays, where free aminoluciferin must be liberated by catalysis [25]. Increased signal intensity, at steady state light emission, for both substrates was observed in a concentration-dependent manner for UCHL3. As previously demonstrated for Z-RLRGG-AML, then, reactions containing both enzyme and Ub-AML resulted in the generation of free aminoluciferin substrate as a product of Ub-AML cleavage. Although the dynamic range displayed by these two reagents were roughly equivalent, achieving this S/B for Z-RLRGG-AML required approximately 6000 times more UCHL3 than for Ub-AML (compare legends for Figure 2, Panels B and D), revealing the extremely sensitive nature of Ub-AML relative to Z-RLRGG-AML. Ub-AML and Z-RLRGG-AML were then tested in parallel for assessing DUB activity with USP2core, USP7, USP8 and UCHL3 as a reference control (Figure 3, Panel A). Similar to previous reports with UCHL3, the linear range for Z-RLRGG-AML was approximately 10- and 1000-fold above the no enzyme control. The detection range for this reagent allowed activity measurements for UCHL3 ranging from micro- to nanomolar concentrations. In stark contrast, the use of Ub-AML resulted in greatly decreased lower limits of detection for UCHL3 (S/B equal to ~6 at sub-femtomolar concentrations), with S/B values of 10 to 1000 over a dose response of at least three orders of magnitude. Similar results were observed for USP2core and USP8, with a dynamic range for activity measurements ranging from approximately nano- to femtomolar concentrations. To achieve a dynamic range comparable to Ub-AML, reactions containing Z-RLRGG-AML required >1000-fold higher concentrations of USP2core and USP8. For USP7, activity measurements with Ub-AML yielded a linear dose response indistinguishable from USP8 and USP2core. However, comparisons between Ub-AML and Z-RLRGG-AML revealed a dramatic difference in the ability of these two substrates to assess USP7 activity. Even at micromolar concentrations of this DUB, Z-RLRGG-AML was a very poor substrate, as S/B ratios did not reach 3. This difference may likely be due to the requirement of full-length ubiquitin binding to USP7 for full catalytic competency [26]. As with other DUBs[20, 27], USP7 binding to its substrate includes extended interactions away from the catalytic site, with these interactions driving conformational changes in the enzyme that ultimately lead to increased stability of a transition state intermediate. These changes are not likely recapitulated through binding of smaller, peptide substrates, such as Z-RLRGG-AML. By extension, the nature of the full length Ub-AML is also likely to contribute to the increased sensitivity of this reagent for USP8 and USP2core as well. Previous reports indicate that Z-RLRGG derivatives have a relatively poor affinity for DUBs[20] compared to full-length ubiquitin substrates which would explain the requirement for much higher concentrations of both enzyme and substrate in this assay. Although luciferin-based assays preclude the dissection of individual kinetic parameters, differences in the overall kinetic efficiency between the two substrates should manifest itself through these comparative studies. To highlight the contributions of the luciferase/luciferin assay versus mechanistic differences in substrate-enzyme recognition between Ub-AML and Z-RLRGG-AML, we compared these reagents to Ub-AMC and Ub-Rho110 (Figure 3, Panel B). For these substrates, S/B values of 3 to 40 were observed at concentrations of enzyme spanning 0.3 to 5nM. Below this concentration range, signal did not exceed background (data not shown). As expected, these reagents possess lower limits of detection than found for DUB-Glo™, yet they do not display both the sensitivity and dynamic range of Ub-AML. The enhanced sensitivity of Ub-AML compared to other substrates appears to result from the combination of increased binding affinity/catalytic efficiency compared to Z-RLRGG, and the enhanced sensitivity of luciferase technology over fluorogenic substrates. Perhaps the most important advantage of increased sensitivity, from a drug discovery perspective, is the ability to optimize substrate amounts in such a way as to increase the probability of detecting inhibitor molecules at lower enzyme concentrations. In addition, the use of medicinal chemistry to increase compound potency creates the need for assays that provide adequate S/B at very low enzyme concentrations. In any case, having the widest possible dynamic range in an assay is beneficial.
Figure 2. Monitored progression of luminescence for Ub-AML and Z-RLRGG-Glo.
