Abstract
The [Fe]-hydrogenase is an ideal system for studying the electronic properties of the low spin iron site that is common to the catalytic centres of all hydrogenases. Because they have no auxiliary iron-sulfur clusters and possess a cofactor containing a single iron centre, the [Fe]-hydrogenases are well suited for spectroscopic analysis of those factors required for the activation of molecular hydrogen. Specifically, in this study we shed light on the electronic and molecular structure of the iron centre by XAS analysis of [Fe]-hydrogenase from Methanocaldococcus jannashii and five model complexes (Fe(ethanedithiolate)-(CO)2(PMe3)2, [K(18-crown-6)]2[Fe(CN)2(CO)3], K[Fe(CN)(CO)4], K3[Fe(iii)(CN)6], K4[Fe(ii)(CN)6]). The different electron donors have a strong influence on the iron absorption K-edge energy position, which is frequently used to determine the metal oxidation state. Our results demonstrate that the K-edges of Fe(ii) complexes, achieved with low-spin ferrous thiolates, are consistent with a ferrous centre in the [Fe]-hydrogenase from Methanocaldococcus jannashii. The metal geometry also strongly influences the XANES and thus the electronic structure. Using in silico simulation, we were able to reproduce the main features of the XANES spectra and describe the effects of individual donor contributions on the spectra. Thereby, we reveal the essential role of an unusual carbon donor coming from an acyl group of the cofactor in the determination of the electronic structure required for the activity of the enzyme.
Introduction
Molecular hydrogen was one of the earliest energy sources available on earth. As a consequence, many microorganisms continue to use it as electron donor.1 To do so, these organisms rely on the hydrogenase enzymes, which catalyze the production and oxidation of molecular hydrogen. Their active sites exploit first-row transition metals exhibiting turnover rates comparable to that of platinum.2,3 Nature’s use of the most abundant metal on earth, iron, is obviously attractive,4 but synthetic iron catalysts typically are not highly active toward hydrogen.5 It is therefore of obvious value to fully characterize the catalytic centres that mediate H2 heterolysis under mild conditions. This knowledge may lead to new biomimetic methods for the generation and utilization of H2.6
Although the three known classes of hydrogenases are not phylogenetically related, their active sites share striking similarities.2,7 [NiFe]- and [FeFe]-hydrogenases feature binuclear 3d-transition metal centres, complemented by iron-sulfur clusters.8 The third class, the [Fe]-hydrogenases, lacks FeS clusters.9 This enzyme has been found exclusively in some methanogenic archea and it exploits only one metal-ion incorporated in FeGP-cofactor.2,10 In all three hydrogenases, this iron centre is ligated by at least one thiolate sulfur and three potential π-accepting ligands, consisting of CO and CN− molecules in the [NiFe]- and [FeFe]-hydrogenases and of two COs and an acyl-carbon in case of [Fe]-hydrogenases.2,11,12 These commonalities point towards convergent evolution, indicating specific ligand structures that facilitate H2 activation by iron.
The first step of enzymatic H2 activation is heterolysis, giving a proton and a hydride, which is bound to the metal. This transformation is well known to proceed with the help of low-spin iron centres.13,14 Although it is understood that ligands on Fe must stabilize the low-spin ferrous state, specific electronic roles of the various ligands in facilitating hydrogen activation are not well understood.
Even though extensive studies over the past decade have advanced our understanding of the electronic structure and the catalytic mechanism of [NiFe]- and [FeFe]-hydrogenases,8 our understanding of the mononuclear [Fe]-hydrogenase is still rather primitive. This situation is due to the fact that only recently its active site structure has been described (Fig. 1).2,15 Due to the presence of a single metal ion and the absence of other metal chromophores, this enzyme is an ideal target for studying the relationship between electronic structure of the low-spin iron-site and hydrogen activation. Furthermore, the FeGP-cofactor,16–17 can be removed from the enzyme and examined independently (see below). [Fe]-hydrogenase is also named H2-forming methylenetetrahydromethanopterin (methylene-H4MPT) dehydrogenase (Hmd), because its function is the reversible reduction of methenyl-H4MPT+ (acceptor) to methylene-H4MPT. This conversion, a methenyl-H4MPT+ hydrogenation, occurs instead of H2 oxidation, which is typical for other hydrogenases.9,15,18 The reaction is reversible, Hmd also catalyzes the release of H2 from methylene-H4MPT. This class of hydrogenases only transfers hydride and it is independent of electron transfer by FeS clusters. According to a recently proposed catalytic mechanism,19 the molecular hydrogen is heterolytically cleaved by the functions of Fe(ii) and the carbocation of the acceptor and the hydride produced is transferred to the acceptor.
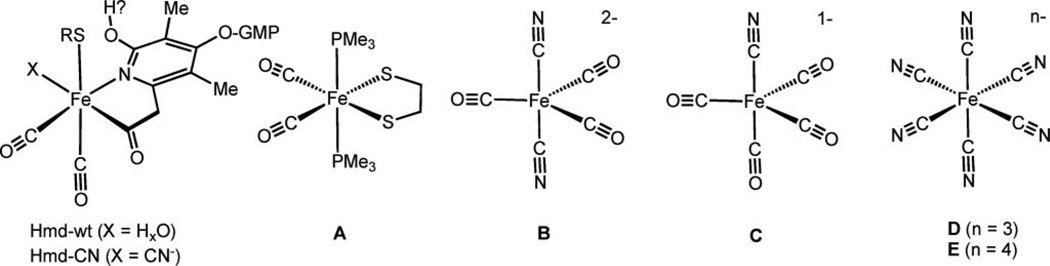
Fig. 1.
On the left: structure of the [Fe]-hydrogenase octahedral metal binding site as modelled by EXAFS analysis.1 The iron is coordinated to: the Cys176-sulfur, two CO and the pyridinol-sp2-hybridized nitrogen and the acyl-carbon of the cofactor. An “unknown” donor, here represented as X, is associated to the iron trans to the acyl carbon. This is considered the vacant position ready for H2 binding. The model complexes are represented in A–E: Fe(ii)(edt)(CO)2(PMe3)2 (A); K2[Fe(0)(CN)2(CO)3] (B); K[Fe(0)(CN)(CO)4] (C); K3[Fe(iii)(CN)6] (D); K4[Fe(ii)(CN)6] (E).
