Abstract
Structural analogs are evaluated as peptide internal standards for protein quantification with liquid chromatography-multiple reaction monitoring mass spectrometry (LC-MRM); specifically, single conservative amino acid replacements (SCAR) are performed to create tagged standards that differ by the addition or subtraction of a single methylene group in one amino acid side chain. Because the performance of stable isotope-labeled standards (SIS) has been shown to be superior to structural analogs, differences in both development and quantitative performance between assays based on SIS and SCAR peptides are explored. To establish an assay using the structural analogs, analysis of endogenous, SCAR, and SIS peptides was performed to examine their ion signal, fragmentation patterns, and response in LC-MRM. Performance of SCAR and SIS peptides was compared for quantification of epidermal growth factor receptor from lung cancer cell lysates and immunoglobulin M in the serum of multiple myeloma patients.
Keywords: Peptide Standards, Quantitative Mass Spectrometry, Multiple Reaction Monitoring, Epidermal Growth Factor Receptor, Immunoglobulins
Introduction
Liquid chromatography-multiple reaction monitoring mass spectrometry (LC-MRM) has emerged as a versatile platform for quantification of protein expression. 1–3 Relative quantification can be accomplished without the use of internal standards.4,5 However, LC-MRM approaches to develop biomarkers and to mechanistically explore biological processes will require calculation of protein copy number per cell, per given weight of tissue, or per volume in biofluids. These measurements, termed absolute quantification,2 are achieved though the use of synthetic peptide standards injected at known concentrations with each sample. Selection and evaluation of these internal standards, including stable isotope labeled standards (SIS), structural analogs, and other molecules for peptide quantification, has been reviewed.6–8 Currently, optimal methods employ the use of stable isotope-labeled standard (SIS) peptides.9,10 An SIS peptide incorporating an amino acid labeled with the stable isotopes, 13Cx and 15Ny, is synthesized for each endogenous peptide of interest, creating an internal standard that differs in mass, but not in other chemical or physical properties. Multiplexed experiments monitoring panels of proteins11–13 or post-translational modifications14–16 require a number of standards with a cost as high as $1,000 (USD) each.5 Therefore, evaluation of alternative strategies for the design of standard peptides may provide additional tools for LC-MRM assessment of protein biomarkers and dissemination of assays to multiple laboratories.
Drawing on the literature for quantification of other types of molecules, different types of internal standards have already been explored as alternatives to SIS peptides, and the utility of these standards improves with increasing similarity between the standard and the endogenous molecule.17,18 Assay performance using structural analogs as standards is also improved when additional purification steps are used for sample preparation.19,20 Multiple stable isotope-labeled peptide internal standards have been concatenated into a single “protein” to enable evaluation of variability in both proteolytic digestion and instrument performance.21 Relative quantification has been achieved using one labeled reference peptide (LRP) for the normalization of all peptides in the LC-MRM analysis; statistical analysis indicated that relative quantification with matched SIS peptides resulted in the lowest CV values, but the LRP method was able to effectively quantify relative protein expression levels. 5 Based on extensive use in small molecule quantification, structural analogs have also been used as internal standards for peptide quantification (e.g. using a standard containing norleucine to quantify an endogenous peptide containing valine or exchanging cysteine residues for alanines in a standard).17, 22–26 In addition, commercially available naturally-occurring peptides with similar sequences or lengths have also been used as standards for bioactive peptide quantification.27–30 Sequence modification has also been explored as a strategy for creating peptide standards. A minimally permutated analog (MIPA) peptide is synthesized by switching the sequence position of two (usually adjacent) amino acids in the endogenous peptide. 31 Although the composition remains the same, the sequence modification produces peptides that have a shift in retention time and a change in the fragment ion masses detected. Quantification was reported by comparing either the total peak areas for peptides with retention time differences or comparing the intensities of fragment ion pairs that had mass differences due to the sequencechange for co-eluting peptides.31
Single conservative amino acid replacement (SCAR) peptides are another option for protein quantification using structural analogs. This strategy changes one amino acid side chain to incorporate the addition or subtraction of a methylene group and introduce a mass difference of 14 Da (e.g., glycine to alanine or vice versa). This type of strategy (using the difference of a methylene group) has been previously employed in chemical derivatization techniques for relative quantification in proteomics.32 Because these SCAR peptides are chemically similar to the endogenous peptides, they elute at similar retention times in reversed phase liquid chromatography and have similar fragmentation patterns. Because this strategy has been used in this laboratory primarily to supplement data from assays reliant on SIS peptides, the methods for assay development using SCAR peptides are reported with two examples to illustrate aspects of SCAR peptide design and explore the utility and limitations of these structural analogs. Quantification using SCAR peptides produces values for protein expression that are not significantly different from those obtained using assays based on SIS peptides for expression of epidermal growth factor receptor (EGFR) in digests of whole cell lysate and immunoglobulin M (IgM) in digests of serum acquired from multiple myeloma patients.
Materials and Methods
Peptide synthesis reagents were purchased from Protein Technologies, Inc. (Tucson, AZ), with the exception of N-Methyl-2-pyrrolidone (Applied Biosystems, Carlsbad, CA) and the stable isotope-labeled amino acids (Sigma-Aldrich, Milwaukee, WI). HPLC grade solvents were purchased from Burdick and Jackson (Honeywell, Muskegon, MI) and all other chemicals were acquired from Sigma-Aldrich (Milwaukee, WI).
