Abstract
Genetic variability has been shown to affect statin responsiveness. Participants from the Utrecht Cardiovascular Pharmacogenetics (UCP) studies were enrolled from a population-based registry of pharmacy records linked to hospital discharge records (PHARMO) to investigate tagging SNPs within candidate genes involved in the cholesterol lowering pathway for modification of the effectiveness of statins in reducing the risk of myocardial infarction (MI). Patients who received a prescription for an antihypertensive drug and/or had hypercholesterolemia were selected from the PHARMO database. We designed a nested case-control study in which cases were hospitalized for MI and controls were not. Patients were contacted through their community pharmacies. For this study, only hypercholesterolemic participants were selected. Logistic regression analysis was used to investigate pharmacogenetic interactions. The Heart and Vascular Health Study (HVH) was used to replicate findings from UCP.
The study population included 668 cases and 1217 controls. We selected 231 SNPs of which 209 SNPs in 27 genes passed quality control. Ten SNPs in eight genes were found to influence the effectiveness of statins in UCP, of which the most significant interaction was found with SCARB1 rs4765615. Other genes that reached statistical significance (p<0.05) included two SNPs in PCSK9 (rs10888896 and rs505151 (E670G)), two SNPs in ABCG5 (rs4245786 and rs1864815), LIPC rs16940379, ABCA1 rs4149264, PPARG rs2972164, LRP1 rs715948, and SOAT1 rs2493121. None of the total of 5 SNPs that were available for replication in HVH reached statistical significance.
In conclusion, ten SNPs were found to modify the effectiveness of statins in reducing the risk of MI in the UCP study. Five were also tested in the HVH study, but no interactions reached statistical significance.
Keywords: pharmacogenetics, statin, case-control study, cholesterol, myocardial infarction, PCSK9, SCARB1
Introduction
To reduce the risk of cardiovascular events, statins are among the most prescribed drugs worldwide. Although the efficacy has been well established in clinical trials [1], interindividual differences in response exist [2]. Besides non-genetic factors such as age, concomitant drug use, and co-morbidities, it has been well recognized that variability in statin-related genes contribute to differences in response to statins. These include both genes in the lipid and non-lipid pathways [2].
Statins’ foremost pharmacological action is the competitive inhibition of HMG-CoA reductase, the first enzyme and rate-limiting step in the cholesterol biosynthesis cascade. Subsequently, there is an increase in hepatic low-density lipoprotein (LDL) receptors resulting in increased LDL clearance from the blood stream. Although the HMGCR gene, encoding HMG-CoA reductase, is an important candidate gene for the pharmacogenomics of statins, a range of other cholesterol pathway related genes may be of importance for statin responsiveness. These include genes that are involved in the hepatic cholesterol metabolism or metabolism and transport of plasma lipoproteins. Well known examples of such genes that have previously been subject of pharmacogenomic research are LDLR, encoding LDL receptor, CETP, encoding cholesteryl ester transfer protein and APOE, encoding apolipoprotein E [2].
Most pharmacogenetic studies have investigated the cholesterol lowering response to statins as opposed to clinically important outcomes such as myocardial infarction (MI). Therefore, the aim of this study was to investigate the genetic influence of tagging SNPs within candidate genes involved in the cholesterol lowering pathway of statins on the effectiveness of statins in reducing the risk of MI.
Methods
Design and Setting
Participants from the Utrecht Cardiovascular Pharmacogenetics (UCP) studies were enrolled from the population-based Pharmaco-Morbidity Record Linkage System (PHARMO, www.pharmo.nl). PHARMO links drug dispensing histories from a representative sample of Dutch community pharmacies to the national registration of hospital discharges (Dutch National Medical Registry).
First, patients who received a prescription for an antihypertensive drug [3], and/or had hypercholesterolemia (prescription for a cholesterol-lowering drug or total cholesterol >5.0 mmol/l) [4], were selected from the PHARMO database for pharmacogenetic studies on antihypertensive drugs [3] and statins [4] respectively. From this cohort, a nested case-control study was designed using hospital discharge records. Patients hospitalized for MI (International Classification of Diseases (ICD)-9 code 410) were included as cases if they were registered in PHARMO for at least one year and were older than 18 years. The index date was defined as the date of hospitalization for the first MI. Controls met the same eligibility criteria as the cases, but had not developed MI.
Participants were contacted through community pharmacies, where they received a letter in which the purpose of the study was explained. They were asked to return an informed consent form and a filled-out questionnaire. After the participant had consented to participate in the study, he/she was sent material for saliva collection. All participants were explicitly asked to consent for the collection, storage and genotyping of the DNA material. Approval for this study was obtained from the Medical Ethics Committee of the University Medical Center Utrecht, The Netherlands.
For this study, all hypercholesterolemic (prescription for a cholesterol-lowering drug, total cholesterol>5.0 mmol/l, or self-reported hypercholesterolemia) participants were selected. In detail, the case-control ratios for sampling from the nested case-control study on the antihypertensive drugs [3] and statins [4] was one to one and one to three respectively.
Ascertainment of exposure to statins (and other drugs)
Coded pharmacy records were used to ascertain exposure to statins (and other drugs) before the index date. In PHARMO, complete pharmacy records were available as of 1991, including the day of delivery, daily dose, and durations of therapy. To define exposure to statins, we assessed the association of different cumulative defined daily doses (DDD) (cumulative DDD cut-off points of 90, 180, 360, and 720 DDD) with the risk of MI. The DDD is the dose per day for a drug used for its main indication in adults. Our data showed that statins were not effective in reducing the risk of MI in patients exposed to a cumulative dose of 180 DDD or less. The effectiveness of statins in patients exposed for more than 180 days but less than 360 days did not differ from a cumulative exposure for more than 360 or 720 days. Therefore, participants were considered exposed when the cumulative DDD of statin use was more than 180, whereas participants with the cumulative DDD of 180 or less (including 0 DDDs) were considered as the reference group.
