Abstract
Biofilms cause much of all human microbial infections. Attempts to eradicate biofilm-based infections rely on disinfectants and antibiotics. Unfortunately, biofilm bacteria are significantly less responsive to antibiotic stressors than their planktonic counterparts. Sublethal doses of antibiotics can actually enhance biofilm formation. Here, we have developed a non-invasive microscopic image analyses to quantify plasmid conjugation within a developing biofilm. Corroborating destructive samples were analyzed by a cultivation-independent flow cytometry analysis and a selective plate count method to cultivate transconjugants. Increases in substrate loading altered biofilm 3-D architecture and subsequently affected the frequency of plasmid conjugation (decreases at least two times) in the absence of any antibiotic selective pressure. More importantly, donor populations in biofilms exposed to a sublethal dose of kanamycin exhibited enhanced transfer efficiency of plasmids containing the kanamycin resistance gene, up to tenfold. However, when stressed with a different antibiotic, imipenem, transfer of plasmids containing the kanR+ gene was not enhanced. These preliminary results suggest biofilm bacteria “sense” antibiotics to which they are resistant, which enhances the spread of that resistance. Confocal scanning microscopy coupled with our non-invasive image analysis was able to estimate plasmid conjugative transfer efficiency either averaged over the entire biofilm landscape or locally with individual biofilm clusters.
Keywords: Biofilm plasmid transfer, Non-invasive biofilm diagnostics, Antibiotic selection pressure, Confocal microscopy
Introduction
Nosocomial (hospital acquired) infections are the fourth leading cause of death in the US with two million cases annually (or ~10 % of American hospital patients) leading to more than $30 billion in added medical cost per annum (Klevens et al. 2007; Scott 2009). About 60–70 % of nosocomial infections are associated with some type of implanted medical device. It is estimated that over five million medical devices or implants are used per annum in the US alone. The medical profession has attempted to eradicate biofilm-based infections by resorting to disinfectants and antibiotics; mostly synthetic compounds selected for their ability to inactivate or kill suspended bacteria. Biofilm bacteria are significantly less responsive to antibiotics and antimicrobial stressors than planktonic organisms of the same species (Gilbert et al. 2002; Stewart 2002). Further, recent studies have shown that sublethal doses of antibiotics can actually enhance biofilm formation. It was reported that genes coding for alginate biosynthesis were induced by exposure to the β-lactam antibiotic, imipenem, which caused structural changes in the biofilm, e.g., an increased biofilm volume and alginate polymer matrix (Bagge et al. 2004). Similarly, subinhibitory concentrations of tobramycin induced biofilm formation in Pseudomonas aeruginosa and Escherichia coli (Hoffman et al. 2005). We have also observed this enhanced biofilm formation response for Pseudomonas putida biofilms exposed to sublethal doses of kanamycin, ciprofloxacin, and norfloxacin (Ma and Bryers 2010). These results suggest enhanced biofilm formation in the presence of antibiotics may be one universal defense mechanism of bacteria in avoiding the lethal effects of antibiotics. A further complication is that antibiotic resistance genes typically reside on plasmid DNA. Being bound within a biofilm may affect plasmid gene expression and frequency of pDNA conjugative transfer between bacteria. This is important since plasmid-bearing bacteria, in a biofilm, may transfer genes and phenotypes (e.g., disinfectant and antibiotic resistance) more readily to neighboring bacteria (Huang et al. 1993, 1994).