Peptide Synthesis and Evaluation
Peptides were chosen for internal standard analysis from previous LC-MRM screening experiments. Human epidermal growth factor receptor (EGFR_HUMAN) and Immunoglobulin M (IGHM_HUMAN), were selected for comparison of quantification achieved using different types of internal standards. All of the selected peptides were between 7 and 25 amino acids in length with no cysteine or methionine residues. Stable isotope standards were synthesized using either heavy leucine (for EGFR) or heavy proline (for IgM). SCAR peptides were created by replacing amino acids, such as an alanine with glycine or a glutamic acid with aspartic acid (data not shown). All synthetic peptides were checked for uniqueness within the SwissProt human proteome database using the iPIP program, which was written in-house.33
Peptides were synthesized at the 25 μmole scale (Symphony, Protein Technologies, Tucson, AZ) using standard FMOC chemistry.34 Three peptides were synthesized for each analysis: the endogenous species, and two internal standard peptides, one using a stable isotope-labeled amino acid and a second using a single conservative amino acid replacement. All peptides were purified and characterized as previously described.35 The concentration of each peptide solution was determined by amino acid analysis using 6N HCl-phenol hydrolysis, derivatization, and detection after reversed phase chromatography (AminoQuant, HP).
Mass spectrometry evaluation for each of the synthetic peptides was performed on a triple quadrupole mass spectrometer (TSQ Quantum Ultra, Thermo, San Jose, CA). Peptide solutions (1μM) were infused at 300 nl/minute in 30% aqueous acetonitrile with 0.1% formic acid. The scan time was set for 1 second with Q1 resolution at 0.4 and Q3 resolution at 0.7. For full scan MS/MS, the collision energy was theoretically calculated using Equation 1,
| Equation 1 |
where CE is collision energy and m/z represents the doubly charged mass-to-charge ratio. CE values were then tuned manually as necessary to optimize fragmentation. Data were analyzed using QuanBrowser (Xcalibur 2.0.7, Thermo, San Jose, CA). Peak m/z and intensity values were extracted for each daughter ion (y1 to y(n-1) and b1 to b(n-1)) every 10 seconds of data acquisition time after Gaussian smoothing was applied to the spectra. To compare the distribution of N- and C-terminal charge retention, all of the daughter ion intensities were summed and the percentages of b- and y-ions were calculated. Because these experiments used trypsin for digestion, the y-ions were then further analyzed by calculating the percentage of the total y-ion signal detected for each y ion. Both of these calculations were statistically analyzed by ANOVA to determine if the SIS and SCAR peptides presented similar fragmentation patterns to the endogenous species (α = 0.05). After initial data acquisition, optimization of collision energies was run on all fragment ions for each peptide, these values were compared between the endogenous, SCAR, and SIS peptides; then, the same optimized collision energy was used for all three peptides. Fragment ions (n = 3 to 6) were selected for monitoring based on intensity and selectivity.
Preparation of HCC827 Cell Lysates and Serum for LC-MRM
Cells (n = 107) from the EGFR mutant human non-small-cell lung cancer cell line HCC827 were lysed with aqueous 8 M urea buffered in 100 mM ammonium bicarbonate on ice, sonicated, and centrifuged at 14, 000 x g for 10 minutes. After lysis in denaturing buffer, proteins were reduced with 2 mM tris(2-carboxyethyl) phosphine and alkylated with 20 mM iodoacetamide. The denatured cell lysate was then diluted (10-fold) in ammonium bicarbonate and digested overnight with trypsin at 37¼C. The final digest of HCC827 lysate was then diluted to 4 different concentrations in 2% aqueous acetonitrile containing 0.1% formic acid and spiked with both the SIS and SCAR peptides for EGFR, such that 10 fmol is injected for each analysis. The samples were analyzed in triplicate.
According to the standard of care for myeloma patients, human serum samples were acquired from excess remaining after completion of diagnostic and prognostic measurements, as approved by the Institutional Review Board at the University of South Florida. Serum proteins were denatured, reduced, alkylated, and digested in solution, as described above. Samples were further diluted with 2% aqueous acetonitrile containing 0.1% formic acid and spiked with both the SIS and SCAR internal standards for IgM, such that 0.5 nl of serum and 10 fmol of standard peptides were injected for each analysis.