The emerging model for the iron coordination in Hmd from Methanocaldococcus jannashii (jHmd) (Fig. 1) comprises five to six ligands: two carbon monoxides, the sulfur of Cys176, the nitrogen and the acyl-carbon of the pyridinol derivative, and an unknown ligand modelled as a “solvent-oxygen” in agreement with EXAFS and a recent X-ray diffraction model inspired by the crystal structure of theC176Amutated [Fe]-hydrogenase.15,20 This oxygen atom occupies the position where the hydrogen molecule (substrate) is proposed to bind. The iron coordination of the active site appears to be very similar to that of isolated (protein-free) cofactor. The sulfur now appears to be supplied by 2-mercaptoethanol used in the extraction of FeGP-cofactor.15,21 Mössbauer spectroscopy studies on FeGP-cofactor revealed low spin iron with an oxidation state of 2+ or 0, indicative of the highly covalent nature of the Fe-ligand bonding.22 Recently reported infrared analyses of model compounds are consistent with a low-spin Fe(ii) oxidation state of the iron centre.23,24
XANES (X-ray Absorption Near Edge Spectroscopy) is a powerful technique to study the electronic structure of metal centres. The shape and position of the X-ray absorption edge serve as fingerprints of a specific binding motif and, if good reference systems are available, allow the determination of oxidation states, number of ligands and symmetry of the coordination sphere.25 In fact, both edge position and electronic structure of a system depend on the nature of the donor groups.26
Herein, we compare the XANES of Hmd with a variety of model complexes that exhibit a range of oxidation states and contain ligands similar to the ones found in the enzyme. Specifically, we have analyzed the K-edge XANES of iron in jHmd wild type (jHmd-wt), in a cyanide inhibited form of jHmd (jHmd-CN) and in Fe(edt)(CO)2(PMe3)2, [K(18-crown-6)]2[Fe(CN)2(CO)3], K[Fe(CN)(CO)4]; K3[Fe(iii)(CN)6], and K4[Fe(ii)(CN)6] (Fig. 1). The program FEFF8.2 was used for ab initio self-consistent field, full multiple scattering calculations, allowing us to reproduce the major features of the edge profiles.27
Based on the present analysis, recent EXAFS analysis20 and high resolution crystal structures2,15 we identify the dominating features that dictate the electronic structure of [Fe]-hydrogenases manifested in XANES spectra of Hmd.
Results and discussion
Since the iron coordination in Hmd is unique, its XANES can not be compared with spectra of known metalloproteins. Thus, we selected model compounds as anchor points for assessing the influence of iron oxidation state and its coordination sphere on the spectroscopic features. These model compounds are octahedral or five-coordinate iron centres with ligands similar to the donor groups in the enzyme. Here, we present data from Fe(ii)(edt)(CO)2(PMe3)2 (A, edt = 1,2-ethanedithiolate), [K(18-crown-6)]2[Fe(0)(CN)2(CO)3] (B), K[Fe(0)(CN)(CO)4] (C), K3[Fe(iii)(CN)6] (D) and K4[Fe(ii)(CN)6] (E). The two iron hexa-cyanides, D and E, were selected to enlighten the influence of metal oxidation states on the absorption edge profiles. Both formal Fe(0)-compounds are trigonal bipyramidal and highlight the differences between CN and CO groups on the electronic structure of the metal ion, whereas compound A is square pyramidal and shows the influence of a P/S donor.
These five compounds, jHmd-wt, and jHmd-CN were analysed by XANES. The analysis of the EXAFS based on the crystallographic coordinates and symmetry proved sample integrity of the model complexes and provided average metal donor distances comparable to the enzyme data (electronic supplementary information, ESI, Tab. S1‡).
Model complexes
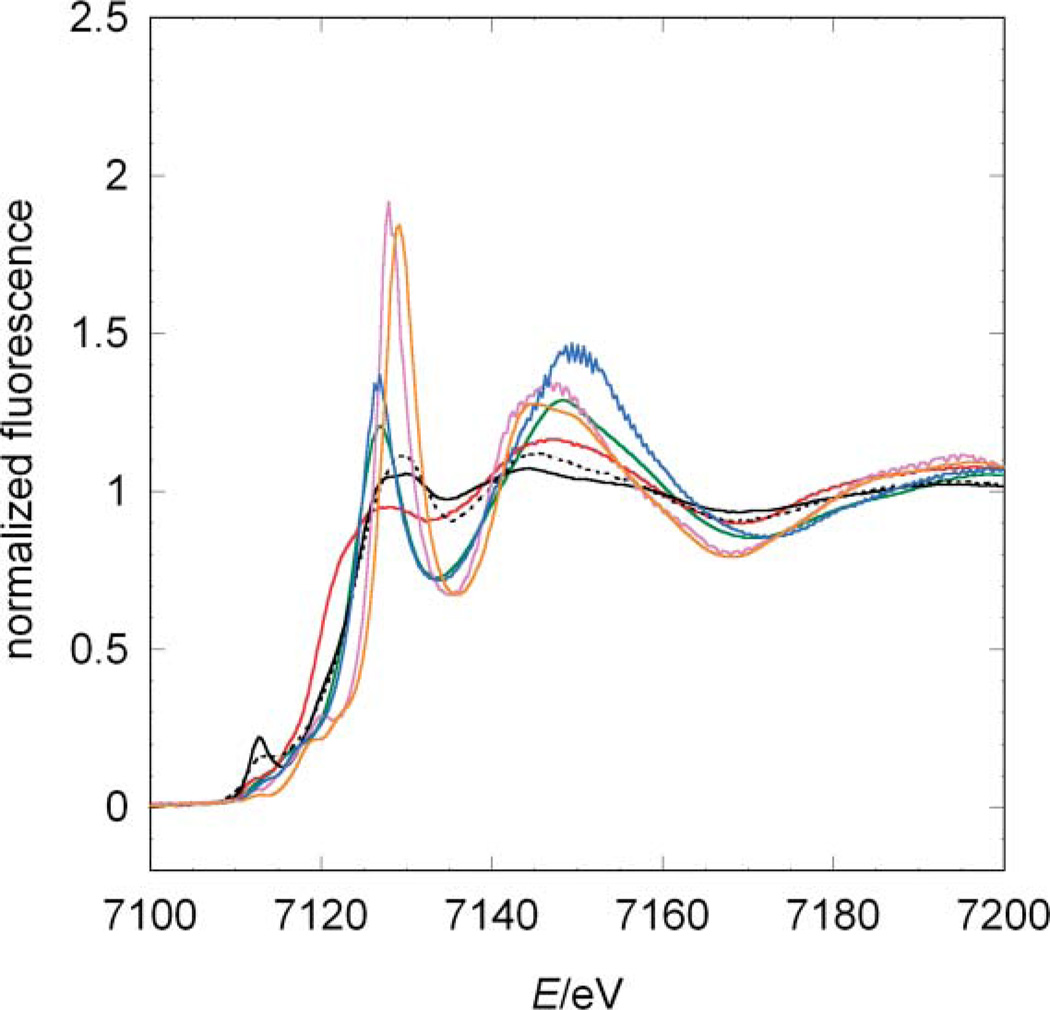
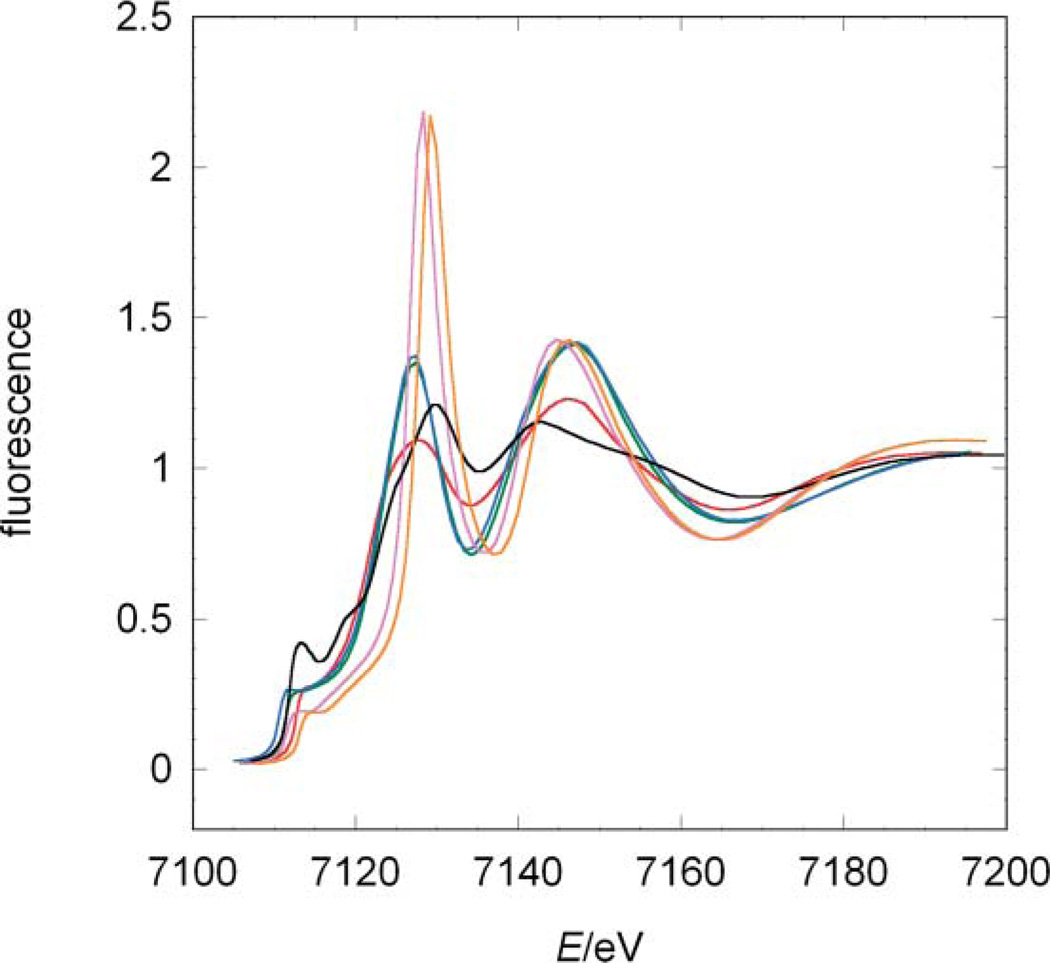