To adjust for potential confounding, we identified all prescriptions for concomitant drug use for each patient. The projected end date of a prescription was calculated using information on the daily dose instruction and the quantity dispensed. We considered a patient a current user when the index date was between the start and end date of a prescription. Past users were patients who were not current users, but had used the drug prior to the index date.
Assessment of potential confounding factors and effect modifiers
Questionnaires were used to assess cardiovascular disease (CVD) risk factors such as smoking, hypertension, hypercholesterolemia, diabetes mellitus, use of alcohol, diet, history of CVDs, family history of CVDs, weight and height. Furthermore, information from the general practitioner files and laboratory registrations were available for part of the population. In case of a discrepancy between community pharmacy data and questionnaire data, community pharmacy data was the primary source for defining hypercholesterolemia and diabetes status. Ischemic heart disease (IHD) was defined as “yes” if a participant was hospitalized for an IHD or ever used nitrates.
DNA collection and DNA extraction
Part of patients were send three cotton swabs and tubes containing buffer to collect buccal cell samples as described elsewhere [5]. Other participants were sent an Oragene collection kit and donor instructions provided by the manufacturer (DNA Genotek, Ottawa, Canada) [4]. DNA was extracted according to the manufacturer’s instructions (http://www.dnagenotek.com/techsupport_documents.htm). Samples with a DNA concentration higher than 100 ng/µl were diluted to the Illumina Golden-Gate assay required 50 ng/µl.
Candidate gene selection and SNP selection
We selected a total of 231 SNPs in 27 genes that are involved in the cholesterol lowering pathway of statins. We selected common tagging SNPs within 200 bp (up- and downstream) with a minor-allele frequency (MAF) higher than 0.2 (based on a power calculation with 80% power to detect a SI of 2 or 0.5) and a r2>0.8 using a web-based tool called QuickSNP version 1.1 (HapMap release 21 [6], U.S. residents with northern and western European ancestry (CEPH individuals)).[7] Additionally, dbSNP [8] nonsynonymous coding SNPs (MAF>0.2) and previously (pharmaco)genetically associated SNPs were included. Illumina SNP designability scores lower than 0.4 (1.1=best validated) or failure codes (http://www.illumina.com/documents/products/technotes/technote_goldengate_design.pdf) were either substituted with a SNP in linkage disequilibrium (LD) or, if unavailable, removed from the SNP list, resulting in a final set of 231 SNPs.
Genotyping
For each individual participating in the study, SNPs were genotyped using the custom GoldenGate assay on an Illumina BeadStation 500 GX (Illumina Inc. San Diego, CA, USA). Genotype calls of all SNPs were individually examined for their resulting quality. SNPs with a low signal, poor clustering, deviation from Hardy–Weinberg equilibrium (HWE) (≤0.01) or a high number of missing genotypes (>10%) were excluded.
Replication study (Heart and Vascular Health Study (HVH))
The setting for the replication study was a large integrated health care system in Washington State, called Group Health Cooperative (GHC). The data were from an ongoing case-control study of incident MI and stroke cases with a shared control group and has been described elsewhere [9, 10]. The study was approved by the human subjects committee at GHC, and all study participants provided an informed consent.
All study participants were GHC members aged 30–79 years. We selected MI cases and controls if they had a prescription for a cholesterol-lowering drug or total cholesterol measurement of >5.0 mmol/l. Cases were hospitalized for a non-fatal incident MI, identified from computerized hospital discharge abstracts and billing records [9, 10]. Controls were a random sample of GHC members frequency matched to MI cases on age, sex, and calendar year of identification. The index date for MI cases was the date of admission for the first acute MI, whereas controls were assigned a computer-generated random date within the calendar year for which they had been selected. Medication use was ascertained using computerised GHC pharmacy records. Definitions of drug exposure matched the definitions from the UCP study. Eligibility and risk factor information were collected by trained medical record abstractors from a review of the GHC medical record using only data available prior to the index date and through a telephone interview.
A venous blood sample was collected from all consenting subjects, and DNA was extracted from white blood cells using standard procedures. Genotype data was available from two sources. Part of the genotype data was available from the Illumina 370CNV BeadChip system. Imputation was performed using BIMBAM with reference to HapMap CEU using release 22, build 36 using one round of imputations and the default expectation-maximization warm-ups and runs. In addition, genotype data was available from a Illumina (Illumina Inc, San Diego California) GoldenGate custom panel using BeadArray® technology.
SNPs that showed a significant interaction (p<0.05) with statin treatment in UCP were identified in the HVH study genotype data. Five of these SNPs had genotype data available from the Illumina 370CNV BeadChip system and/or the Illumina GoldenGate custom panel. For these SNPs, if a subject had genotype results available from both Illumina methods, the GoldenGate panel results were preferentially selected. One SNP had genotype data available only from the Illumina GoldenGate panel (n=865). All remaining SNPs had genotype data available only from the Illumina 370CNV BeadChip system (n=2446). SNPs from the Illumina 370 CNV BeadChip system were chosen with a lower cut-off for the RSQR (or OEvar) score of 0.6. The RSQR denotes the average of the observed-to-expected variance ratio of any SNP, which indicates deviation from Hardy-Weinberg equilibrium and quality of imputation.
Statistical methods
The same analysis was applied to the UCP and HVH study. Logistic regression (LR) analysis was used to study the association between statins and the risk of MI, and to adjust for potential confounders. Matching variables --- age, sex, region, and index date --- were included in our statistical model. The inclusion of potential confounders in the LR model was motivated by the assessment of the influence of each potential confounder on the OR for the association between use of statins and risk of MI. The potential confounding factors that we considered were: Use of different cardiovascular drugs (antihypertensive drugs, platelet aggregation inhibitors, anticoagulants, other cholesterol-lowering drugs, and organic nitrates), use of alcohol, physical activity, family history of CVD, and other factors assessed by the questionnaire. Only covariates IHD and the use of calcium channel blockers showed at least a 5% change in the regression coefficient (beta) for statin use; therefore, they were included in the LR model. We estimated the multiplicative synergy index (SI), which is the ratio of the OR in those with the variant to the OR in those without the variant. For the significant (unadjusted or adjusted) pharmacogenetic associations, ORs were calculated separately in the strata defined by genotype. Heterozygotes and homozygotes for the variant allele of the PCSK9 E670G (rs505151) polymorhpism were combined because of a low frequency homozygous variant allele carriers. For each SNP, HWE was tested using a Χ2 goodness-of-fit test. Analyses were performed using SPSS version 16.0. Subsequently, q-values (the positive false discovery rate (pFDR) analogue of the p-value) were calculated for each gene-treatment interaction that was tested in UCP to account for multiple testing [11].