LC-MRM Data Acquisition and Analysis
A dilution series was created for all of the synthetic peptides ranging in concentration from 100 attomoles to 25 femtomoles per injection. Samples were run in triplicate on a nanoLC system (EasynLC, Proxeon, Denmark) interfaced with a triple quadrupole mass spectrometer (Vantage, Thermo Scientific, San Jose CA) fitted with a nanospray ion source. Separations were achieved with C18 reversed phase chromatography using a trap column (100 μm ID and 2 cm in length packed with 5 μm particles, Nanoseparations, The Netherlands) and an analytical column (75μm ID and 15 cm in length packed with 3 μm particles, Nanoseparations, The Netherlands). A 20 minute gradient was set up from 5% to 50% B solvent using the following solvent system: A) aqueous 2% acetonitrile with 0.1% formic acid and B) aqueous 90% acetonitrile with 0.1% formic acid). The following instrument parameters were used 2,500 V spray voltage, 10 V skimmer offset, and 210 ¼C transfer tube temperature. Peptide precursors were selected with a Q1 resolution of 0.4; the resulting fragment ions were selected in Q3 at a resolution of 0.7. The scan width was set to 0.002, and each transition was acquired for 20 milliseconds. The resulting data was analyzed using Skyline 1.1.36 Total peak area for each peptide was extracted for comparison. The ratio of the endogenous peptide to each internal standard (SIS or SCAR) was calculated to determine the concentration of the endogenous peptide. The results were statistically analyzed comparing the endogenous concentration obtained by SIS vs. SCAR for each peptide first by an f-test to determine the presence of equal variances and then by a two-tailed t-test (α = 0.05).
Results
Comparison of Reversed Phase Retention of Endogenous, SIS, and SCAR Peptides
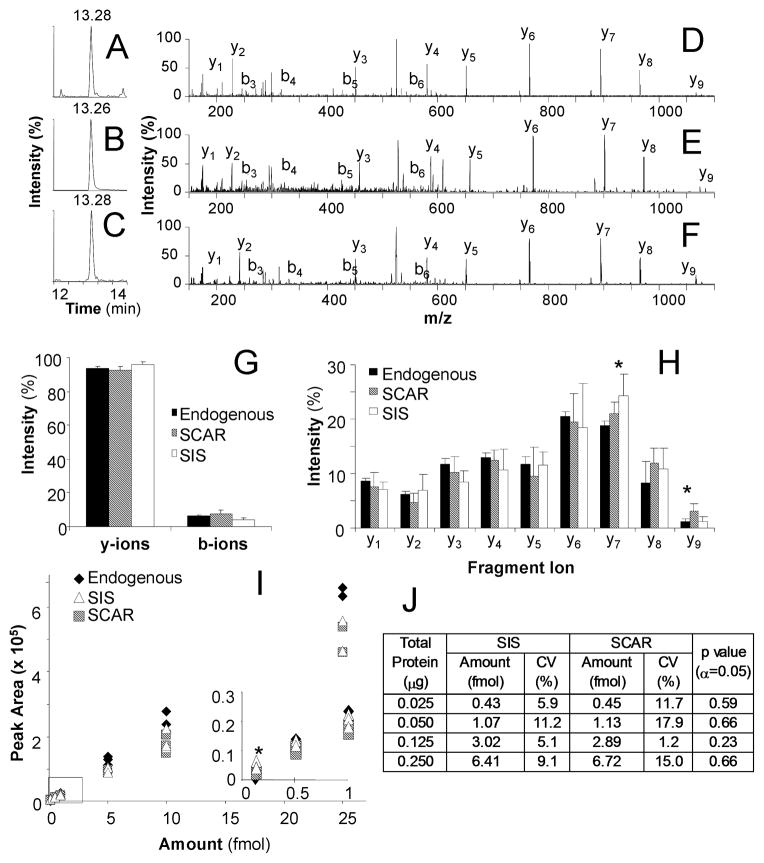
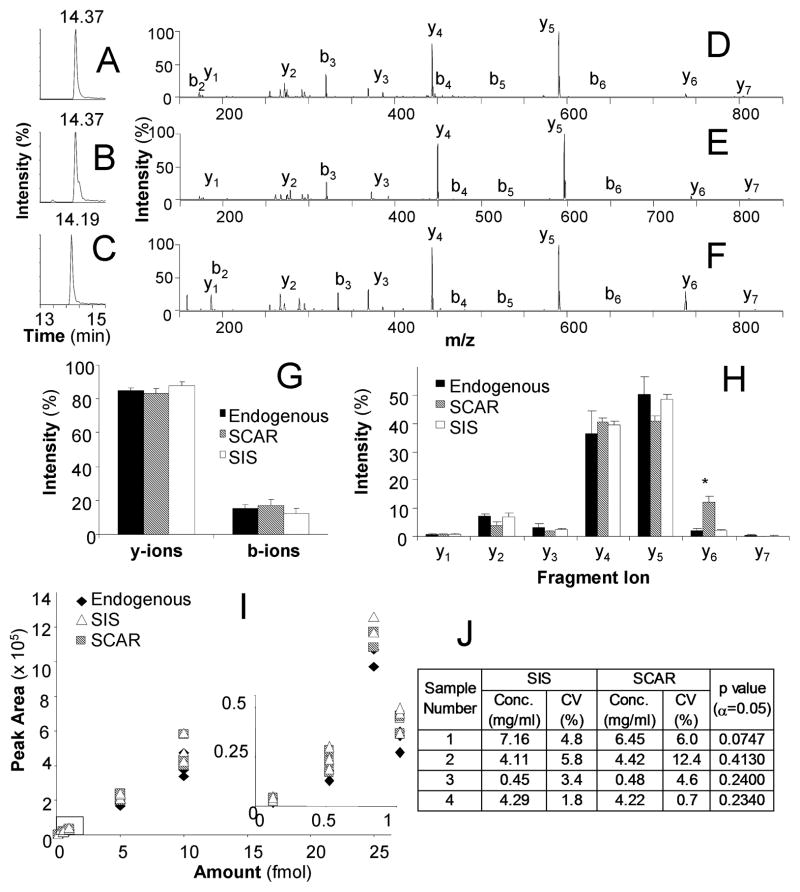
All peptides were analyzed by LC-MRM using samples containing 100 attomoles to 25 femtomoles. Similar to deuterated standards,37 the SCAR peptides often have differences in reversed phase HPLC retention time from the endogenous peptide. The average retention times for the EGFR peptides were as follows: 13.25 minutes for the endogenous peptide, 13.25 minutes for the SIS peptide, and 13.27 for the SCAR peptide (Figure 1A–C). Average retention times for the IgM peptides were 14.36 minutes for the endogenous peptide, 14.36 minutes for the SIS peptide, and 14.21 minutes for the SCAR peptide (Figure 2A–C). In both cases, the eluting SCAR peptide peaks overlap with the endogenous and SIS peptides. The peptides’ retention times remain similar even with longer gradients. Using 60 minute gradients, the difference in elution between the EGFR peptides was 0.1 minutes and between the IgM peptides was 0.6 minutes. The differences increased to 0.4 minutes (EGFR) and 0.9 minutes (IgM), when using 120 minute gradients. If the endogenous and standard peptides are monitored as a pair, the window for scheduled LC-MRM would need to be expanded to accommodate these differences in retention time, resulting in a decrease in peak capacity. If each is scheduled independently, no decrease in peak capacity occurs. However, if the user wants to detect the endogenous peptide and the structural analog over the same time window, the strategy that would least effect the peak capacity would center the window between the elution times of the two peptides.