The XANES spectra for compounds A–E reveal strong differences in rising edge position and shape (Fig. 2). Compounds D and E are highly symmetrical and only differ in the formal oxidation state of iron, which results in highly similar spectra. The small shift in their absorption edges reflects the slightly different effective charges at the iron ion. In line with the high symmetry of these iron centres, the white-lines [Footnote: In XAS the white-line denotes the main peak formed by the rising edge and the following minimum] of complexes D and E are very sharp.28 In accordance with the formal charge, the ferrocyanide compound (E) has an edge position at lower energies than the ferricyanide D, 7127 eV and 7127.5 eV, respectively (orange and purple lines in Fig. 2).
Fig. 2.
Comparison among the experimental XANES of the model systems and jHmd-wt. Black line: jHmd-wt; black broken line: jHmd-CN; red line: Fe(ii)(edt)(CO)2(PMe3)2 (A); blue line: [K(18-crown-6)]2[Fe(0)(CN)2(CO)3] (B); green line: K[Fe(0)(CN)(CO)4] (C); orange line: K3[Fe(iii)(CN)6] (D); purple line: K4[Fe(ii)(CN)6] (E).
In compounds B and C, iron is in the formal oxidation state 0. These pentacoordinate complexes are structurally similar, differing only in the relative ratio of CO/CN− donors. This similarity is reflected by their XANES (green and blue lines in Fig. 2). Compared to the data for D and E, the white-lines of B and C are less sharp, indicative of their lower symmetry. As expected, both edge positions are at lower energies with respect to D and E. The differences observed in the peak intensities might be ascribed to the exchange of one CO by CN−: CO is a good σ donor and a strong π acceptor, while CN− is a good σ donor and poor π acceptor, which influences in this case only the matrix element for transitions into the unoccupied orbitals, but not their energy levels.13
In compound A, the Fe(ii) ion is bound to two CO ligands, two sulfur and two phosphine donors. The S and P donor ligands mimic to some extent the sulfur ligand bound to Fe in Hmd. The low-symmetry of the iron coordination results in a broadened white-line. Interestingly, the edge is positioned at the lowest energy, despite the formal oxidation state of the iron (red line in Fig. 2). This shift is ascribed to the influence of the phosphine and sulfur atoms. Thus, the nature of the ligands has for these complexes a stronger influence on the absorption edge position than the formal oxidation state.
The model compounds are chosen to illustrate the two competing effects determining the edge position and the metal–ligand distances: metal oxidation state and ligand type. An additional effect becomes apparent: the shape of the XANES depends on the homogeneity of the electron donors. The more cyanides are present in the sample the sharper is the white-line. A confirmation of the trend comes from compound A. Its coordination is heterogeneous and thus the white-line is small.
jHmd-wt and jHmd-CN
Intriguingly, the XANES-spectrum of jHmd-wt (full black line in Fig. 2) revealed characteristics very similar to compound A, with smooth maxima and minima. This similarity makes the latter the spectroscopic model closest to jHmd. Nevertheless, the spectra of jHmd-wt and A do not exhibit similar rising edges. The spectrum of jHmd-CN (broken black line in Fig. 2) shares with the wild type the position of the pre-edge peak as well as the maxima and minima above the edge, but not the relative intensities. jHmd-wt and jHmd-CN also share most of the rising edge. Thus, the electronic structures for wild type and the cyanide-inhibited form are similar, consistent with the similarity of the IR spectra in the νCO region.12 The higher white-line intensity in jHmd-CN is indicative of a higher symmetry. This is in line with the pre-edge peak area that is much smaller for jHmd-CN than for jHmd-wt (Tab. 1). This feature is frequently used as a measure of the symmetry and homogeneity of the iron coordination.29 Deviation from an ideal octahedral coordination increases the 1 s to 3d/4p transition probability and thus the pre-edge peak intensity.
Table 1.