Results
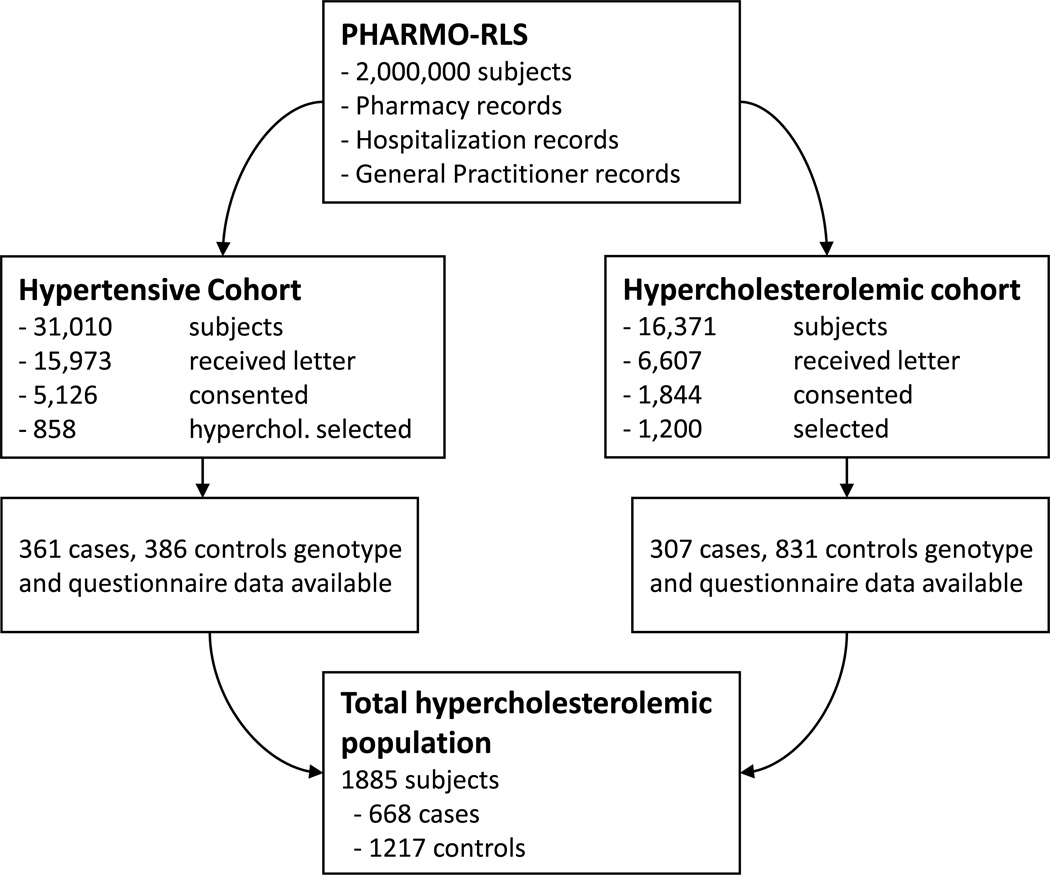
The data collection procedure for this study has previously been published [4]. Briefly, figure 1 summarizes how the final number of participants was arrived at. For the hypercholesterolemic cohort 9,764 patients could not be approached for various reasons (death of a patient, pharmacy did not participate, amount of controls per case was decreased, or patient was untraceable due to change in the community pharmacy computer information system). For the genotyping assay, out of 1,844 consenting subjects, all cases were selected (n = 315), accompanied by the matched controls and a random sample of unmatched controls, to bring the total population to 1,200 (approximately three controls per case). After exclusion of patients that donated an insufficient amount of DNA, patients of which the genotyping results did not pass quality control (QC), and patients with a self-reported ethnicity other than Caucasian, 307 cases and 831 controls were included from the hypercholesterolemic cohort. From the hypertensive cohort, we selected 429 hypercholesterolemic cases and 429 hypercholesterolemic controls. After excluding patients already included from the hypercholesterolemic cohort study, patients for whom the genotyping did not pass QC, and patients with a self-reported ethnicity other than Caucasian, we were able to amass 361 cases and 386 controls.
Figure 1.
Flow diagram of study population.
The total UCP study population included 1885 individuals, of which 668 were MI cases and 1217 controls. Table 1 describes the clinical characteristics of the population according to case control status. The well known cardiovascular risk factors smoking (current), a BMI of more than 30 kg/m2, and the presence of IHD were more frequently seen in cases compared to controls. Current use of other non-statin cholesterol-lowering drugs was associated with a decreased risk of MI. Current use of beta-blockers and calcium channel blockers was more frequently seen in cases than controls, which is due to oversampling of nonantihypertensive users in the control group as described in the methods section [4].
Table 1.