Figure 1. Quantification of Endogenous EGFR in HCC827 NSCLC Cells using SIS and SCAR Peptides.
LC-MRM total ion chromatograms for the endogenous peptide (A), SIS peptide (B), and SCAR peptide (C). MS/MS data averaged over one minute of acquisition time for the endogenous peptide (D), SIS peptide (E), and SCAR peptide (F). The distribution of charge retention is plotted using b vs. y-ion signal for the three peptides (G), then y-ion data were further analyzed to compare individual transitions. All transitions were similar except for y7 in which the SIS peptide showed higher peak intensity, and y9 in which the SCAR peptide showed higher intensity (H). A dilution series of the peptide set shows similar peak areas for all three peptides, resulting in accurate absolute quantification by each internal standard (I). EGFR expression in lysate from the HCC827 lung cancer cell line was analyzed by both internal standards; no significant difference was found between the two calculations (J).
Figure 2. Quantification of Endogenous IgM in Serum using SIS and SCAR Peptides.
LC-MRM total ion chromatograms for the endogenous peptide (A), SIS peptide (B), and SCAR peptide (C). MS/MS data averaged over one minute of acquisition time for the endogenous peptide (D) SIS peptide (E) and SCAR peptide (F). The distribution of charge retention is plotted using b vs. y-ion signal for the three peptides (G), and y-ion data were further analyzed to compare individual transitions. All transitions were similar except for y6 in which the SCAR peptide showed higher intensity and y5 for which the SCAR peptide had lower intensity (H). A dilution series of the peptide set shows similar peak areas for all three peptides, resulting in accurate absolute quantification by each internal standard (I). The expression of IgM was measured by both internal standards in serum samples from 4 multiple myeloma patients; no difference was found between the results calculated using the SIS and SCAR peptides (J).
QqQ MS and MS/MS of Infused Peptides
The first step in comparison of synthetic endogenous, SIS, and SCAR peptides is infusion on the triple quadrupole mass spectrometer. For the peptides reported here, MS analysis detected the same charge state distributions for the endogenous, SIS, and SCAR peptides in each case. For the peptides from EGFR and IgM, the doubly protonated species was detected as the only peak. In addition, the amount of ion signal observed for each peptide was similar for both the EGFR and IgM test cases.
Product ion scans acquired during direct infusion indicated similarities and differences in the fragmentation of the endogenous, SIS, and SCAR peptides. In these examples, little difference is noted between the fragmentation pattern of SCAR peptides when compared to both endogenous and SIS peptides. When differences are noted, they are usually restricted to the fragment ions created by cleavage of the bonds adjacent to the amino acid replacement. Tandem mass spectra are shown for the sequences representing the endogenous EGFR peptide, GSTAENAEYLR, the SIS peptide, GSTAENAEY(13C615N)LR, and the SCAR peptide, ASTAENAEYLR, are shown in Figure 1D–F, respectively. Tandem mass spectra are shown for the endogenous IgM peptide, DGFFGNPR, the SIS peptide, DGFFGN(13C515N)PR, and the SCAR peptide, DAFFGNPR, in Figure 2D–F, respectively.