Edge positions, pre-edge peak positions and pre-edge peak areas for jHmd-wt, jHmd-CN and the model complexes. For simplicity, only the distances for the donor groups carbon monoxide, cyanide and sulfur from the iron ion are given here. In parentheses the error margins on the last digit are given
| Sample | edge position eV |
pre-edge peak position eV |
1s→3d pre-edge peak area 10−2 eV |
|---|---|---|---|
| jHmd-wt | 7119.2(2) | 7112.7(2) | 60(1) |
| jHmd-CN | 7117.0(2) | 7112.7(2) | 28(2) |
| A | 7116.0(2) | 7112.0(2) | 9(2) |
| B | 7124.6(2) | 7112.9(2) | 8(1) |
| C | 7123.8(2) | 7112.6(2) | 18(2) |
| D | 7127.4(2) | 7112.3(2) | 2(1) |
| E | 7126.6(2) | 7111.9(2) | 1(1) |
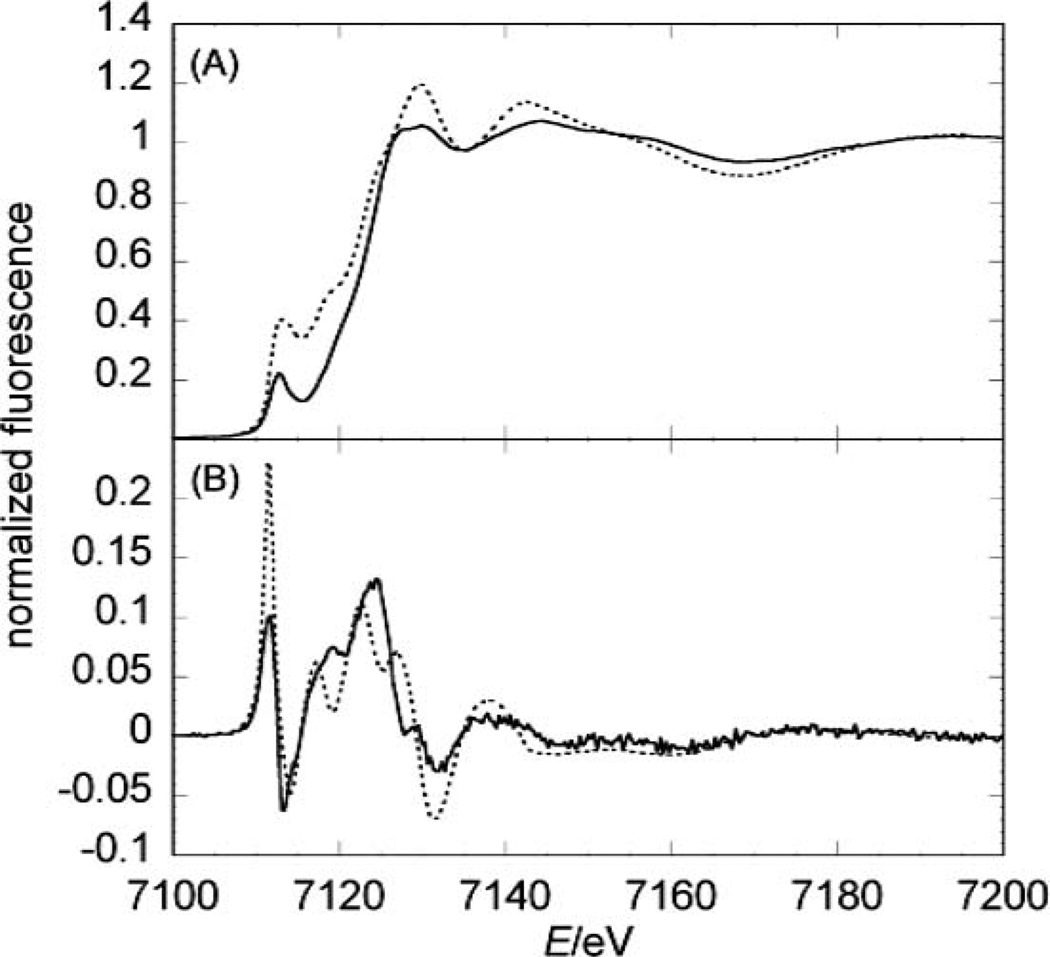
The first derivative of the XANES spectra of the wild type (full line in Fig. 3B) helps identifying features that are smoothed by the energy resolution of the monochromator (~4000 at 7.2 keV; note that its accuracy is ~0.1–0.2 eV). The jHmd-wt derivative is characterized by the splitting of the second peak into three peaks representing unoccupied orbitals (white-line range: from 7115 to 7130 eV, Fig. 3B). This feature is absent in the inhibited form (Fig. S1‡). Thus, the presence of the characteristic can be considered as an indicator of the active form of jHmd.
Fig. 3.
Comparison between experimental and simulated spectra of jHmd-wt. (A) the XANES and in (B) its 1st derivative is shown. The full lines represent the experimental data, while the broken lines represent the simulations.
Comparison of the absorption edges
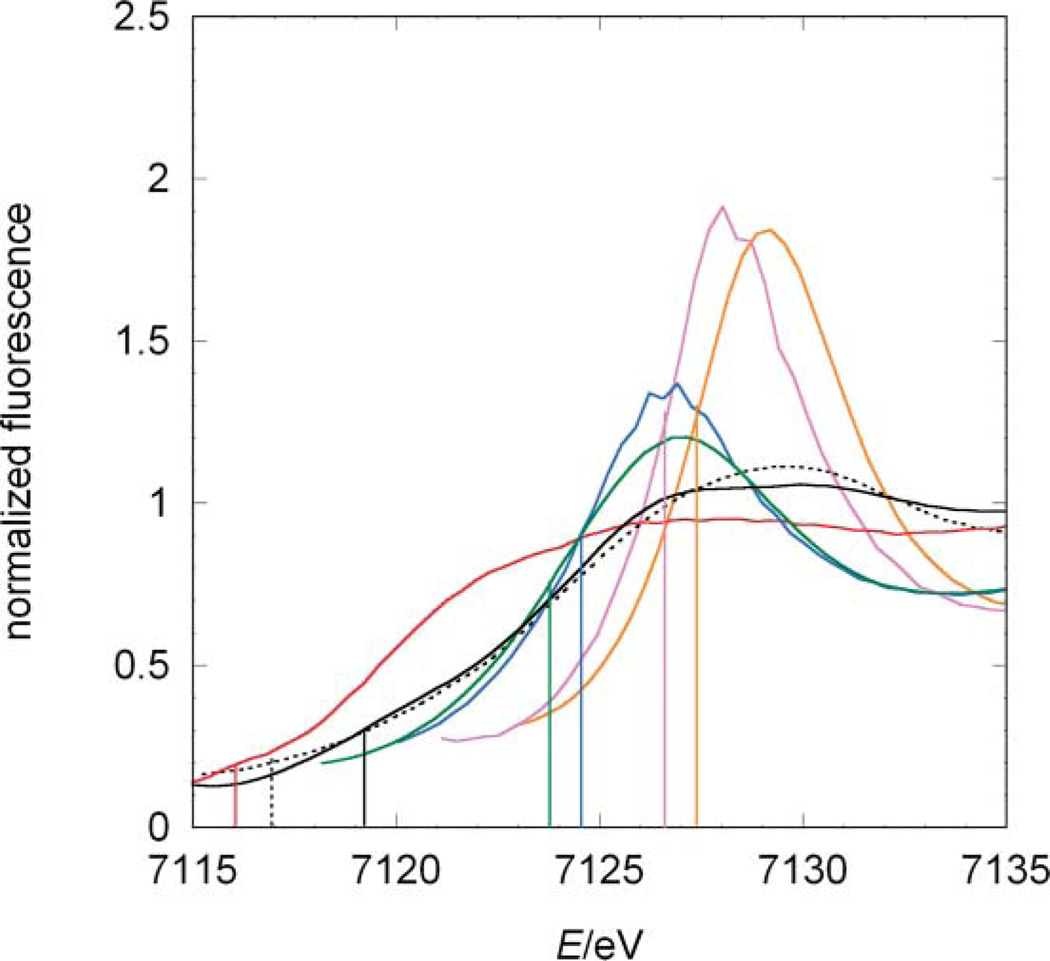
The absorption edges reflect the influence of both the formal oxidation state of the iron centre and its ligands (Fig. 4). The edge positions have the following order: 7118.8 (jHmd-wt), 7119.0 (jHmd-CN), 7119.5 (A), 7124.0 (C), 7125.0 (B), 7127.0 (E), and 7127.5 eV (D). For the complexes B, C, E and D, with only diatomic ligands and oxidation states 0, 0, 2+ and 3+, respectively, the edges shift towards lower energies upon reduction. Interestingly, the edges of B and C differ by 1 eV from each other, although they are both Fe(0). This difference indicates the sensitivity of the effective charges at the metal ion to the donor types. E and D, which share the same coordination sphere but differ in oxidation state, are separated along the entire rising edges indicating that the corresponding molecular orbitals are only shifted in energy and the matrix elements for the transition from the 1 s orbital are unaffected (Fig. 2, 4). In contrast, compound A shows the profound influence of the ligand type. Here, the donors P and S strongly affect the edge position, which is shifted towards lower energies by 7.5 eV from the other ferrous compound E. In accordance, the edge positions of jHmd-wt and jHmd-CN are similar to that of A, suggesting a comparable effective charge at the iron ion.