Clinical characteristics UCP by case control status.
| Case n=668 |
% | Control n=1217 |
% | p | ||||
|---|---|---|---|---|---|---|---|---|
| Gender | Female | 171 | 25.6% | 285 | 23.4% | 0.290 | ||
| Age (years) | Mean (sd) | 63.0 (9.9) | 62.3 (9.4) | 0.097 | ||||
| Body Mass Index at ID | >30 kg/m2 | 140/607 | 23.1% | 193/1104 | 17.5% | 0.005 | ||
| Familial History CVD | Yes, <60 | 135/637 | 21.2% | 214/1167 | 18.3% | 0.103 | ||
| Yes, >60 | 266/637 | 41.8% | 464/1167 | 39.8% | ||||
| Diabetes Status | Diabetes, no medication | 71/658 | 10.8% | 90/1203 | 7.5% | 0.052 | ||
| Diabetes, medication | 75/658 | 11.4% | 144/1203 | 12.0% | ||||
| Smoking Status | Current | 152/624 | 24.4% | 195/1135 | 17.2% | 0.001 | ||
| Past | 283/624 | 45.4% | 573/1135 | 50.5% | ||||
| Alcohol Status (consumptions) | No use | 118/646 | 18.3% | 162/1188 | 13.6% | 0.062 | ||
| <=1 | 237/646 | 36.7% | 432/1188 | 36.4% | ||||
| >1 – <2 | 170/646 | 26.3% | 328/1188 | 27.6% | ||||
| >2 | 121/646 | 18.7% | 266/1188 | 22.4% | ||||
| Physical Activity | > 4 hrs a week | 491/645 | 76.1% | 939/1191 | 78.8% | 0.180 | ||
| Cumulative DDD use statins | >180 DDD | 218 | 32.6% | 646 | 53.1% | <0.001 | ||
| Type of Statin | Atorvastatin | 44/218 | 20.2% | 164/646 | 25.4% | <0.001 | ||
| Pravastatin | 35/218 | 16.1% | 110/646 | 17.0% | ||||
| Simvastatin | 125/218 | 57.3% | 335/646 | 51.9% | ||||
| Other | 14/218 | 6.4% | 37/646 | 5.7% | ||||
| Ischemic Heart Disease | Yes | 211 | 31.6% | 268 | 22.0% | <0.001 | ||
| Antihypertensives | ||||||||
| Calcium Channel Blockers | Current use | 142 | 21.3% | 193 | 15.9% | 0.003 | ||
| Diuretics | Current use | 82 | 12.3% | 171 | 14.1% | 0.279 | ||
| Beta Blockers | Current use | 275 | 41.2% | 415 | 34.1% | 0.002 | ||
| Ace Inhibitors | Current use | 140 | 21.0% | 273 | 22.4% | 0.459 | ||
| AT2 Receptor Antagonists | Current use | 48 | 7.2% | 110 | 9.0% | 0.165 | ||
| Other drugs | ||||||||
| Non-statin Cholesterol Lowering drugs | Current use | 14 | 2.1% | 46 | 3.8% | 0.046 | ||
| Insulin | Ever use | 30 | 4.5% | 54 | 4.4% | 0.957 | ||
| Oral Antidiabetics | Current Use | 52 | 7.8% | 96 | 7.9% | 0.936 | ||
| Platelet Aggregation Inhibitors | Current Use | 219 | 32.8% | 438 | 36.0% | 0.162 | ||
| Coumarins | Current Use | 37 | 5.5% | 76 | 6.2% | 0.537 | ||
Abbreviations: ATII = Angiotensin II; DDD = Defined Daily Dosage
Out of the 231 selected SNPs, 209 passed quality control and were tested for an interaction with statin treatment. The LR analysis revealed ten SNPs in eight genes that significantly (p<0.05) interacted with statin treatment (table 2), either with or without adjustment for the additional confounding factors or both. SCARB1 rs4765615 showed the most significant interaction, with a more beneficial effect of statins for GG and AG carriers (OR 0.30, 95% confidence interval (CI) 0.22–0.42 and OR 0.30, 95%CI 0.18–0.50 respectively) as compared to AA carriers (OR 0.64, 95%CI 0.41–0.98). The PCSK9, and ABCG5 gene were both represented by two SNPs among the significant interactions. The only nonsynonymous SNP that was found to interact with statin treatment, was PCSK9 rs505151, for which variant allele carriers had no significant benefit from statin treatment (OR 0.63, 95%CI 0.30–1.32) compared to homozygous wildtype carriers who did benefit (OR 0.36, 95%CI 0.28–0.45). The five other SNPs that appeared to be implicated in the pharmacogenetics of statins were found in the LRP1, LIPC, ABCA1 SOAT1, and PPARG gene. The q-value for the interaction with SCARB1 rs4765615 and PCSK9 rs10888896 was 0.19 and 0.24 respectively, whereas the q-value of the interactions with the other eight SNPs within the significant results was 0.57. The SIs for all of the SNPs can be found in Table II of the Supplementary data.
Table 2.
Significant interactions in UCP and replication results from the HVH study.