Increasing concentrations (indicated) of UCHL3 were incubated with either 500nM Ub-AML (A) or 20μM Z-RLRGG-AML(C) with immediate and continuous monitoring of luminescence (RLUs). Calculated S/B values (see methods) are shown for Ub-AML (B) and Z-RLRGG-AML(D), with the dashed line drawn to illustrate S/B = 3. Data are representative of triplicate measurements.
Figure 3. Determination of assay dynamic range for various DUB substrates.
Panel A: Indicated concentrations of UCHL3 (▲/△), USP2core (◆/◇), USP7 (●/○) and USP8 (■/□) were incubated with either 250nM Ub-AML (closed symbols) or 20μM Z-RLRGG-AML (open symbols) for 30min prior to RLU measurement and S/B calculation (see methods). Panel B: Indicated concentrations of USP2core (◆/◇), USP7(●/○), and USP8 (■/□) were incubated with 250nM of either Ub-AMC (closed symbols) or Ub-Rho110 (open symbols) with immediate and continuous monitored fluorescence. S/B values were calculated from 30m time points (see methods). Data are representative of triplicate measurements.
3.3 Ub-AML reveals DUB activity not detectable with Z-RLRGG-AML or Ub-AMC
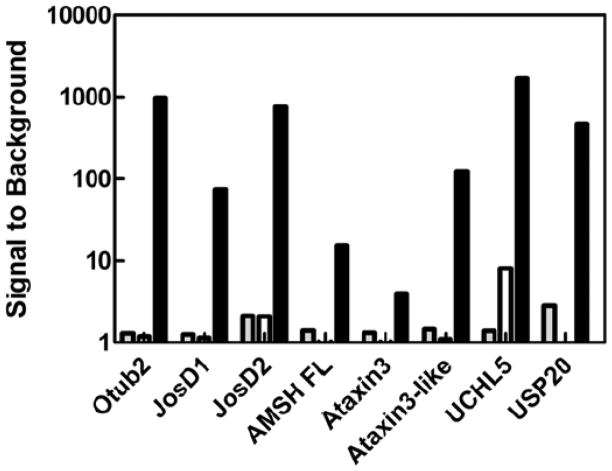
Although numerous reports exist of small molecule screening of isopeptidases with HTS amenable reagents [13, 17, 28, 29], many DUBs remain relatively unexplored as drug discovery targets. Even though a DUB may be described as active towards polyUb chains and/or in the context of a cytosolic environment, the difficulty in translating this activity (e.g. through SDS-PAGE or LC-MS) to HTS is obvious. In some cases, even the use of luciferin based technology alone may not suffice, as Z-RLRGG-AML has been demonstrated to perform for only a subset of these DUBs[16]. Given the apparent increased sensitivity and lower limits of detection observed for Ub-AML over both Ub-AMC and Ub-Rho110, we sought to test Ub-AML and Z-RLRGG-AML against a panel of DUBs that typically yield low S/B for Ub fluorogenic derivatives. A titration curve for each DUB (nano- to micromolar) was carried out against Ub-AML and Z-RLRGG-AML in parallel. Each DUB was also tested with Ub-AMC and Ub-Rho110 as reference substrates. For most DUBs, achieving S/B levels greater than 3-fold with either Ub-AMC or Z-RLRGG-AML required micromolar concentrations of enzyme (data not shown). Strikingly, though, for Ub-AML all DUBs (with the exception of Ataxin3) tested at 10nM yielded S/B ratios exceeding 10 after 30m (Figure 4 and Table 1). To our knowledge, this is the first demonstration of a homogenous assay for many of these DUBs. USP14 has previously been shown to be active against Ub-AMC, but only in the presence of affinity-purified proteasomal machinery from human cells[28]. Considering the enhanced sensitivity generally observed in these studies with Ub-AML, it seemed possible that this reagent might detect USP14 activity, even in the absence of proteasomal components. Remarkably, a dose response was observed for this DUB, as well; although higher concentrations of USP14 (approximately 300nM) were required, S/B levels approached 10 after 2h.
Figure 4. The use of Ub-AML with a panel of “difficult-to-detect” DUBs.
S/B determinations (see methods) for indicated DUBs (10nM) incubated with 250nM Ub-AML (black bars) or 20μM Z-RLRGG-AML (white bars) for approximately 30m prior to RLU measurement. For reference, S/B values for these DUBs (10nM) with Ub-AMC (250nM, 30m) are illustrated with gray bars.