Fig. 4.
Comparison between the edge positions of the model compounds, jHmd-wt and jHmd-CN. Black line: jHmd-wt; black broken line: jHmd-CN; red line: Fe(ii)(edt)(CO)2(PMe3)2 (A); blue line: [K(18-crown-6)]2[Fe(0)(CN)2(CO)3] (B); green line: K[Fe(0)(CN)(CO)4] (C); orange line: K3[Fe(iii)(CN)6] (D); purple line: K4[Fe(ii)(CN)6] (E). For each sample, the edge position—first maximum of the first derivative in the rising edge—is indicated.
Comparison of pre-edge peak intensities
The pre-edge peak intensity correlates with the symmetry of metal binding site: for octahedral sites and homogenous donor types it is lower than for tetrahedral sites and inhomogeneous donor types.29 In line with their octahedral iron-coordination and their homogenous ligand sphere, the hexacyanides D and E possess the smallest pre-edge peak intensity of about 1 and 2 × 10−2 eV, respectively. Complex A exhibits a pre-edge peak intensity of about 9 × 10−2 eV, which reflects the heterogeneity of its octahedral ligand set. Complexes B and C are square pyramidal and as a consequence, the pre-edge peak areas are higher: complex C, has an area of about 18 × 10−2 eV. An interesting exception to the rule is complex B with an area of only 9 × 10−2 eV. This indicates higher symmetry potentially caused by the compensation of π-accepting and π-donating ligands. For jHmd-wt and jHmd-CN the largest pre-edge peak intensities are observed. The heterogeneity and asymmetry of the coordination sphere and likely the steric constraints by the protein environment result in a highly distorted geometry at the iron site. Interestingly, jHmd-wt pre-edge peak intensity (60 × 10−2 eV) is twice of that of the inhibited form (28 × 10−2 eV) and considerably higher than that of the isolated cofactor (44 × 10−2 eV).21 These differences suggest a correlation between distortions of the active site geometry and catalytic activity of Hmd. Moreover also a partial occupation of the “oxygen position” can increase the intensity of the pre-edge peak. In fact, our EXAFS refinements without a sixth ligand are only slightly worse than the ones for six-fold coordination (Fig. S4‡). Partial occupancy may indicate that ligands at this site are labile. We can conclude that octahedral geometry is consistent with low spin ferrous state and with all experimental results from protein crystallography and spectroscopy. This does not exclude a transient pentacoordination.
Metal-donor distances
The metal-carbon distances of the model systems do not differ significantly for Fe(ii) and Fe(0) complexes (Tab. S1); the values are similar to those reported for other iron ions ligated by carbon monoxide.23,30 The Fe-CO distances observed in Hmd fall within this region as well (Tab. S1). The iron-sulfur distances obtained in the EXAFS refinement for jHmd-wt (2.33 Å) is closer to the ones of compound A and related models with nitrogen donors in place of phosphines (2.31 Å),31 than to those calculated by molecular modelling for [Fe]-hydrogenases32 (2.38Å) and the ones measured for the [FeFe]-hydrogenases (2.25Å).33 In general, Fe(ii)-S distances are poorly sensitive to oxidation state, but highly sensitive to spin state.34,35 For example, the Fe–S distance in Et3NH[Fe(0)(SPh)(CO)4] (2.332(5)Å)34 is almost identical to that in Fe(ii)H(SPh)(CO)2[P(OPh)3]2 (2.343(3)Å).35
XANES analysis
To gain further insights into the electronic structure of the iron ion in [Fe]-hydrogenases, in silico simulations of the XANES spectra of the model complexes and jHmd-wt were carried out with the program FEFF8.2 (see experimental). We analyzed both the XANES and their first derivatives in order to highlight homologies and differences among spectra and withdrawn qualitative information about the iron ion electronic structure.
Structure-based EXAFS refinement
The coordinates used in our simulations were extracted from EXAFS refinements based on crystal structure models reflecting the different geometries of the iron sites (see Fig. 1). The X-ray diffraction coordinates were read into Excurv98 as starting parameters for the refinement of the EXAFS data. Interestingly, the solvent oxygen of jHmd-wt is for this geometry slightly better modelled at 2.49 Å (Fig. S3‡), instead of 2.05 Å as presented in Hiromoto et al. 2009 for ideal octahedral coordination.15 Attempts to model the EXAFS with an oxygen contribution at other distances led to poorer fit indexes (Fig. S3‡) in line with the periodicity/interference of the EXAFS signal. Besides, we tried modelling this contribution by other elements (e.g. phosphorus, sulfur, fluorine, bromine, iodine) and obtaining comparable results for fluorine and bromine (Fig. S4‡). These results are indicative for the presence of an atom/molecule at the “solvent oxygen” position but do not allow an unambiguous identification. Thus, we favor oxygen for consistecy with the crystallographic model.
XANES simulations of the model systems
Simulations of the XANES for the model systems served as a proof of principles and defined the required cluster size in this study (first two to three shells). The principal features of the spectra have been reproduced: the curves trend matches the experimental data and the sequence of the rising edge positions is reflected by the simulations (Fig. 5). Moreover, the white-lines correlate with the symmetry and homogeneity of the coordination sphere.
Fig. 5.
Comparison among the simulated XANES of the model systems and jHmd-wt. Black line: jHmd-wt; red line Fe(ii)(edt)(CO)2(PMe3)2 (A); blue line: [K(18-crown-6)]2[Fe(0)(CN)2(CO)3] (B); green line: K[Fe(0)(CN)(CO)4] (C); orange line: K3[Fe(iii)(CN)6] (D); purple line: K4[Fe(ii)(CN)6] (E).
In our simulations, the pre-edge peak regions are not reproduced very well. This problem is connected to the inaccurate calculation of transition probabilities into pre-edge states. Thus, the over intense pre-edge area characteristics are considered as artefacts.
XANES simulations of jHmd-wt
The positions of maxima and minima and the general shape of the experimental data are reproduced indicating the agreement between the electronic states in jHmd-wt and the simulation (Fig. 3A and B). Only the relative intensities and thus the magnitude of the transition matrix element are not adequately modelled. In the first derivative of the simulated spectrum a distinctive three-fold split white-line peak is preserved, highlighting the existence of three narrow energy levels.