| UCP | HVH | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | G | # | HWE | p | q | OR*(95% CI) | OR**(95% CI) | RSQR | # | HWE | p | OR* (95% CI) | OR** (95% CI) |
| SCARB1 | rs4765615 | AA | 452 | 0.80 (0.53–1.20) | 0.63 (0.41–0.97) | 0.23 | ||||||||
| AG | 837 | 0.54 | 0.001 | 0.19 | 0.33 (0.24–0.46) | 0.29 (0.21–0.41) | ||||||||
| GG | 411 | 0.38 (0.24–0.61) | 0.31 (0.19–0.51) | |||||||||||
| -- | 185 | |||||||||||||
| PCSK9 | rs10888896 | CC | 119 | 1.44 (0.65–3.18) | 1.38 (0.60–3.16) | 0.18 | ||||||||
| CG | 658 | 0.12 | 0.003 | 0.24 | 0.37 (0.26–0.54) | 0.32 (0.22–0.47) | ||||||||
| GG | 1105 | 0.40 (0.31–0.53) | 0.34 (0.25–0.45) | |||||||||||
| -- | 3 | |||||||||||||
| ABCG5 | rs4245786 | AA | 1099 | 0.56 (0.43–0.73) | 0.46 (0.35–0.61) | 0.08 | ||||||||
| AG | 692 | 0.23 | 0.016 | 0.57 | 0.26 (0.18–0.37) | 0.24 (0.16–0.34) | ||||||||
| GG | 93 | 0.49 (0.19–1.24) | 0.38 (0.13–1.12) | |||||||||||
| -- | 1 | |||||||||||||
| ABCG5 | rs1864815 | AA | 834 | 0.60 (0.44–0.81) | 0.48 (0.35–0.67) | 0.68 | 1071 | 0.96 (0.70–1.33) | 0.80 (0.57–1.13) | |||||
| AT | 835 | 0.61 | 0.022 | 0.57 | 0.29 (0.21–0.40) | 0.25 (0.18–0.36) | 1104 | 0.646 | 0.82 | 0.99 (0.72–1.36) | 0.83 (0.60–1.17) | |||
| TT | 198 | 0.38 (0.20–0.74) | 0.42 (0.21–0.84) | 271 | 1.23 (0.62–2.44) | 0.98 (0.47–2.04) | ||||||||
| -- | 18 | 0 | ||||||||||||
| LIPC | rs16940379 | GG | 986 | 0.47 (0.35–0.62) | 0.38 (0.28–0.51) | 0.82 | 168 | 0.88 (0.66–1.18) | 0.69 (0.51–0.95) | |||||
| CG | 749 | 0.68 | 0.031 | 0.57 | 0.32 (0.23–0.45) | 0.28 (0.20–0.40) | 939 | 0.637 | 0.45 | 1.15 (0.81–1.63) | 1.02 (0.70–1.46) | |||
| CC | 149 | 0.83 (0.38–1.82) | 0.81 (0.36–1.79) | 1339 | 1.38 (0.54–3.53) | 1.43 (0.52–3.93) | ||||||||
| -- | 1 | 0 | ||||||||||||
| ABCA1 | rs4149264 | CC | 1192 | 0.52 (0.40–0.68) | 0.44 (0.34–0.58) | |||||||||
| CG | 607 | 0.67 | 0.034 | 0.57 | 0.27 (0.19–0.40) | 0.23 (0.15–0.34) | 0.35 | |||||||
| GG | 82 | 0.54 (0.19–1.51) | 0.50 (0.16–1.58) | |||||||||||
| -- | 4 | |||||||||||||
| PPARG | rs2972164 | GG | 585 | 0.42 (0.26–0.65) | 0.35 (0.22–0.57) | 1.00 | 507 | 1.06 (0.73–1.54) | 0.96 (0.64–1.43) | |||||
| AG | 897 | 0.14 | 0.034 | 0.57 | 0.33 (0.24–0.45) | 0.28 (0.20–0.39) | 1176 | 0.48 | 0.89 | 1.00 (0.73–1.37) | 0.79 (0.57–1.11) | |||
| AA | 395 | 0.60 (0.42–0.85) | 0.49 (0.34–0.72) | 763 | 1.02 (0.64–1.64) | 0.84 (0.50–1.39) | ||||||||
| -- | 8 | 0 | ||||||||||||
| PCSK9 | rs505151 | AA | 1632 | 0.42 (0.33–0.52) | 0.36 (0.28–0.45) | NI† | 788 | 0.69 (0.46–1.04) | 0.61 (0.39–0.94) | |||||
| G | 158 | 0.23 | 0.038 | 0.57 | 0.91 (0.46–1.79) | 0.63 (0.30–1.32) | 76 | 0.621 | 0.56 | 1.47 (0.38–5.71) | 1.05 (0.18–6.26) | |||
| -- | 95 | 1 | ||||||||||||
| LRP1 | rs715948 | GG | 908 | 0.17 (0.08–0.37) | 0.17 (0.08–0.37) | 1.05 | 271 | 0.95 (0.70–1.29) | 0.79 (0.57–1.09) | |||||
| AG | 794 | 0.7 | 0.04 | 0.57 | 0.49 (0.36–0.67) | 0.44 (0.31–0.60) | 1011 | 0.058 | 0.93 | 1.06 (0.76–1.47) | 0.90 (0.63–1.28) | |||
| AA | 181 | 0.45 (0.33–0.60) | 0.36 (0.26–0.49) | 1164 | 1.12 (0.54–2.31) | 0.86 (0.39–1.86) | ||||||||
| -- | 2 | 0 | ||||||||||||
| SOAT1 | rs2493121 | AA | 217 | 0.20 (0.11–0.39) | 0.19 (0.10–0.38) | |||||||||
| AT | 832 | 0.75 | 0.047 | 0.57 | 0.49 (0.36–0.68) | 0.43 (0.31–0.60) | 0.51 | |||||||
| TT | 824 | 0.44 (0.32–0.61) | 0.36 (0.25–0.50) | |||||||||||
| -- | 12 | |||||||||||||
For each gene and SNP, the number of subjects for each genotype are given, together with the HWE. The SI was used to test for the interaction between the genotype and statin treatment. The p value for each interaction denotes whether there is an overall difference in the effectiveness of statins between the three genotype groups (with two degrees of freedom). The q value gives an estimate of the proportion of false discoveries among the statistically significant results. For each genotype stratum, the OR reflects the effectiveness of statins within the specific genotype group. The RSQR is a measure for the quality of an imputed SNP in the HVH (cut-off=0.6).