To examine the fragmentation patterns more quantitatively, the fraction of ions produced with N-terminal and C-terminal charge retention is calculated for each peptide, using the total signal of the b- and y-ions observed in 6 tandem mass spectra. For EGFR (Figure 1G) and IgM (Figure 2G) peptides, no significant differences were found in the overall distribution of fragment ion charge retention (p > 0.05). Because the b-ion signals had low intensity in these examples and no b-ions would be selected for LC-MRM, further quantitative analysis of fragmentation patterns was only performed for y-ions. For the EGFR peptides, one significant difference in y-ion signal distribution was observed when the SCAR peptide was compared to either the endogenous or SIS peptides (Figure 1H). The y9 fragment ion signal was higher intensity in the SCAR peptides, when compared with the endogenous peptide (2.5-fold, p = 0.0056) and the SIS peptide (2.6-fold, p = 0.0087). When comparing the SIS to the endogenous peptide, one significant difference was noted for the level of the y7 fragment ion (1.3-fold higher signal, p = 0.007). The other fragment ions were not significantly different. The SCAR peptides for IgM had decreased ion signal for the y5 fragment ion (p = 2.7E-5) and increased signal for the y6 fragment ion (p = 8.0E-5) when compared to endogenous and SIS peptides, as shown in Figure 2H. No differences were noted between the endogenous and SIS peptides for IgM. Selection of transitions should balance the increases and decreases in fragment ion signals to reduce or eliminate these effects.
All y-ion transitions with m/z values greater than that of the doubly protonated peptide were selected for further monitoring. Optimized collision energy values were the same for the endogenous, SIS, and SCAR peptides in these examples. For the EGFR peptides, five fragment ions were monitored. These transitions corresponded to 60.6 ± 1.5% (endogenous), 65.0 ± 4.5% (SIS), and 66.4 ± 5.1% (SCAR) of the total y-ion signal. These values were not found to be significantly different (p = 0.06). For the IgM peptides, three transitions were monitored, which corresponded to 88.8 ± 2.5% (endogenous), 90.0 ± 1.4% (SIS), and 92.0 ± 1.5% (SCAR) of the total y-ion signal. The differences in these values had minimal statistical significance (p = 0.04). In both cases, a correction factor based on the ratio of percentages of total y-ion signal between the endogenous and SCAR peptide could be used to normalize the ion signals, if desired. However, no correction factor is used in these examples.
LC-MRM Calibration Curves
In both EGFR (Figure 1I) and IgM calibration curves (Figure 2I), all three peptides were detected in all samples and no differences were noted for limit of detection or limit of quantification. Total peak area was derived from the sum of each area under the curve for all of the transitions monitored. Using ANOVA, the endogenous peptide peak area was compared with the SIS peak area and the SCAR peak area at each point in the calibration curve. For the EGFR peptides, only one point on the calibration curve, 100 amol, was significantly different between the three peptides; the endogenous peptide had lower intensity than both the SIS and SCAR peptides (p < 0.003). This variation in peak area was most likely due to the low signal-to-noise ratios observed at that injected amount. For the IgM peptides, no significant differences were found in the total peak areas of the three peptides (p > 0.05) at any amount injected.
Implementation in Biological Samples
EGFR expression levels were quantified in cell lysate from HCC827 lung cancer cells. Different amounts of digested lysate ranging from 0.025 to 0.25 μg of total protein were spiked with SIS and SCAR peptides, and analyzed with LC-MRM. Quantification using the SIS and SCAR internal standards produced similar results for EGFR expression in fmol for each sample (p > 0.05), as listed in Figure 1J. On average, CV values for the assay based on SCAR peptide were 1.4-fold higher than for the SIS peptide.
The concentration of IgM was determined using both SIS and SCAR peptides in four separate samples acquired from multiple myeloma patients (Figure 2J). In all cases, no significant differences were noted the values for the concentration (in mg/ml) of endogenous peptide calculated using either SIS or SCAR peptides (p > 0.05). When compared with the use of SIS peptides, the average CV value for IgM measurements was 1.3-fold higher when using assays dependent on SCAR peptides.
Discussion
Assessment of Advantages and Limitations in the SCAR Strategy
The use of SCAR peptides has some advantages. It enables straightforward automated synthesis, whereas stable isotope-labeled amino acids are manually inserted into the peptide sequence. In certain cases, the reduction of the side chain length can improve the results of synthesis of peptides with hydrophobic regions. This strategy also allows for the synthesis of peptide standards that can be used for estimation of protein copy number in conjunction with stable isotope-labeling strategies for relative quantification (e.g., stable isotope-labeling with amino acids in cell culture, SILAC38) or with stable isotope-labeled protein standards spiked into the biological matrix to control for all sample preparation and processing steps. Because of the 14 Da mass tag, the SCAR peptides will not overlap with the endogenous or the stable isotope-labeled peptides. Also, evaluation of these changes in peptide sequence gives some insight into the potential differences between peptides with highly similar sequences (e.g. mutated sequences or highly homologous peptides). We are also exploring the use of two SCAR standards to bracket the endogenous peptide peak area to provide more accurate quantification than single point intensity calibration methods.
The use of SCAR peptides can be limited in certain cases, because they are structural analogs and because the mass tags are incorporated using naturally-occurring amino acids. Structural analogs may not have the same stability or degradation as the endogenous peptides, as previously noted during the comparison of assays for angiotensin IV.17 They may also differ in ionization efficiency, but differences in ion signal have not been noted for the peptide analogs used in this study. If they do not co-elute in LC-MRM; the endogenous and SCAR peptides may have differences in ion suppression due to the background matrix, which may explain the differences between the CV values for the assays based on the SIS and SCAR peptides. In these two examples, the endogenous proteins have fairly high abundance (> 0.1% of the total protein), and the peptides have overlapping retention profiles; both of these factors may be reasons why the assay based on the structural analog does not lose much in performance when compared to the SIS peptide.