Impact of individual donors on the electronic structure
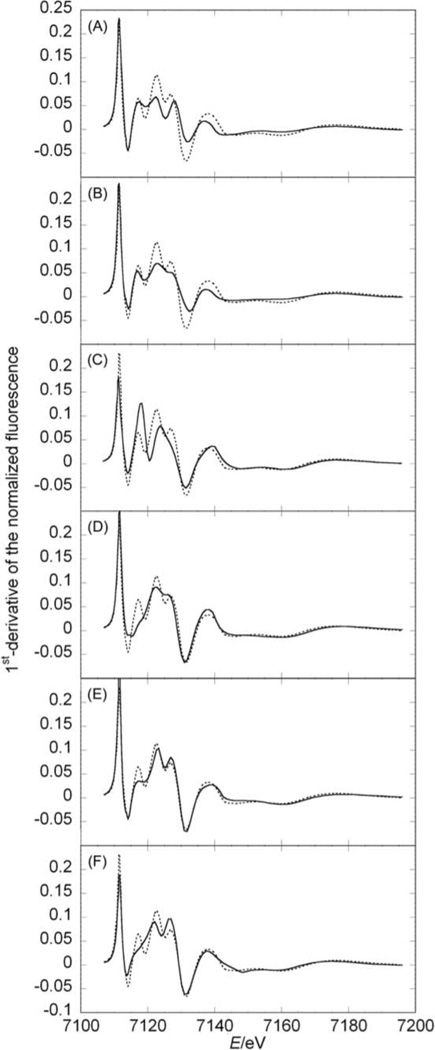
In order to evaluate the impact of individual donors on the electronic structure of Hmd-wt, we successively omitted one of them in the simulations. Now we focused on first derivatives to highlight the impact on the electronic structure induced by systematically removing one metal donor (Fig. 6). Omitting one of the two CO ligands (CO1 and CO2) has a strong influence on most of the XANES spectrum, in the energy interval from 7117 to 7167 eV, in line with their intense multiple scattering contributions (Fig. 6A, B). The differences between these simulations show the importance of the geometry. In the XANES simulations this geometry is taken into account by multiple scattering contributions (from the central iron ion to a donor, continuing via the central iron atom to the donor in trans position and back to the iron ion); for CO1 this multiple scattering pathway includes the sulfur (2.3 Å away from the iron ion) and for CO2 the nitrogen atom opposite to CO2 (2.0 Å away from the metal ion) resulting in different phase and amplitude of these contributions (Fig. 1). Interestingly, in both spectra the white-line peak splitting is conserved, indicating that these ligands can not strongly contribute to the identity of the three unoccupied orbitals that give rise to this rising edge.
Fig. 6.
Comparison between the first derivative of jHmd-wt XANES simulation in the full coordination (dotted lines) and simulations omitting one donor at the time (full lines): (A) CO1 omitted, (B) CO2 omitted, (C) acyl-C of the cofactor omitted, (D) pyridol-N of the cofactor omitted, (E) Cys176-S omitted, (F) oxygen omitted. The comparisons for the XANES simulations are shown in Fig. S4.‡
The donors from the pyridinol derivative significantly influence the region from 7115 to 7150 eV, corresponding to the white-line till the following minimum (Fig. 6C, D). The characteristics at 7155 and 7175 eV remain unchanged. Pyridinol-N and acyl-C omissions change the structure of the white-line peak, indicating their function in fine tuning the electronic structure. The sulfur contribution to the white-line feature is rather mild. The region from 7115 to 7120 eV is appreciably affected (first transition to an unoccupied orbital in the XANES) (Fig. 6E). It seems that here the COs and the other ligands give a contribution stronger than found for typical metal binding enzymes, thus diminishing the influence of sulfur. Removing the “solvent-oxygen” in the simulation affected the entire rising edge (Fig. 6F from 7114 to 7128 eV). This influence is small except for the first characteristic of the white-line peak at 7117 eV, which is lost.
The above results highlight the distinctive contribution from each of the six ligands. The differing contributions of the two CO ligands suggest that they play a key electronic role, as expected because of their strong tendency to back-bond.
The comparison of the jHmd-wt XANES with the model complexes identified the ferrous complex A as a mimic for the enzyme’s electronic structure. Moreover, jHmd-wt and complex A share the CO and sulfur ligand types. These similarities and the identical absorption edge positions concludes that the enzyme harbours a ferrous ion. The analysis of the pre-edge peak intensities shows that in contrast to complex A the octahedral symmetry is much more distorted in the enzyme, pointing to a specific tuning of the electronic structure (and thus most likely of the reactivity) by the protein environment, namely by the pockets harbouring the CO groups (Fig. S7‡). This hypothesis is supported by the much smaller pre-edge peak intensity observed for the isolated FeGP-cofactor.21 An appreciable difference in the symmetry of the iron site in jHmd-wt and in the isolated cofactor could be inferred also from the electric field gradient sensed by the 57-Fe Mössbauer nucleus and the corresponding quadrupole splitting of the Mössbauer spectra (0.65 mm s−1 in wt-Hmd and 0.43 mm s−1 for FeGP).22 The jHmd-CN experimental XANES showed that the CN− binding alters significantly the electronic structure of the iron ion. This change, as well as the saturation of the coordination sphere of Fe(ii), is relevant to its inhibitory effect.
In the XANES simulations the essential contributions of the acyl-carbon and of the pyridinol nitrogen to the electronic structure were visualized by changes in the three unoccupied orbitals of the rising edge. Indeed, the identity of the feature representing the white-line transitions to unoccupied orbitals vanished when one of these donors was omitted. The ligand trans to H2 is known to have a strong influence on hydrogen back donation, the intra molecular distance and metal-hydrogen bond length.13 Herein, we show that this is likely for jHmd-wt, where the acyl-carbon is trans to the supposed H2 binding site. Interestingly, the omission of the oxygen smoothed down the first transition.
Conclusions
A formal 2+ oxidation state is consistent with all the available data on jHmd-wt: Mössbauer spectroscopy22 and our experimental data. The octahedral geometry is compatible with the 2+ oxidation state but not with Fe(0).36 In addition, the rising edge of jHmd-wt’s XANES overlaps nicely with that for the HoxC subunit of the hydrogen-sensing [NiFe]-hydrogenase from Ralstonia eutropha, which is accepted as Fe(ii) (Fig. S6‡).37 The experimental and simulated spectra of jHmd-wt are consistent with the recently proposed acyl-carbon ligand. It is interesting that nature has chosen a very low symmetry site for H2 activation. While the two CO ligands ensure low-spin state, the arrangement of the other four donors tune the reactivity toward H2, which is activated at the labile site trans to the acyl ligand reminiscent of the CO ligand trans to the hydride binding site in the [FeFe]- and [NiFe]-hydrogenases.8
Our analysis demonstrates that in the resting enzyme, Fe has an octahedral geometry with a five- to six-fold coordination. This geometry, which was indicated in our recent C176-mutated and DTT-bound enzyme,15 is incompatible with Fe(0), but is expected for low spin ferrous iron. Collectively, all these findings help to define the features that lead to efficient utilization of H2, especially the specific positioning of the three π-accepting ligands. The results of this work may provide a useful guide to the design of new catalysts aiming at cheaper and greener energy processing.
Experimental
Preparation of the samples for XAS analysis
All the samples were homogenized with degassed BN and filled in Hesar glass® cuvettes covered with Kapton® film. After freezing, the samples were stored in liquid nitrogen for the data collection.
Protein samples
All protein samples were prepared in anaerobic condition as described by Pilak et al.38 The holoenzyme was reconstituted from the apoenzyme and FeGP-cofactor isolated from Hmd purified from Methanothermobacter marburgensis as described by Shima et al.2
Model system samples
The model complexes A, B, C were prepared under anaerobic conditions following our published procedures.39 Complexes D and E were obtained from commercial sources (Sigma-Aldrich).