Horizontal gene transfer (HGT) is considered to be the most dominant mechanism for bacteria to rapidly adapt to changing environments by spreading genetic information (Ashelford 1998; Dahlberg et al. 1998; Ravatn et al. 1998). Effects of environmental selection pressure on conjugational plasmid transfer have gained considerable interest recently because it is now generally realized that “selection-free” situations are hard to achieve (Top et al. 1995; Hohnstock et al. 2000; Pinedo and Smets 2005; Slater et al. 2008). It has been suggested that environmental selection stress should increase plasmid conjugational transfer (Pinedo and Smets 2005). However, both positive and negative correlations between selection stresses and plasmid conjugational transfer frequencies have been observed in experimental systems exposed to different selective pressures (e.g., antibiotics, ethanol, or sodium dodecyl sulfate; Edwards et al. 1999; Slater et al. 2008, 2010). This stimulation or depression of plasmid conjugation is most likely due to the different transcription levels of gene expression (Goh et al. 2002). Increasing attention is being paid to the impact of subinhibitory levels of antibiotics on conjugational resistance gene transfer in natural environment, since long-term persistence of antibiotics in polluted environment may have the potential to increase biofilm formation, enhanced gene transfer, and promote that spread of antibiotic resistant phenotypes (Hausner and Wuertz 1999; Ingerslev et al. 2001). Similarly, recalcitrant hazardous wastes (at concentrations that would negatively impact cell growth) could affect the transfer of plasmids that carry catabolic genes or the transfer of plasmids that carry heavy metal resistance genes could be enhanced in metal-laden environments. Recent studies have quantified those parameters that may accelerate the degradation of specific pollutants by bioremediation or bioaugmentation, using recombinant bacteria containing plasmids with genes coded for the degradation of pollutants, such as the TOL plasmid used in this study, containing genes coded for the degradation of toluene and benzyl alcohol (Boon et al. 2002; Nancharaiah et al. 2003). To date, little quantitative data exists regarding plasmid retention, transfer, and expression in mixed culture biofilm ecosystems. Classical selective plate counting approaches to quantify plasmid transfer in planktonic cells fail miserably when applied to biofilm communities since they provide only biofilm-averaged information, not spatially dynamic data.
To quantify the local instantaneous frequency of plasmid conjugative transfer, we developed non-invasive microscopic analyses that can estimate plasmid conjugational transfer in optical “slices” taken horizontally (x–y plane) with depth (z-plane) across either an entire biofilm landscape or an individual biofilm cluster (Edwards et al. 1999). TOL conjugational plasmid transfer efficiency was quantified as a function of different limiting nutrient concentrations or in the presence of selection pressures. The non-invasive method developed in this study provided quantitative data of local and overall conjugational plasmid transfer in mixed culture.
Materials and methods
Bacterial strains, plasmids, and media
P. putida KT2442 strains were used throughout this study. All strains are listed in Table 1. P. putida KT2442 contains the mini-Tn7 transposon system, expressing different mutant variants of the green fluorescent protein (GFP), using the E. coli lac promoter, PA1/04/03. All other remaining strains were donated courtesy of Dr. Barth Smets (Technical University Denmark). Two of these P. putida KT2442 strains express two different fluorescent proteins, GFP and DsRed. The TOL (pWWO) plasmid is a well-characterized plasmid that codes for the degradeation of certain organic compounds, including toluene and benzyl alcohol (De Kievit et al. 2001; Ingerslev et al. 2001). It is a constructed narrow host range, mobilizable plasmid of large size (117 kb) and also contains a resistance marker for the antibiotic, kanamycin. Finally, TOL (pWWO) constitutively expresses the gene encoding for GFP in sufficient quantities to enable visualization of host donor cells, using epifluorescent or confocal laser scanning microscopy (CLSM; Klevens et al. 2007). Luria–Bertini (LB) broth (bacto tryptone 10 g/L, bacto yeast extract 5 g/L, NaCl 4 g/L) and modified fastidious anaerobe broth (FAB) chemically defined medium (supplemented with glucose 20 or 200 mg/L)were used for batch suspended cell cultures (Heydorn et al. 2000). Both GFP and DsRed are very stable in both complicated and chemically defined medium. For suspended culture inocula, a sample of one colony from the streak plate was collected by sterile loop and added to 25 ml of 10 g/L LB broth, then incubated at 30 °C overnight.
Table 1.