-- = missing genotype
OR = odds ratio
SI = synergy index
= adjusted for age, sex, region, index date
= adjusted for age, sex, region, index date, use of calcium channel blockers, and ischemic heart disease
G = genotype
# = number of participants
HWE = Hardy-Weinberg equilibrium
lower = lower limit of the 95% confidence interval
upper = upper limit of the 95% confidence interval
p = p-value for the interaction
q = q-value for the interaction
NI = not imputed
RSQR = the average of the observed by expected variance ratio
= HVH study data only available from the Illumina GoldenGate pan
For the HVH study, 10,860 (2,976 cases and 7,884 controls) subjects were initially eligible. 551 controls were excluded because of a prior MI (2,976 cases and 7,333 controls). Subsequently, 1,097 normocholesterolemic subjects were excluded (2,835 cases and 6,377 controls). An additional 6,766 subjects were exluded because no genotyping results were available resulting in a total study population of 1,182 cases and 1,264 controls. The PCSK9 SNP was genotyped using the Illumina GoldenGate panel only and the other nine were imputed using the Illumina 370CNV BeadChip system genotype data only. Four of the nine had an RSQR score more than 0.6, and five had an RSQR score less than 0.6 and were therefore not included in the analysis. None of the five interactions tested in HVH showed a significant interaction (table 2). Nonetheless, similarly to the results from the UCP study, PCSK9 rs505151 variant allele carriers had no significant benefit from statin treatment (OR 1.05, 95%CI 0.18–6.26), whereas homozygous wildtype carriers did (OR 0.61, 95%CI 0.39–0.94). In addition, the point estimates and directionality of the non-significant HVH results for the LIPC rs16940379 interaction resemble the UCP results in which homozygous wildtype allele carriers appear to respond better to statin treatment compared to homozygous variant allele carriers.
Discussion
In this population-based retrospective case-control study, we tested 209 SNPs in 27 genes involved in the cholesterol-lowering pathway and found ten SNPs in eight genes to influence the effectiveness of statins in UCP, of which the most significant interaction was found with SCARB1 rs4765615. Also genetic variability within the PCSK9, LIPC, LRP1, ABCG5, ABCA1, PPARG, and SOAT1 genes were found to affect statin effectiveness. Five out of ten statistically significant SNPs were available in the HVH study for replication but failed to reach statistical significance. In both studies, carriers of the PCSK9 rs505151 variant allele had no benefit from statin treatment, although the formal test for interaction was not statistically significant in the HVH study.
The highly significant interaction with SCARB1 rs4765615 showed that homozygous carriers of the A allele did not benefit from statin treatment compared to those carrying one or two G alleles. Scavenger receptor class B member 1, encoded by SCARB1, functions as a receptor for high-density lipoprotein (HDL) and plays an important role in the reverse cholesterol transport (RCT). Recently, a variant other than SCARB1 rs16940379 was shown to affect the LDL cholesterol (LDLc) response to atorvastatin [12]. Despite the unknown underlying mechanism of this gene treatment interaction, also our study indicates a role for SCARB1 in the response to statins. Genetic variability of SCARB1 should therefore be investigated in future studies. The imputation score of the HVH study data for SCARB1 rs16940379 and four other SNPs was too poor and were not used for further analysis. Except for the interaction with SCARB1 rs16940379 and PCSK9 rs10888896 (which will be discussed hereafter), these interactions are generally characterized by high q-values, no well defined known functional SNP that is in LD with the interacting tagging SNP, and/or models that lack of gene-dose effect, suggesting that these findings may be false positives.
PCSK9, encoding proprotein convertase subtilisin/kexin type 9, was found to be the third locus involved in autosomal dominant hypercholesterolemia (ADH) [13]. PCSK9 promotes the degradation LDLR and gain-of-function mutations have been shown to lead to higher LDLc levels, whereas loss-of-function mutations have been shown to result in lower LDLc [14] and protection against CHD [15]. The mutations that cause severe hypercholesterolemia are rare, but also common mutations have been shown to affect lipid levels and possibly the response to statins. Such a common nonsynonymous polymorphism is PCSK9 E670G (included in the current study), which has been shown to be a marker for higher plasma LDLc levels in several [16–19] but not all studies [20–22]. Also an association between PCSK9 E670G and increased carotid artery intima media thickness was found [18], although others did not show an association with CAD/CHD/vascular disease risk [17, 21, 22]. In turn, it has been suggested that individuals carrying PCSK9 loss-of-function polymorphisms have an increased lipid response to statins [23], whereas gain-of-function polymorphisms have been shown to result in a decreased lipid response to statin therapy [24, 25].
Three studies investigated the PCSK9 E670G with respect to statin responsiveness. The PROSPER trial including almost 6000 elderly subjects (mean age 75 years) could not reveal a significant difference in LDLc response or CHD risk reduction between carriers and noncarriers of the variant [21]. Among 49 SNPs in nine candidate genes, the PCSK9 E670G variant was also included in a pharmacogenetic study in the PROVE IT-TIMI 22 trial. Lipid response of 1378 hypercholesterolemic post ACS subjects randomized to pravastatin or atorvastatin did not differ among PCSK9 E670G genotype strata [31]. In the Treating to New Targets (TNT) trial, carriers of the 670G allele were found to have a significantly smaller decrease in LDLc levels in response to statin treatment [26]. Similar to the results of the TNT study and the observations that PCSK9 gain-of-function variants have a deleterious effect on statin responsiveness [24, 25], we show that carriers of the 670G allele have a better response to statin treatment in UCP (significant) and HVH (not significant). Nonetheless, the magnitude of the observation by the TNT study (1.8 mg/dl less LDLc reduction PCSK9 670G carriers) does not reflect the large effect of PCSK9 E670G found in this study, suggesting a partially lipid independent mechanism behind this gene treatment interaction.
Mechanistically, statins decrease the endogenous cholesterol biosynthesis by inhibition of 3-hydroxy-3-methylglutaryl–coenzyme A, which leads to transcriptional activation of both LDLR and PCSK9 [14]. Although PCSK9 counteracts the statin induced increased LDLR activity, the net result of statin treatment is reduction in plasma LDL cholesterol. In the case of PCSK9 E670G polymorphism, it can be hypothesized that the net result of statin treatment is no longer beneficial due to its gain-of-function nature.
Additionally, our tagging SNP approach revealed a second – more significant – PCSK9 SNP that affected the response to statins. PCSK9 rs10888896 resides in the first intron and has not been extensively researched. No effect of this SNP on LDLc reduction was found in the Treating to New Targets (TNT) trial [26]. Possibly, the variant allele of the PCSK9 is in LD with a (recessive) gain-of-function polymorphism, because our results suggest that homozygous wild-type variant carriers have no benefit from statin treatment. Although this SNP was unavailable in the HVH population, the PCSK9 gene is of great interest for statin responsiveness, and the effect of PCSK9 rs10888896 should therefore be assessed in future studies.