XAS measurements
XAS data collection of the enzyme samples is described by Korbas et al.21 XAS data for the model systems were collected in transmission mode at D2 beamline of the EMBL Outstation Hamburg at DESY, Germany. A Si(111) double-crystal monochromator scanned X-ray energies around Fe K-edge (6.8–8.1 keV). Harmonic rejection was achieved by a focusing mirror (cut-off energy at 20.5 KeV) and a monochromator detuning to 50% of its peak intensity. Sample cells were mounted in a two-stage Displex cryostat and kept at about 20 K. For each of these samples three scans were collected and averaged to ensure good statistics. Spectrometer energy was calibrated by recording Bragg reflections from a static Si(220) crystal in back-reflection geometry during each scan.40 Data reduction, such as background removal, normalization and extraction of the fine structure, was performed with KEMP21 assuming a threshold energy E0,Fe = 7120 eV. EXAFS data were analysed in an energy range from 3 Å−1 to 14.5 Å−1 with Excurv98.41
X-ray absorption near edge structure (XANES) analysis
Qualitative analysis and comparison of the XANES was carried out using the software ATHENA.42 The intensities and energies of the 1s→3d pre-edge features of the normalized Fe K-edge absorption spectra were quantified using the program WinXAS.43 Spectra were fitted over 13 eV around the pre-edge features and modelled by pseudo-Voigt peak shapes. Energy positions, full width at half maximums (FWHMs) and heights were refined. The background underneath the pre-edge features was modelled as well by a pseudo-Voigt function. The total intensity of the 1s→3d pre-edge feature was assigned to the area of the pseudo-Voigt peak at about 7112.5 eV. Features in the rising edge were not included. The absolute edge positions were identified as the first maximum of the first derivatives within the rising edge (Fig. S2‡).
FEFF simulations
The XANES spectra were simulated by ab initio self-consistent field full multiple scattering calculations with FEFF8.2.27 We used as input for the calculations the atomic coordinates extracted from EXAFS modelling, which was carried out with the program Excurve98 on the basis of the available structures.2,15 Attempts to directly use X-ray diffraction coordinates led to less accurate results (data not shown). No corrections or exchange potentials were introduced in the simulation. Attempts to impose a given ionicity on the metal ion yield results where the edge position remains unchanged and the intensities varies less than 2.5%. Thus the simulations of our samples are not sensitive to this feature and this possibility was ignored here. All simulated spectra have been shifted by 3 to 6 eV towards lower energy for comparison with experimental data. This is standard and reflects small inaccuracies in the calculation of the 1s-continuum transitions. The simulations were aligned with respect to the white-line peak positions of the XANES and their first derivatives.
Acknowledgements
SV and SS were financed by an emeritus grant to Rudolf K. Thauer from the Max Planck Society. We also thank him for fruitful discussions and his continuing interest. Moreover, we thank Eckhard Bill (Max-Planck-Institute für Bioanorganische Chemie, Mülheim a. d. Ruhr, Germany) for fruitful discussions. TR was financed by the US Department of Energy, Office of Science.
Footnotes
All the graphs were created using the program KaleidaGraph 4.0 (http://www.synergy.com) and structure 1 with Accelrys DS Visualizer v2.0.1.7347.
Electronic supplementary information (ESI) available: Supplementary material. See DOI: 10.1039/b922557a
Notes and references
- 1.Stephenson M, Stickland LH. Biochem. J. 1931;25:205–214. doi: 10.1042/bj0250205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shima S, Pilak O, Vogt S, Schick M, Stagni MS, Meyer-Klaucke W, Warkentin E, Thauer RK, Ermler U. Science. 2008;321:572–575. doi: 10.1126/science.1158978. [DOI] [PubMed] [Google Scholar]
- 3.Cammack R, Frey M, Robson R. Hydrogen as a fuel: Learning from nature. London: Taylor & Francis; 2001. [Google Scholar]; Hoffmann P, Harkin T. Tomorrows energy. MIT press; 2001. [Google Scholar]
- 4.Enthaler S, Junge K, Beller M. Angew. Chem., Int. Ed. 2008;47:3317–3321. doi: 10.1002/anie.200800012. [DOI] [PubMed] [Google Scholar]
- 5.von Wangelin AJ. Iron Catalysis in Organic Chemistry. Reaction and Applications. Wiley-VCH; 2008. Oct 13, p. 2008. [Google Scholar]
- 6.Armstrong FA, Fontecilla-Camps JC. Science. 2008;321:498–499. doi: 10.1126/science.1161326. [DOI] [PubMed] [Google Scholar]; Gloaguen F, Rauchfuss TB. Chem. Soc. Rev. 2009;38:100–108. doi: 10.1039/b801796b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vignais PM, Billoud B, Meyer J. FEMS Microbiol. Rev. 2001;25:455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 8.Fontecilla-Camps JC, Volbeda A, Cavazza C, Nicolet Y. Chem. Rev. 2007;107:4273–4303. doi: 10.1021/cr050195z. [DOI] [PubMed] [Google Scholar]
- 9.Zirngibl C, Vandongen W, Schwörer B, Von Bünau R, Richter M, Klein A, Thauer RK. Eur. J. Biochem. 1992;208:511–520. doi: 10.1111/j.1432-1033.1992.tb17215.x. [DOI] [PubMed] [Google Scholar]
- 10.Shima S, Thauer RK. Chem. Rec. 2007;7:37–46. doi: 10.1002/tcr.20111. [DOI] [PubMed] [Google Scholar]
- 11.Pierik AJ, Roseboom W, Happe RP, Bagley KA, Albracht SPJ. J. Biol. Chem. 1999;274:3331–3337. doi: 10.1074/jbc.274.6.3331. [DOI] [PubMed] [Google Scholar]
- 12.Lyon EJ, Shima S, Boecher R, Thauer RK, Grevels FW, Bill E, Roseboom W, Albracht SPJ. J. Am. Chem. Soc. 2004;126:14239–14248. doi: 10.1021/ja046818s. [DOI] [PubMed] [Google Scholar]
- 13.Kubas GJ. Chem. Rev. 2007;107:4152–4205. doi: 10.1021/cr050197j. [DOI] [PubMed] [Google Scholar]