Because the amino acid replacement strategy is flexible in the site of tag incorporation, the SCAR strategy method has been effective for the development of more than 60 assays in this laboratory. However, specific elements of peptide design must be taken into account, as described below. First, SCAR standards must be carefully selected for peptides from a protein group that has high sequence homology, like the Src family kinases. LIEDNEYTAR (Fyn, Lck, Src, and Yes) and VIEDNEYTAR (Hck and Lyn) differ only by the N-terminal amino acid, which would be similar to one potential SCAR labeling strategy. However, the alanine in LIEDNEYTAR can be replaced with glycine, creating a peptide (LIEDNEYTGR) with the same molecular weight as VIEDNEYTAR, but different y-ion transitions. BLAST searching potential sequences for each standard peptide against the proteome eliminates this issue; this analysis is parallel to the determination of uniqueness for endogenous peptides prior to assay development. Second, the SCAR strategy would have to account for known sequence variants. As an example, the SCAR peptide for the lower mass endogenous variant could be reduced in mass, while the SCAR peptide for the higher mass endogenous variant could be increased in mass to create standards that would not interfere with the endogenous peptides. In a case where the variant had not been observed before, the SCAR peptide could have same sequence as a naturally-occurring variant (e.g., changing glycine to alanine could occur with single point mutation altering the mRNA codon from GGX to GCX, where X indicates any ribonucleotide). The peptide representing that endogenous variant could be measured using the transitions designated for the SCAR peptide, but it would be difficult to accurately quantify by subtraction of the signal for the standard. If the SCAR standard was spiked in at levels significantly higher than the endogenous expression; a small amount of ion signal corresponding to the endogenous variant peptide would likely go unnoticed as a minor variation in the standard peak area. Third, SCAR peptides incorporating a tag that increases their mass would be inappropriate for monitoring peptides containing methylation sites, because the standard would be isobaric with the modified peptide, which could create interference. Therefore, SIS peptides would be more appropriate for monitoring methylation. Finally, the performance of the SCAR peptide has to be compared to the endogenous peptide for each analytical step. SCAR peptides can be evaluated without changes to our current workflow for assay development, which includes infusion for collision energy optimization and calibration curves to estimate limits of detection and quantification. Other experiments would need to be added when the workflow involves additional processing (e.g., strong cation exchange chromatography in f-MRM39 or immunoprecipitation in SISCAPA40 or iMALDI41). In immunoprecipitation, the effect of amino acid replacement could be highly variable depending on the importance of the residue in the antigenicity; a SCAR strategy could be devised to minimize the impact of the amino acid replacement.42
Conclusions
The aim of this study was to assess the utility of single conservative amino acid replacements (SCAR) as mass tags for creating peptide standards for protein quantification experiments. Because this approach has been used in our laboratory to supplement data generated by quantification based on SIS peptides, a method for evaluation of peptide standards and compensation for differences in ion signal in MS and fragmentation pattern in MS/MS has been developed for SCAR peptides. In two examples monitoring proteins in cell lysate and serum, the measured values for protein expression were similar for assays developed using SCAR and SIS peptides, but the CV values were 1.3 to 1.4-fold higher for the structural analogs when compared to the SIS peptides. Finally, the use of SCAR peptides as secondary standards can augment experiments using SILAC or SIS proteins and allow for measurement of protein expression levels.
Acknowledgments
Funding for this study was received from the National Cancer Institute, R21-CA141285, and a Career Development Project from the Moffitt Lung SPORE, P50-CA119997. This work and the Moffitt Proteomics Facility are supported by the US Army Medical Research and Materiel Command under Award No. DAMD17-02-2-0051 for a National Functional Genomics Center, the National Cancer Institute under Award No. P30-CA076292 as a Cancer Center Support Grant, and the Moffitt Foundation. The Bankhead-Coley Cancer Research program of the Florida Dept. of Health for purchase of the liquid chromatographs and the triple quadrupole mass spectrometers (06BS-02-9614 and 09-BE04-SIG). Amino acid analysis was performed by Virginia Johnson and Lawrence Dangott at the Texas A&M University Protein Chemistry Laboratory. The authors would like to thank Jiannong Li and Eric Haura for the HCC827 lung cancer cells and Karin Burgess and Kaaron Benson for their help in acquiring serum samples.. Multiple myeloma patients’ serum samples were acquired and analyzed under a protocol approved by the Institutional Review Board at the University of South Florida. The authors would like to thank Eric Welsh for preparation of high resolution figures for publication.