Table 1.
| Otub2 | JosD1 | JosD2 | AMSH | Ataxin3 | Ataxin3-like | UCHL5 | USP20 | USP14 | |
|---|---|---|---|---|---|---|---|---|---|
| Ub-AMC | 1.2 | 1.1 | 2.1 | 1.0 | 0.9 | 1.1 | 8.1 | N.D.4 | 1.0 |
| Ub-Rho110 | 1.7 | 1.5 | 8.2 | 1.2 | 1.3 | 1.6 | 103 | N.D. | N.D. |
| DUB-Glo™ | 1.3 | 1.3 | 2.1 | 1.4 | 1.3 | 1.5 | 1.4 | 2.9 | 2.0 |
| Ub-AML | 980 | 74 | 760 | 16 | 4.0 | 120 | 1700 | 480 | 7.13 |
Signal-to-Background = (maximum signal − background signal)/background. background signal measured in the absence of DUB
DUB concentration equal to 10nM.
[USP14] = 330nM.
N.D. = not determined
3.5 Demonstration of Selectivity with hSUMO2- and NEDD8-AML
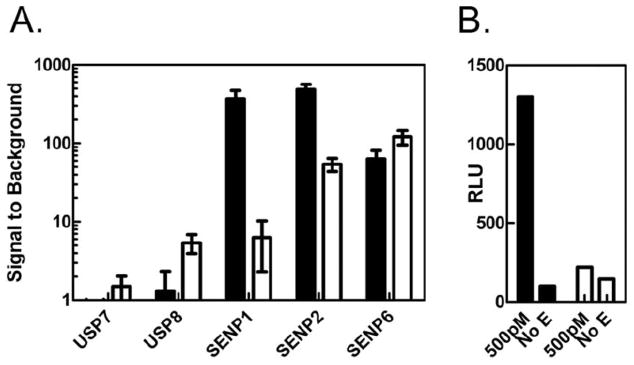
In order to provide additional Ubl-AML reagents for screening, hSUMO2-AML and NEDD8-AML were generated and compared to Z-RLRGG-AML. The use of hSUMO2-AML was associated with robust S/B after 30 min for SENP1, SENP2, and SENP6 at 200pM enzyme (Figure 5, Panel A). For SENP1 these S/B values were 10 to 50-fold higher than Z-RLRGG-AML. Interestingly, this difference was not as large for the other desumoylases tested, perhaps implying that SENP1 activity might be more stringent in its requirement for extended substrate interactions than either SENP2 or SENP6. As control reactions for specificity, USP7 and USP8 were also tested for their ability to cleave hSUMO2-AML. At 200pM neither of these DUBs showed significant signals for the hSUMO2-AML, although activity was detected with Z-RLRGG-AML. Both DUBs were tested at much higher concentrations (up to 50nM) to see if any cleavage was possible. No activity was detected for USP7; however, for USP8 appreciable signals were observed at these higher concentrations (data not shown). The significance of this was not investigated. To place this observation in context, though, comparable signals when using Ub-AML required 50-fold lower concentrations of USP8. The substrate NEDD8-AML also yielded higher signals than Z-RLRGG-AML when used to measure the activity of DEN1 (Figure 5, Panel B), demonstrating proof-of-concept for this substrate, as well. Taken together, the data for hSUMO2- and NEDD8-AML demonstrate that these luciferin conjugated reagents are sensitive reagents for desumoylases and deneddylases, respectively, and demonstrates selectivity for Ub/Ubl-AML substrates.
Figure 5. The use of hSUMO2- and NEDD8-AML for measuring isopeptidase activity.
Panel A: Indicated isopeptidases (50nM to 70pM) were incubated with hSUMO2-AML (250nM, black bars) or Z-RLRGG-AML (20μM, white bars) for 30m prior to RLU measurement and S/B calculation according to methods. At 200pM isopeptidase (shown) hSUMO2-AML displays clear selectivity for desumoylases over DUBs. Errors bars indicate S.D. from triplicate measurements. Panel B: RLUs were measured from NEDD8-AML (250nM, black bars) or Z-RLRGG-AML (20μM, white bars) after 30m incubation in either the presence or absence of 500pM Den1.