None of the results from the UCP study showed a statistically significant interaction in the HVH study (table 2). Besides PCSK9 E670G, only the interaction with LIPC rs16940379 showed a similar trend in the HVH study as was found in UCP study. Hepatic lipase, encoded by LIPC, may affect statin responsiveness through its involvement in modulation of LDL size and density, which in turn is has been shown to affect the risk of CHD [27]. Although no data are available on the role of rs16940379 on hepatic lipase activity, our results indicate a role for LIPC in the pharmacogenomics of statins.
The present study has several limitations. For statistical power reasons, we assessed only SNPs with a MAF cut-off of 0.2, the consequences of which are that we are likely to miss potentially important SNPs. Nevertheless, our study covers the common genetic variability within the selected candidate genes completely. Furthermore, we considered all statins and all dosage regimes as a homogenous group. Although all statins share the primary working mechanism by which they lower cholesterol, it has been shown that there are differences between the different statins [28, 29]. The sample size of the current study does not allow to study the interaction between individual statins and genetic variability. Also, our replication study has two limitations. First, five out of ten SNPs that were found to interact with statins in UCP were not available in the HVH study, or had a low imputation score. Second, imputation of rs1864815, rs16940379, and rs2972164 gives uncertainty about the true genotype and thereby lowers the statistical power to detect an interaction.
A strength of the current study is the availability of a replication study that used the same study design (case-control, exposure definition, outcome, data analysis) as the UCP study. In addition, a centralized system to define exposure (availability of the community pharmacy records) and outcomes (hospital records) was available. Statin exposure was defined based on pharmacy records, which validity to measure drug exposure has been shown to be good [30]. Testing a large number of variables, the possibility of chance findings (spurious associations) increases. We addressed the issue of multiple testing by calculation of q-values that suggested that a large proportion of our significant interaction were false discoveries. Finally, our study assessed the impact of gene-treatment interactions on the clinically important (endpoint) outcome MI, instead of surrogate parameters.
We show that the PCSK9 E670G polymorphism may be of great importance for the effectiveness of statins in reducing the risk of MI because carriers of one or two variant alleles do not seem benefit from statin treatment. PCSK9 gain-of-function variant carriers have been shown to have high untreated and treated cholesterol levels, but a similar percentage LDLc fall from statin treatment compared to wild-type carriers [24]. Therefore, from a clinical perspective, carriers of PCSK9 670G variant allele may benefit from more aggressive lipid-lowering treatment and could especially benefit from novel hypercholesterolemic therapy strategy of PCSK9 inhibition.
In conclusion, variant allele carriers PCSK9 E670G polymorphism do not seem to benefit from statin treatment and confirmation of the pharmacogenetic associations with LIPC rs16940379, SCARB1, rs16940379, and PCSK9 rs10888896 should be subject of future research to pinpoint possible causal variants that affect statin responsiveness.
Supplementary Material
Acknowledgments
Funding:
Anke-Hilse Maitland-van der Zee is funded by a Veni grant from the Netherlands Organization for Scientific Research (NWO)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest section: The division of Pharmacoepidemiology & Pharmacotherapy employing authors Bas Peters, Olaf Klungel, Anthonius de Boer and Anke-Hilse Maitland-van der Zee has received unrestricted funding for pharmacoepidemiological research from GlaxoSmithKline, Novo Nordisk, the private-public funded Top Institute Pharma (www.tipharma.nl, includes co-funding from universities, government, and industry), the Dutch Medicines Evaluation Board, and the Dutch Ministry of Health. The other authors declare no conflict of interest.
REFERENCES
- 1.Cheung BM, Lauder IJ, Lau CP, Kumana CR. Meta-analysis of large randomized controlled trials to evaluate the impact of statins on cardiovascular outcomes. Br J Clin Pharmacol. 2004;57(5):640–651. doi: 10.1111/j.1365-2125.2003.02060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters BJ, Klungel OH, de Boer A, Maitland-van der Zee AH. Genetic determinants of response to statins. Expert Rev Cardiovasc Ther. 2009;7(8):977–983. doi: 10.1586/erc.09.83. [DOI] [PubMed] [Google Scholar]
- 3.van Wieren-de Wijer DB, Maitland-van der Zee AH, de Boer A, Kroon AA, de Leeuw PW, Schiffers P, et al. Interaction between the Gly460Trp alpha-adducin gene variant and diuretics on the risk of myocardial infarction. J Hypertens. 2009;27(1):61–68. doi: 10.1097/hjh.0b013e328317a74d. [DOI] [PubMed] [Google Scholar]
- 4.Peters BJM, Rodin AS, Klungel OH, van Duijn CM, Stricker BHC, Slot Rvt, et al. Pharmacogenetic interactions between ABCB1 and SLCO1B1 tagging SNPs and the effectiveness of statins in the prevention of myocardial infarction. Pharmacogenomics. 2010;11(8):1065–1076. doi: 10.2217/pgs.10.81. [DOI] [PubMed] [Google Scholar]
- 5.van Wieren-de Wijer DB, Maitland-van der Zee AH, de Boer A, Stricker BH, Kroon AA, de Leeuw PW, et al. Recruitment of participants through community pharmacies for a pharmacogenetic study of antihypertensive drug treatment. Pharm World Sci. 2008 doi: 10.1007/s11096-008-9264-x. [DOI] [PubMed] [Google Scholar]