References
- 1.Barr JR, Maggio VL, Patterson DG, Jr, Cooper GR, Henderson LO, Turner WE, Smith SJ, Hannon WH, Needham LL, Sampson EJ. Isotope dilution-mass spectrometric quantification of specific proteins: model application with apolipoprotein A-1. Clinical Chemistry. 1996;42:1676–1682. [PubMed] [Google Scholar]
- 2.Gerber SA, Rush J, Stemman M, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proceedings of the National Academy of Sciences. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnidge DR, Dratz EA, Martin T, Bonilla LE, Moran LB, Lindall A. Absolute quantification of the G protein-coupled receptor rhodopsin by LC/MS/MS using proteolysis product peptides and synthetic peptide standards. Anal Chem. 2003;75:445–451. doi: 10.1021/ac026154+. [DOI] [PubMed] [Google Scholar]
- 4.Bluemlein K, Rasler M. Monitoring protein expression in whole-cell extracts by targeted label- and standard-free LC-MS/MS. Nature Protocols. 2011;6:859–869. doi: 10.1038/nprot.2011.333. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Liu Q, Zimmerman LJ, Ham AL, Slebos RJC, Rahman J, Kikuchi T, Massion PP, Carbone DP, Billheimer D, Leibler DC. Methods for protein and peptide quantification by liquid chromatography-multiple reaction monitoring mass spectrometry. Molecular and Cellular Proteomics. 2011;10:M110.006593. doi: 10.1074/mcp.M110.006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John H, Walden M, Schäfer S, Genz S, Forssmann WG. Analytical procedures for quantification of peptides in pharmaceutical research by liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2004;378:883–897. doi: 10.1007/s00216-003-2298-y. [DOI] [PubMed] [Google Scholar]
- 7.van den Broek I, Sparidans RW, Schellens JH, Beijnen JH. Quantitative bioanalysis of peptides by liquid chromatography coupled to (tandem) mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;872:1–22. doi: 10.1016/j.jchromb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Vogeser M, Seger C. Pitfalls associated with the use of liquid chromatography-tandem mass spectrometry in the clinical laboratory. Clin Chem. 2010;56:1234–1244. doi: 10.1373/clinchem.2009.138602. [DOI] [PubMed] [Google Scholar]
- 9.Kettenbach AN, Rush J, Gerber SA. Absolute quantification of proteins and post-translational modification abundance with stable isotope-labeled synthetic peptides. Nature Protocols. 2011;6:175–186. doi: 10.1038/nprot.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott MH, Smith DS, Parker CE, Borchers C. Current trends in quantitative proteomics. Journal of Mass Spectrometry. 2009;44:1637–1660. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- 11.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–88. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Kuzyk MA, Smith D, Yang J, Cross TJ, Jackson AM, Hardie DB, Anderson NL, Borchers CH. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics. 2009;8:1860–77. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Gruidl M, Remily-Wood E, Liu RZ, Eschrich S, Lloyd M, Nasir A, Bui MM, Huang E, Shibata D, Yeatman T, Koomen JM. Quantification of b-catenin signaling components in colon cancer cell lines, tissue sections, and microdissected tumor cells using reaction monitoring mass spectrometry. J Proteome Res. 2010;9:4215–4227. doi: 10.1021/pr1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G, Fang B, Liu RZ, Lin H, Kinose F, Bai Y, Oquz U, Remily-Wood ER, Li J, Altiok S, Eshrich S, Koomen J, Haura EB. Mass spectrometry mapping of epidermal growth factor receptor phosphorylation related to oncogenic mutations and tyrosine kinase inhibitor sensitivity. J Proteome Res. 2011;10:305–319. doi: 10.1021/pr1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin LL, Tong J, Prakash A, Peterman SM, St-Germain JR, Taylor P, Trudel S, Moran MF. Measurement of protein phosphorylation stoichiometry by selected reaction monitoring mass spectrometry. J Proteome Res. 2010;9:2752–2761. doi: 10.1021/pr100024a. [DOI] [PubMed] [Google Scholar]
- 17.Lanckmans K, Sarre S, Smolders I, Michotte Y. Use of a structural analogue versus a stable isotope labeled internal standard for the quantification of angiotensin IV in rat brain dialysates using nano-liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:1187–1195. doi: 10.1002/rcm.2950. [DOI] [PubMed] [Google Scholar]
- 18.Stokvis E, Rosing H, Beijnen JH. Stable isotopically labeled internal standards in quantitative bioanalysis using liquid chromatography/mass spectrometry: necessity or not? Rapid Commun Mass Spectrom. 2005;19:401–407. doi: 10.1002/rcm.1790. [DOI] [PubMed] [Google Scholar]
- 19.Fu I, Woolf EJ, Matuszewski BK. Effect of the sample matrix on the determination of indinavir in human urine by HPLC with turbo ion spray tandem mass spectrometric detection. J Pharm Biomed Anal. 1998;18:347–357. doi: 10.1016/s0731-7085(98)00048-x. [DOI] [PubMed] [Google Scholar]
- 20.Sancho JV, Pozo OJ, López FJ, Hernández F. Different quantitation approaches for xenobiotics in human urine samples by liquid chromatography/electrospray tandem mass spectrometry. Rapid Commun Mass Spectrom. 2002;16:639–645. doi: 10.1002/rcm.617. [DOI] [PubMed] [Google Scholar]