It is conceivable that the selectivity observed for Ub/Ubl-AMLs would be advantageous during Ub/Ubl protease drug discovery, presumably in a manner analogous to Ub/Ubl-AMC or –Rho110. Given the extended interactions away from the active site determined to exist between Ub/Ubls and their cognate proteases [20–22, 30], and the observation that DUB conformation and/or catalytic competency is altered upon complex formation with Ub [26], it stands to reason that the use of a full-length Ub/Ubl substrate for HTS would provide the best chance for identifying “hit” compounds during primary screening that might also demonstrate selectivity for different isopeptidases. Although, Z-RLRGG-AML has been demonstrated to have utility by displaying comparable reactivity across a broad range of DUBs, desumoylases, and deneddylases[16], in certain circumstances the ability to distinguish between the proteolytic activity of Ub/Ubl proteases might be advantageous. It should be noted that one report does exist in which an HTS campaign employing Z-RLRGG-AMC was able to identify a viral protease inhibitor with apparent selectivity at the P4 position[29]. Nevertheless, if enzyme specificity is not observed with Z-RLRGG- based reagents, such as DUB-Glo™, it is possible that the use of these reagents might be susceptible to the identification of inhibitors that lack selectivity, as well. Therefore, we suggest that Ub/Ubl-AML provides a valuable alternative to DUB-Glo™ for determining Ub/Ubl protease activity through bioluminescence.
3.6 Ub-AML is a robust reagent for HTS
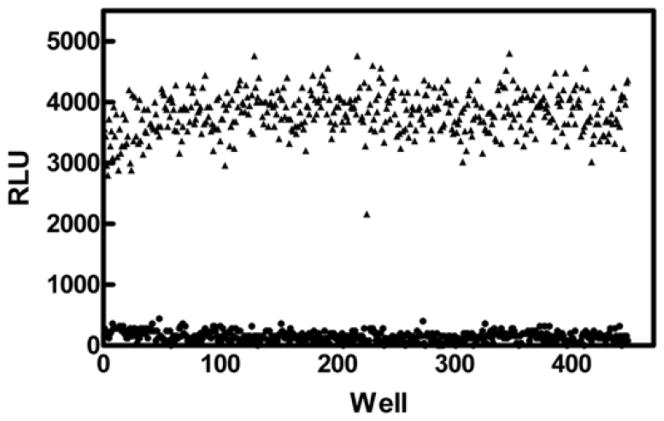
We used Ub-AML to investigate a number of experimental parameters that typically impact the decision to employ a given reagent for HTS, including flexibility, utility, cost, and robustness. The use of these Ub/Ubl-AML substrates has been miniaturized to 30μl in low volume, 384-well plates. Through the use of a representative USP, both enzyme and Ub-AML concentrations were titrated to achieve assay parameters in a manner similar to that shown for UCHL3 (Figure 2). Optimal assay parameters were determined that 1) maximized assay window time at “steady-state” of light generation, 2) established an acceptable S/B window, and 3) minimized reagent consumption. Under these conditions, a representative sampling of assay plates were processed in the presence (maximum signal, 3% DMSO) or absence (minimum signal) of DUB, and the Z-factor [31] was calculated for the assay (Figure 6). During this run, fourteen independent 384-well plates were assayed after 1h of incubation in the presence of the DUB. Overall S/B was determined to be 24-fold with a Z′ = 0.67, demonstrating the robust nature of Ub-AML for HTS drug discovery.
Figure 6. Screening parameters for Ub-AML.
RLUs for signal maximum (3% DMSO vehicle, ▲) and signal minimum (no enzyme, ●) were determined from (14) independent 384-well plates. Overall S/B was determined to be 24-fold, with a Z-factor of 0.67, demonstrating the robust nature of this assay format for screening isopeptidases.
Conclusions
Ub/Ubl-AML substrates yield enhanced sensitivity through luciferase/luciferin biology, while mimicking the full length Ub/Ubl biological substrate. Compared to Ub-AMC and Ub-Rho110 (e.g. UCHL3, USP2core), Ub-AML (250–500nM) displayed 100 to 1000-fold lower limits of detection for many DUBs, with S/B levels above 10 even with femtomolar enzyme concentrations. The use of Ub-AML for a panel of DUBs yielded S/B of 10 to 500 with nanomolar concentrations of enzyme, while these DUBs did not yield appreciable activity with Ub-AMC, Ub-Rho110, or DUB-Glo™. Notably, USP14 activity (S/B = 5 to 10 at 300nM) can be detected, even in the absence of additional proteosomal components. Thus, this reagent provides the potential for a homogenous in vitro assay for some DUBs for the first time. The generation of hSUMO2- and NEDD8-AML allowed the detection of Ub/Ubl protease activity that is not only sensitive, but also selective for its respective Ub/Ubl protease. Ub/Ubl-AML reagents are amenable to miniaturization and automation, with a Z factor of >0.6 being determined for Ub-AML. Ub/Ubl-AML substrates display enhanced sensitivity and flexibility compared to Ub-AMC and –Rho110, while providing a powerful alternative to DUB-Glo™ by providing a full length substrate displaying up to 1000-fold lower limits of detection for Ub/Ubl protease activity.