P. putida strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source and referenceb |
|---|---|---|
| P. putida | ||
| KT2442 | Rifr | ATCC47054 |
| TUM-PP10 | KT2442::dsRed; Rifr Nalr Tcr | Nancharaiah et al. (2003) |
| TUM-PP12 | TUM-PP10 (pWWO::gfpmut3b); Rifr Kmr Nalr Tcr | Nancharaiah et al. (2003) |
| AKN44 | KT2442::miniTn7PA1/03/04ecfp-a; Gmr | Lambertsen et al. (2004) |
| AKN70 | KT2442::miniTn7PA1/03/04egfp-a; Gmr | Lambertsen et al. (2004) |
| AKN145 | KT2442::miniTn7PA1/03/04DsRedexpss, Gmr | Lambertsen et al. (2004) |
| AKN-PP11 | AKN44(pWWO::gfpmut3b); Rifr Kmr | This study |
| Plasmids | ||
| pWWO (TOL) | Self-transmissible; approx. 117 kb; narrow host range | Christensen et al. (1998) |
| TOL-gfpmut3b | Integration of PA1/04/03::gfpmut3b from pJBA28 into TOL | Christensen et al. (1998) |
Rifr rifampin resistance, Nalr nalidixic acid resistance, Kmr kanamycin resistance, Tcr tetra- cycline resistance, Gmr gentamicin resistance
ATCC American Type Culture Collection, Rockville, MD, USA
Fluorescent microscopy and image analysis
A Zeiss confocal laser scanning microscope LSM510 (Jena, Germany) mounted on an Axiovert 100 M inverted microscope was used for all optical sectioning. A Plan APO 100X oil objective with a numerical aperture of 1.4 or a C-APO 40× water immersion lens with a numerical aperture of 1.2 were used for obtaining cell images expressing green and red fluorescent proteins.
A excitation wavelength of 488 nm (argon laser) with a 505–530 nm band pass emission filter and a 543 HeNe laser with a 590 long pass emission filter were used to detect GFP- and DsRed-expressing bacteria, respectively. All image analysis related to biofilm fluorescent density calibration was carried out using “Image J” version 1.30v software (Research Services Branch, National Institute of Mental Health, Bethesda, Maryland; http://rsb.info.nih.gov/ij/) The COMSTAT software (Center for System Microbiology, Arne Heydorn, Oxford, England; Heydorn et al. 2000), a powerful tool for classifying biofilm 3-D structure, was used to quantify the relationship between the architecture of biofilms (defined by different COMSTAT-generated features of the 3-D biofilms) and conjugational plasmid transfer.
As described previously, a non-invasive method that based on a calibration of cell fluorescence to known cell number concentrations was carried out to determine the local bacterial concentrations within a biofilm (Ma and Bryers 2010). To satisfy our goal of quantifying cell numbers from fluorescent densities, all of the control parameters (such as background and size ranges) in the image analysis software were maintained constant between each image.
Non-invasive determination of conjugational plasmid transfer
Conjugation experiments were carried out using a biofilm reactor system as described previously (Ma and Bryers 2010). TOL plasmid transfer was quantified in biofilms accumulated at 30 °C in the once-through parallel plate flow cell system above. Plasmid-bearing cells of P. putida TUM-PP12 are designated as “donors”, the plasmid-free cells P. putida KT2442 are called “recipients”, P. putida KT2442 that have obtained TOL plasmids are called “transconjugants”, and any P. putida TUM-PP10 plasmid-free daughter cells that arise by random segregational loss from parent donors are called “segregants”. The following shorthand T=transconjugants, R=recipients, D=donors, and S=segregants will be used throughout all experimental procedures and analyses. To quantify the effects of nutrient concentrations on the frequency of TOL plasmid transfer, a protocol in which recipient cells were introduced to the established donor cell biofilms was chosen since this protocol generated biofilm clusters with very distinct regions or boundaries between populations of T, R, and D for the best results of imagine quantification (Fig. S1C). The donor strain (P. putida TUM-PP12) was cultivated in 10 g/L LB medium supplemented with 50 μg/ml kanamycin for overnight. Then, 0.5 ml overnight culture collected at early stationary phase, which minimizes the effects of growth on observed plasmid transfer kinetics, was added to 25 ml fresh medium. The cell pellet was washed with phosphate-buffered saline (PBS) three times, centrifuged in between washes, to completely remove supernatant medium before the adhesion study. The cell pellet was then diluted to an OD600 of 0.1–0.2 (a known cell concentration between 106 and 107 cells per milliliter), then delivered by peristaltic pump once through the flow cells continuously for 2–3 h for cell adhesion to the surface. Following this surface inoculation, sterile-modified FAB medium supplemented with glucose and thiamine (10 mg/L), was delivered to the flow cell at a constant flow rate of 120 ml/h for 5 days. Inlet concentration of glucose delivered to flow cell was either 20 or 200 mg/L. An overnight culture of the recipient strain, P. putida KT2442, was cultivated at 30 °C to early stationary phase, then washed once and resuspended in PBS to a final concentration of 106 cells/ml. A 20-ml volume of these recipient cells in PBS was delivered by peristaltic pump into flow cells through the medium inlet for 4–5 min. After 24-h incubation period with the donor cells in static condition, the biofilms were washed by delivering PBS for 30 min to remove any suspended donor cells. In order to investigate nutrient effects on biofilm architectures and plasmid transfer in situ, the biofilms were then cultured with modified FAB medium supplemented with the corresponding concentrations of 20 or 200 mg/L glucose delivered to the flow cells for additional 3 days. Then, the flow cell was clamped at both ends retaining the liquid contents and placed on the microscope stage for imaging. Enumeration of donors, recipients, and transconjugants was carried out by non-invasive CLSM optical sectioning (see above).