- 21.Kito K, Ota K, Fujita T, Ito T. A synthetic approach towards accurate mass spectrometric quantification of component stoichiometry of multiprotein complexes. J Prot Res. 2007;6:792–800. doi: 10.1021/pr060447s. [DOI] [PubMed] [Google Scholar]
- 22.Prokai L, Zharikova AD, Janáky T, Prokai-Tatrai K. Exploratory pharmacokinetics and brain distribution study of a neuropeptide FF antagonist by liquid chromatography/ atmospheric pressure ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2000;14:2412–2418. doi: 10.1002/1097-0231(20001230)14:24<2412::AID-RCM180>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 23.van den Broek I, Sparidans RW, Schellens JH, Beijnen JH. Liquid chromatography/tandem mass spectrometric method for the quantification of eight proteolytic fragments of ITIH4 with biomarker potential in human plasma and serum. Rapid Commun Mass Spectrom. 2008;22:2915–2928. doi: 10.1002/rcm.3695. [DOI] [PubMed] [Google Scholar]
- 24.Yang JZ, Bastian KC, Moore RD, Stobaugh JF, Borchardt RT. Quantitative analysis of a model opioid peptide and its cyclic prodrugs in rat plasma using high-performance liquid chromatography with fluorescence and tandem mass spectrometric detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;780:269–281. doi: 10.1016/s1570-0232(02)00536-6. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi K, Takashima M, Uchimura T, Kobayashi S. Development of a sensitive liquid chromatography-electrospray ionization mass spectrometry method for the measurement of KW-5139 in rat plasma. Biomed Chromatogr. 2000 Apr;14(2):77–81. doi: 10.1002/(SICI)1099-0801(200004)14:2<77::AID-BMC928>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 26.Oosterkamp AJ, Gelpí E, Abian J. Quantitative peptide bioanalysis using column-switching nano liquid chromatography/mass spectrometry. J Mass Spectrom. 1998;33:976–983. doi: 10.1002/(SICI)1096-9888(1998100)33:10<976::AID-JMS710>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Feng WY, Chan KK, Covey JM. Electrospray LC-MS/MS quantitation, stability, and preliminary pharmacokinetics of bradykinin antagonist polypeptide B201 (NSC 710295) in the mouse. J Pharm Biomed Anal. 2002;28:601–612. doi: 10.1016/s0731-7085(01)00697-5. [DOI] [PubMed] [Google Scholar]
- 28.Murphy AT, Witcher DR, Luan P, Wroblewski VJ. Quantitation of hepcidin from human and mouse serum using liquid chromatography tandem mass spectrometry. Blood. 2007;110:1048–1054. doi: 10.1182/blood-2006-11-057471. [DOI] [PubMed] [Google Scholar]
- 29.Fanciulli G, Azara E, Wood TD, Dettori A, Delitala G, Marchetti M. Quantification of Gluten Exorphin A5 in cerebrospinal fluid by liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833:204–209. doi: 10.1016/j.jchromb.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 30.Hatziieremia S, Kostomitsopoulos N, Balafas V, Tamvakopoulos C. A liquid chromatographic/tandem mass spectroscopic method for quantification of the cyclic peptide melanotan-II. Plasma and brain tissue concentrations following administration in mice. Rapid Commun Mass Spectrom. 2007;21:2431–2438. doi: 10.1002/rcm.3106. [DOI] [PubMed] [Google Scholar]
- 31.Winter D, Seidler J, Kugelstadt D, Derrer B, Kappes B, Lehmann W. Minimally permutated peptide analogs as internal standards for relative and absolute quantitification of peptides and proteins. Proteomics. 2010;10:1510–1514. doi: 10.1002/pmic.200900695. [DOI] [PubMed] [Google Scholar]
- 32.Beardsley RL, Reilly JP. Quantitation using enhanced signal tags: a technique for comparative proteomics. J Proteome Res. 2003;2:15–21. doi: 10.1021/pr0255506. [DOI] [PubMed] [Google Scholar]
- 33.IPIP Download. available at http://proteome.moffitt.org/
- 34.Carpino L, Han G. 9-Fluorenylmethoxycarbonyl amino-protecting group. J Org Chem. 1972;37:3404–3409. [Google Scholar]
- 35.Remily-Wood ER, Liu RZ, Xiang Y, Chen Y, Thomas CE, Rajyaguru N, Kaufman LM, Ochoa JE, Hazlehurst L, Pinilla-Ibarz J, Lancet J, Zhang G, Haura E, Shibata D, Yeatman T, Smalley KS, Dalton WS, Huang E, Scott E, Bloom GC, Eschrich SA, Koomen JK. A database of reaction monitoring mass spectrometry assays for elucidating therapeutic response in cancer. Proteomics Clin Appl. 2011;5:383–96. doi: 10.1002/prca.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster LJ, de Hoog CL, Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc Natl Acad Sci USA. 2003;100:5813–18. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 39.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 41.Jiang J, Parker CE, Fuller JR, Kawula TH, Borchers CH. An immunoaffinity tandem mass spectrometry (iMALDI) assay for detection of Francisella tularensis. Anal Chim Acta. 2007;605:70–99. doi: 10.1016/j.aca.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki M, Takeda S, Kato T, Matsuba K. Antigenicities of stem bromelain. Contribution of three-dimensional structure and individual amino acid residues. J Biochem. 1980;87:817–824. doi: 10.1093/oxfordjournals.jbchem.a132811. [DOI] [PubMed] [Google Scholar]