Highlights.
We have generated a novel class of Ub/Ubl-aminoluciferin substrates for DUBs
These reagents display greater sensitivity and selectivity than existing DUB substrates
Ub/Ubl-aminoluciferins are highly amenable to high throughput screening (HTS)
Use of these substrates allows for HTS of DUBs that have low activity for other such reagents
Acknowledgments
This work was supported, in part, by NIH grant R43GM090511.
The authors wish to thank Dr. Christian M. Loch for critical reading of this manuscript and invaluable discussions during its preparation. The authors would also like to acknowledge the contributions of Drs. Mabel Cejas and Yilin Yan during the early stages of this work.
The abbreviations used are
- Ub
ubiquitin
- DUB
deubiquitylase
- HTS
high throughput screening
- AML
aminoluciferin
- USP
ubiquitin specific protease
- UCH
ubiquitin carboxyterminal hydrolase
- SUMO
small ubiquitin-like modifier
- NEDD8
nerounal precursor cell expressed developmently down-regulated 8
- AMC
aminomethylcoumarin
- Rho110
rhodamine 110
Footnotes
Disclosure Statement
Mr. Orcutt and Dr. Strickler are employees of LifeSensors, Inc. which markets and sells reagents for ubiquitin research, including the Ub/Ubl-aminoluciferin product line. Mr. Orcutt and Drs. Wu and Strickler have applied for a patent on Ub/Ubl-aminoluciferin technology [32]. Drs. Eddins, Leach, and Wu are employed at Progenra, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershko A, Ciechanover A. The ubiquitin-proteasome pathway. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piper RC, Lehner PJ. Endosomal Transportation via Ubiquitination. Trends Cell Biol. 2011;21:647–655. doi: 10.1016/j.tcb.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finely D, Peng J. Quantitative Proteomics Reveals the Function of Unconventional Chains in Proteasomal Degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwai K, Tokunaga F. Linear polyubiqutination: a new regulator of NF-kappaB activation. EMBO Reports. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nijman SMB, Luna-Vargas MPA, Velds A, Brummelkamp TR, Dirac AMG, Sixma T, Bernards KR. A Genomic and Functional Inventory of Deubquitinating Enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Reyes Turcu FE, Ventii KH, Wilkinson KD. Regulation and Cellular Roles of Ubiquitin-specific Deubiquitinating Enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 9.Singhal S, Taylor MC, Baker RT. Deubiquitylating enzymes and disease. BMC Biochem. 2008;9:S3. doi: 10.1186/1471-2091-9-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shanmugham A, Ovaa H. DUBs and disease: Activity assays for inhibitor development. Curr Opin Drug Discov and Devel. 2008;11:688–696. [PubMed] [Google Scholar]
- 11.Nicholson B, Marblestone J, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3:191–199. doi: 10.2217/14796694.3.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, Shanks J, Sterner DE, Bernal A, Mattern MR, Wilkinson KD, Butt TR. Characterization of ubiquitin and ubiquitin-like protein isopeptidase activities. J Prot Sci. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian X, Isamiddinova NS, Peroutka RJ, Goldenberg SJ, Mattern MR, Nicholson B, Leach CA. Characterization of selective ubiquitin and ubiquitin-like protease inhibitors using a fluorescence-based multiplex assay format. ASSAY and Drug Dev Technol. 2011;9:165–173. doi: 10.1089/adt.2010.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang L, Melandri CF, Stein MRL. Kinetic and Mechnaistic Studies on the Hydrolysis of Ubiquitin C-terminal 7-Amido-4-Methylcoumarin by Deubiquitinating Enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 15.Hassiepen U, Eidhoff U, Meder G, Jean-Francois Bulber, Hein A, Bodendorf U, Lorthiois E, Martoglio B. A sensitive fluorescence intensity assay for deubiquitinating proteases using ubiquitin-rhodamine110-glycine as substrate. Anal Biochem. 2007;371:201–207. doi: 10.1016/j.ab.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 16.O’Brien MA, Moravec RA, Daily WJ, Scurria MA, Riss TL, Klaubert DH, Cosby N, Bulleit RF. Sensitive, Homogeneous, Bioluminescent Assays for Monitoring Proteases of the Ubiquitin-Proteasome Pathway. Poster 1st Annual Ubiquitin Drug Discovery & Diagnostics; 2009. [Google Scholar]