Estimates of plasmid conjugative transfer efficiency were determined two ways: (1) averaged over an entire horizontal plane of the biofilms using CLSM and our image analysis protocol (above) and (2) estimated from individual biofilm clusters comprising the overall biofilm, using CSLM and our cluster mapping analysis (described below). A ratio of T/D was used to evaluate the plasmid transfer frequencies since donor cells were cultivated on the surface first, and form the initial biofilm structures.
To investigate the antibiotic effects at the subinhibitory levels on plasmid transfer, biofilms of recipient cells (P. putida KT2442) were formed in flow chambers by supplying FAB chemically defined medium with 100 mg/L glucose at a flow rate of 2 ml/min in a once-through manner. Three days later, a suspension of donor P. putida TUM-PP12 cells, at a known concentration of 106 cells/ml, was washed, resuspended in FAB medium containing concentrations of 0 (control, no antibiotics), 0.5 μg/ml kanamycin or 0.1 μg/ml imipenem, introduced into the flow cell, then incubated for another 24 h. Subsequently, FAB chemically defined medium was delivered to the flow cells for an additional 5 days (Fig. S1D). The spatial architecture of the biofilm and the formation of transconjugants in situ were monitored on days 6 and 8. The same non-invasive quantification method was used to enumerate the different cell populations. However, a ratio of T/R was used to evaluate the plasmid transfer frequencies since recipient cells were cultivated on the surface first and host the initial biofilm structures.
Cluster mapping image analysis
A viability mapping method to identify the distinct regions of viable and nonviable bacteria in an oral biofilm was recently described (Hope and Wilson 2003). We have modified this method to measure the geometric distribution of the transconjugants, donors, and recipients within single microcolonies (S1). Images stacks comprised of different regions of transconjugants, recipients, and donors were manually drawn around the edge of the outer viable zone (Fig. S1A). To quantify gene transfer locally within such a heterogeneous biofilm, a viability mapping image analyses tool (Hope and Wilson 2003) was refined to dissect individual clusters into digital cluster geometry (Fig. S1B).The thickness of the outer and inner layers of a microcolony in any x–y plane and the mean angle (θ) of these portions of the biofilm stack was also measured to quantify the effects of local spatial distributions of cells on plasmid transfer (Fig. S1B).