- 6.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grover D, Woodfield AS, Verma R, Zandi PP, Levinson DF, Potash JB. QuickSNP: an automated web server for selection of tagSNPs. Nucleic Acids Res. 2007;35:W115–W120. doi: 10.1093/nar/gkm329. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Psaty BM, Heckbert SR, Atkins D, Lemaitre R, Koepsell TD, Wahl PW, et al. The risk of myocardial infarction associated with the combined use of estrogens and progestins in postmenopausal women. Arch Intern Med. 1994;154(12):1333–1339. [PubMed] [Google Scholar]
- 10.Psaty BM, Heckbert SR, Koepsell TD, Siscovick DS, Raghunathan TE, Weiss NS, et al. The risk of myocardial infarction associated with antihypertensive drug therapies. JAMA. 1995;274(8):620–625. [PubMed] [Google Scholar]
- 11.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerda A, Genvigir FD, Arazi SS, Hirata MH, Dorea EL, Bernik MM, et al. Influence of SCARB1 polymorphisms on serum lipids of hypercholesterolemic individuals treated with atorvastatin. Clin Chim Acta. 2010;411(9–10):631–637. doi: 10.1016/j.cca.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34(2):154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 14.Horton JD, Cohen JC, Hobbs HH. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem Sci. 2007;32(2):71–77. doi: 10.1016/j.tibs.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 16.Evans D, Beil FU. The E670G SNP in the PCSK9 gene is associated with polygenic hypercholesterolemia in men but not in women. BMC Med Genet. 2006;7:66. doi: 10.1186/1471-2350-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu LA, Teng MS, Ko YL, Chang CJ, Wu S, Wang CL, et al. The PCSK9 gene E670G polymorphism affects low-density lipoprotein cholesterol levels but is not a risk factor for coronary artery disease in ethnic Chinese in Taiwan. Clin Chem Lab Med. 2009;47(2):154–158. doi: 10.1515/CCLM.2009.032. [DOI] [PubMed] [Google Scholar]
- 18.Norata GD, Garlaschelli K, Grigore L, Raselli S, Tramontana S, Meneghetti F, et al. Effects of PCSK9 variants on common carotid artery intima media thickness and relation to ApoE alleles. Atherosclerosis. 2008;208(1):177–182. doi: 10.1016/j.atherosclerosis.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 19.Chen SN, Ballantyne CM, Gotto AM, Jr, Tan Y, Willerson JT, Marian AJ. A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. J Am Coll Cardiol. 2005;45(10):1611–1619. doi: 10.1016/j.jacc.2005.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, et al. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78(3):410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polisecki E, Peter I, Robertson M, McMahon AD, Ford I, Packard C, et al. Genetic variation at the PCSK9 locus moderately lowers low-density lipoprotein cholesterol levels, but does not significantly lower vascular disease risk in an elderly population. Atherosclerosis. 2008;200(1):95–101. doi: 10.1016/j.atherosclerosis.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scartezini M, Hubbart C, Whittall RA, Cooper JA, Neil AH, Humphries SE. The PCSK9 gene R46L variant is associated with lower plasma lipid levels and cardiovascular risk in healthy U.K. men. Clin Sci (Lond) 2007;113(11):435–441. doi: 10.1042/CS20070150. [DOI] [PubMed] [Google Scholar]
- 23.Berge KE, Ose L, Leren TP. Missense mutations in the PCSK9 gene are associated with hypocholesterolemia and possibly increased response to statin therapy. Arterioscler Thromb Vasc Biol. 2006;26(5):1094–1100. doi: 10.1161/01.ATV.0000204337.81286.1c. [DOI] [PubMed] [Google Scholar]
- 24.Humphries SE, Whittall RA, Hubbart CS, Maplebeck S, Cooper JA, Soutar AK, et al. Genetic causes of familial hypercholesterolaemia in patients in the UK: relation to plasma lipid levels and coronary heart disease risk. J Med Genet. 2006;43(12):943–949. doi: 10.1136/jmg.2006.038356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naoumova RP, Tosi I, Patel D, Neuwirth C, Horswell SD, Marais AD, et al. Severe hypercholesterolemia in four British families with the D374Y mutation in the PCSK9 gene: longterm follow-up and treatment response. Arterioscler Thromb Vasc Biol. 2005;25(12):2654–2660. doi: 10.1161/01.ATV.0000190668.94752.ab. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JF, Hyde CL, Wood LS, Paciga SA, Hinds DA, Cox DR, et al. Comprehensive wholegenome and candidate gene analysis for response to statin therapy in the Treating to New Targets (TNT) cohort. Circ Cardiovasc Genet. 2009;2(2):173–181. doi: 10.1161/CIRCGENETICS.108.818062. [DOI] [PubMed] [Google Scholar]
- 27.Zambon A, Deeb SS, Pauletto P, Crepaldi G, Brunzell JD. Hepatic lipase: a marker for cardiovascular disease risk and response to therapy. Curr Opin Lipidol. 2003;14(2):179–189. doi: 10.1097/00041433-200304000-00010. [DOI] [PubMed] [Google Scholar]
- 28.Jones P, Kafonek S, Laurora I, Hunninghake D. Comparative dose efficacy study of atorvastatin versus simvastatin, pravastatin, lovastatin, and fluvastatin in patients with hypercholesterolemia (the CURVES study) Am J Cardiol. 1998;81(5):582–587. doi: 10.1016/s0002-9149(97)00965-x. [DOI] [PubMed] [Google Scholar]
- 29.Weitz-Schmidt G. Statins as anti-inflammatory agents. Trends Pharmacol Sci. 2002;23(10):482–486. doi: 10.1016/s0165-6147(02)02077-1. [DOI] [PubMed] [Google Scholar]
- 30.Lau HS, de Boer A, Beuning KS, Porsius A. Validation of pharmacy records in drug exposure assessment. J Clin Epidemiol. 1997;50(5):619–625. doi: 10.1016/s0895-4356(97)00040-1. [DOI] [PubMed] [Google Scholar]
- 31.Mega JL, Morrow DA, Brown A, Cannon CP, Sabatine MS. Identification of genetic variants associated with response to statin therapy. Arterioscler Thromb Vasc Biol. 2009;29:1310–1315. doi: 10.1161/ATVBAHA.109.188474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.