- 17.Baez-Santos YM, Mesecar AD, O’Brien MA. Detecting the Deubiquitin Activity of SARS-CoV PLpro: Turning on the Light with the DUB-Glo™ Protease Assay, Promega Corporation Web Site. 2009 [Google Scholar]
- 18.O’Brien MA, Daily W, Hesselberth JPE, Moravec RA, Scurria MA, Klaubert DH, Bulleit RF, Wood KV. Homogenous, Bioluminescent Protease Assays: Caspase-3 as a Model. J Biomol Screen. 2005;10:137–148. doi: 10.1177/1087057104271865. [DOI] [PubMed] [Google Scholar]
- 19.Monsees T, Geiger R, Miska W. A novel bioluminogenic assay for alpha-chymotrypsin. J Biolumin Chemilumin. 1994;10:213–218. doi: 10.1002/bio.1170100404. [DOI] [PubMed] [Google Scholar]
- 20.Renatus M, Parrado SG, D’Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, Kroemer M. Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reverter D, Lima CD. A Basis for SUMO Protease Specificity Provided by Analysis of Human Senp2 and a Senp2-SUMO Complex. Structure. 2004;12:1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 22.Reverter D, Wu K, Erdene TG, Pan ZQ, Wikinson KD, Lima CD. Structure of a Complex between Nedd8 and the Ulp/Senp Protease Family Member Den1. J of Mol Biol. 2005;345:141–151. doi: 10.1016/j.jmb.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson KD, Gan-Erdene T, Kolli N. Derivitization of the C-Terminus of Ubiquitin and Ubiquitin-like Proteins Using Intein Chemistry: methods and uses. Methods and Enzymol. 2005;399:37–51. doi: 10.1016/S0076-6879(05)99003-4. [DOI] [PubMed] [Google Scholar]
- 24.Shinde R, Perkins J, Contag CH. Luciferin Derivatives for Enhanced in Vitro and in Vivo Bioluminescence Assays. Biochemistry. 2006;45:11103–11112. doi: 10.1021/bi060475o. [DOI] [PubMed] [Google Scholar]
- 25.Fan F, Wood KV. Bioluminescent Assays for High-Throughput Screening. ASSAY and Drug Dev Technol. 2007;5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 26.Hu M, Li P, Li M, Li W, Yao T, Wu J-W, Gu W, Cohen RE, Shi Y. Crystal Structure of a UBP-Family Deubiquitinating Enzyme in Isolation and in Complex with Ubiquitin Aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 27.Reyes Turcu FE, Shanks J, Komander D, Wikinson KD. Recognition of Polyubiquitin Isoforms by the Multiple Binding Modules of Isopeptidase T. Journal of Biological Chemistry. 2008;283:19581–19592. doi: 10.1074/jbc.M800947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee B-H, Lee MJ, Park S, Oh D-C, Elsasser S, Ping-Chung C, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of Proteasome Activity by a Small-Molecule Inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson M, Baker ESC, Ghosh AK, Mesecar AD. A noncovalent class of papain-like protease/deubiquinase inhibitors blocks SARS virus replication. Proc Natl Acad of Sci U S A. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mossessova E, Lima CD. Ulp1-SUMO Crystal Structure and Genetic Analysis Reveal Conserved Interactions and a Regulatory Element Essential for Cell Growth in Yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 31.Zhang JH, Chung TDY, Oldenburg KR. A Simple Statistical Parameter for Use in Evalution and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 32.Orcutt SJ, Yan Y, Wu J, Strickler JE, Butt TR. Bioluminescent detection of protease activity using ubiquitin-luciferase substrate, and screening and diagnostic uses. US Patent Application Pub. No. US 2012/0107857.