- 14.Lubitz W, Reijerse E, van Gastel M. Chem. Rev. 2007;107:4331–4365. doi: 10.1021/cr050186q. [DOI] [PubMed] [Google Scholar]; Henry RM, Shoemaker RK, DuBois DL, DuBois MR. J. Am. Chem. Soc. 2006;128:3002–3010. doi: 10.1021/ja057242p. [DOI] [PubMed] [Google Scholar]
- 15.Hiromoto T, Ataka K, Pilak O, Vogt S, Salomone-Stagni M, Meyer-Klaucke W, Warkentin E, Thauer RK, Shima S, Ermler U. FEBS Lett. 2009;583:585–590. doi: 10.1016/j.febslet.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Lyon EJ, Shima S, Buurman G, Chowdhuri S, Batschauer A, Steinbach K, Thauer RK. Eur. J. Biochem. 2004;271:195–204. doi: 10.1046/j.1432-1033.2003.03920.x. [DOI] [PubMed] [Google Scholar]
- 17.Buurman G, Shima S, Thauer RK. FEBS Lett. 2000;485:200–204. doi: 10.1016/s0014-5793(00)02225-0. [DOI] [PubMed] [Google Scholar]
- 18.Zirngibl C, Hedderich R, Thauer RK. FEBS Lett. 1990;261:112–116. [Google Scholar]
- 19.Hiromoto T, Warkentin E, Moll J, Ermler U, Shima S. Angew. Chem., Int. Ed. 2009;48:6457–6460. doi: 10.1002/anie.200902695. [DOI] [PubMed] [Google Scholar]
- 20.Salomone-Stagni M, Vogt S, Shima S, Meyer-Klaucke W. J. Phys.: Conf. Ser. 2009;190:12197. [Google Scholar]
- 21.Korbas M, Vogt S, Meyer-Klaucke W, Bill E, Lyon EJ, Thauer RK, Shima S. J. Biol. Chem. 2006;281:30804–30813. doi: 10.1074/jbc.M605306200. [DOI] [PubMed] [Google Scholar]
- 22.Shima S, Lyon EJ, Thauer RK, Mienert B, Bill E. J. Am. Chem. Soc. 2005;127:10430–10435. doi: 10.1021/ja051895o. [DOI] [PubMed] [Google Scholar]
- 23.Wang XF, Li ZM, Zeng XR, Luo QY, Evans DJ, Pickett CJ, Liu XM. Chem. Commun. 2008:3555–3557. doi: 10.1039/b805262j. [DOI] [PubMed] [Google Scholar]
- 24.Royer AM, Rauchfuss TB, Gray DL. Organometallics. 2009;28:3618–3620. [Google Scholar]
- 25.Arcovito A, Benfatto M, Cianci M, Hasnain SS, Nienhaus K, Nienhaus GU, Savino C, Strange RW, Vallone B, Della Longa S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6211–6216. doi: 10.1073/pnas.0608411104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Feiters MC, Küpper FC, Meyer-Klaucke W. J. Synchrotron Radiat. 2004;12:85–93. doi: 10.1107/S0909049504027815. [DOI] [PubMed] [Google Scholar]; Risberg ED, Jalilehvand F, Leung BO, Pettersson LG, Sandstrom M. Dalton Trans. 2009:3542–3558. doi: 10.1039/b819257j. [DOI] [PubMed] [Google Scholar]
- 26.Slep LD, Neese F. Angew. Chem., Int. Ed. 2003;42:2942–2945. doi: 10.1002/anie.200301649. [DOI] [PubMed] [Google Scholar]; Pantelouris A, Modrow H, Pantelouris M, Hormes J, Reinen D. Chem. Phys. 2004;300:13–22. [Google Scholar]
- 27.Ankudinov AL, Ravel B, Rehr JJ, Conradson SD. Phys. Rev. B: Condens. Matter Mater. Phys. 1998;58:7565–7576. [Google Scholar]
- 28.Mijovilovich A, Meyer-Klaucke W. J. Synchrotron Radiat. 2002;10:64–68. doi: 10.1107/s0909049502017296. [DOI] [PubMed] [Google Scholar]
- 29.Westre TE, Kennepohl P, DeWitt JG, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 1997;119:6297–6314. [Google Scholar]; Randall CR, Zang Y, True AE, Que L, Charnock JM, Garner CD, Fujishima Y, Schofield CJ, Baldwin JE. Biochemistry. 1993;32:6664–6673. doi: 10.1021/bi00077a020. [DOI] [PubMed] [Google Scholar]; Randall CR, Shu LJ, Chiou YM, Hagen KS, Ito M, Kitajima N, Lachicotte RJ, Zang Y, Que L. Inorg. Chem. 1995;34:1036–1039. [Google Scholar]; Roe AL, Schneider DJ, Mayer RJ, Pyrz JW, Widom J, Que L. J. Am. Chem. Soc. 1984;106:1676–1681. [Google Scholar]
- 30.Wong WK, Chiu KW, Wilkinson G, Galas AMR, Thornton-Pett M, Hursthouse MB. J. Chem. Soc., Dalton Trans. 1983:1557–1563. [Google Scholar]
- 31.Takàcs J, Markó L, Párkányi L. J. Organomet. Chem. 1989;361:109–116. [Google Scholar]
- 32.Yang X, Hall MB. J. Am. Chem. Soc. 2008;130:14036–14037. doi: 10.1021/ja804364p. [DOI] [PubMed] [Google Scholar]
- 33.Löscher S, Schwartz L, Stein M, Ott S, Haumann M. Inorg. Chem. 2007;46:11094–11105. doi: 10.1021/ic701255p. [DOI] [PubMed] [Google Scholar]
- 34.Liaw WF, Kim C, Darensbourg MY, Rheingold AL. J. Am. Chem. Soc. 1989;111:3591–3597. [Google Scholar]
- 35.Wander SA, Reibenspies JH, Kim JS, Darensbourg MY. Inorg. Chem. 1994;33:1421–1426. [Google Scholar]
- 36.Whitmire KH, Kelly AT, Hofmann C. In: Comprehensive Organometallic Chemistry III. Bruce M, editor. vol. 6. Amsterdam: Elsevier; 2006. pp. 1–76. [Google Scholar]
- 37.Löscher S, Zebger I, Andersen LK, Hildebrandt P, Meyer-Klaucke W, Haumann M. FEBS Lett. 2005;579:4287–4291. doi: 10.1016/j.febslet.2005.06.063. [DOI] [PubMed] [Google Scholar]
- 38.Pilak O, Mamat B, Vogt S, Hagemeier CH, Thauer RK, Shima S, Vonrhein C, Warkentin E, Ermler U. J. Mol. Biol. 2006;358:798–809. doi: 10.1016/j.jmb.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 39.Guo YS, Wang HX, Xiao YM, Vogt S, Thauer RK, Shima S, Volkers PI, Rauchfuss TB, Pelmenschikov V, Case DA, Alp EE, Sturhahn W, Yoda Y, Cramer SP. Inorg. Chem. 2008;47:3969–3977. doi: 10.1021/ic701251j. [DOI] [PMC free article] [PubMed] [Google Scholar]; Whaley CM, Rauchfuss TB, Wilson SR. Inorg. Chem. 2009;48:4462–4469. doi: 10.1021/ic900200s. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ruff JK. Inorg. Chem. 1969;8:86–89. [Google Scholar]; Goldfield GA, Raymond KN. Inorg. Chem. 1974;13:770–775. [Google Scholar]
- 40.Pettifer RF, Brouder C, Benfatto M, Natoli CR, Hermes C, Lopez MFR. Phys. Rev. B: Condens. Matter. 1990;42:37–42. doi: 10.1103/physrevb.42.37. [DOI] [PubMed] [Google Scholar]
- 41.Binsted N, Strange RW, Hasnain SS. Biochemistry. 1992;31:12117–12125. doi: 10.1021/bi00163a021. [DOI] [PubMed] [Google Scholar]
- 42.Ravel B, Newville M. J. Synchrotron Radiat. 2005;12:537–541. doi: 10.1107/S0909049505012719. [DOI] [PubMed] [Google Scholar]
- 43.Ressler T. J. Synchrotron Radiat. 1998;5:118–122. doi: 10.1107/S0909049597019298. [DOI] [PubMed] [Google Scholar]