Detection and sorting of plasmid transfer bacteria by flow cytometry
A BD Aria fluorescence-activated cell sorter (FACS; Becton Dickinson Biosciences, San Jose, CA, USA), a high-performance flow cytometer with 13 color and high-speed four-way sorter was used to detect, enumerate, and isolate transconjugant cells from mixed destructive samples of biofilms. These destructive samples were collected from glass tubes connected to the flow cells in a biofilm reactor system as described previously (Ma and Bryers 2010). Prior to the sorting of green fluorescent transconjugant cells, biofilms samples were washed and resuspended in PBS at a desired concentration of 1.5×107 cells/ml in order to obtain a total event rate of 25,000 cells/s, resulting in approximately 1 ml sorting samples within 10 min. Control samples of pure donor and recipient cells were analyzed first to define the regions of the standard gate for subsequent dual strain scatter plots. Analysis of pure cultures of P. putida TUM-PP10 (chromosome tagged with DsRed, without TOL-labeled GFP) was performed through FACS and defined on dot plots as a negative control for false-positive transconjugant-like cells. After mixing, biofilm samples suspended in PBS were applied to FACS flow cytometry. Cell counts for each population were determined based on defined standard gate regions of particle fluorescence and size, using control samples. Each sample was analyzed for 10 min, respectively, in triplicate. Flowjo™ software (Tree Star Inc, Ashland, OR, USA) was used throughout this work for flow cytometry data analysis. In order to verify the specific enumeration of bacterial populations by quantification non-invasively from CLSM images and destructively from flow cytometry, a traditional enumeration method was also used to compare efficiencies of plasmid transfer within biofilms by plating assorted populations on plates with agars containing different antibiotics, growing overnight and counting colony-forming unit (CFU).
Characterization of plasmid transfer bacteria by colony-direct PCR
Sorted populations of various cells collected from FACS flow cytometry were subsequently characterized by multiplex polymer chain reaction (PCR) for conjugative transfer genes. To confirm GFP-based plasmid transfer from donors to recipients and exclude the possibility of false-reading of donors as transconjugants, colony-direct PCR was applied to detect the GFP-containing sequence on the TOL-gfpmut3b plasmid and DsRed-containing sequence on chromosome DNA. Two sets of primers were designed to target a specific sequence of GFP on plasmid or RFP on chromosome: GFP gene specific primer pairs of GFP3b sense (5′-GTCAGTGGAGAGGGTG AAGGT-3′) and GFP3b antisense (5′-TGTGTGGACA GGTA ATGGTTG-3′) facilitated amplification of a 520-bp amplicon on TOL-gfpmub3b plasmid; primer pairs of DsRed-sense (5′-TTGCCATTTGCTTGGGATA-3′) and DsRed-antisense (5′-CTCGGTTCTTTCATACTGCTC-3′) allowed amplification of a 480-bp region of DsRed genes on the donor chromosome. Experiments were repeated three times, and at each time at least 20 random colonies were selected from FACS sortation for the following colony-direct PCR.
A single bacteria colony of each test strain, collected by plating sorted cells on selective LB agar plates, was suspended in 50 μl sterile water, and the cell suspension was boiled at 95 °C for 10 min for lysis. The mixture was then centrifuged at 12,000 rpm to pellet the cell debris, and supernatant was used as template DNA for PCR. Each PCR reaction contained 1 μl bacterial lysate, 0.5 μM of each primer, 50 mM KCl, 30 mM Tris-HCl, 1.5 mM MgCl2, 0.2 mM dNTP, and 1.25 U Taq polymerase. Amplification initially incubated at 95 °C for 1 min denaturation, followed by 35 cycles of 45 s at 92 °C, 45 s at 52 °C, 45 s at 72 °C. Finally, a primer extension reaction was carried out for 10 min at 72 °C. Two negative and two positive controls were performed for PCR reactions. Bacteria colonies of recipient cells (no color) and diH2O were used as DNA template when the mixtures were performed as negative control. PCR reactions of positive controls utilized donor cells (dual labeling) and segregational lost cells (DsRed) as DNA templates. The PCR products were run on 1.2 % agarose gels in 1× TBE buffer for characterization of different populations in plasmid-transfer biofilms.
Statistics
Paired t tests were performed with the observed values of plasmid transfer frequencies within biofilms to identify the statistical significance under different cultural conditions. A P value of 0.05 was used to reveal a significant difference.
Results
Non-invasive estimation of plasmid conjugational transfer frequencies in biofilms as a function of substrate loading
Conjugational plasmid transfer kinetics were quantified non-invasively by CLSM observation of donor, recipient, segregant, and transconjugant cell populations in developing biofilms as a function of limiting substrate loading. We found that Mature biofilms cultivated under a hydrodynamic environment tend to develop heterogeneous architectures comprising numerous microcolonies or clusters secured by cell–cell binding interactions and the surrounding extracellular polymeric matrix (Fig. S1A).
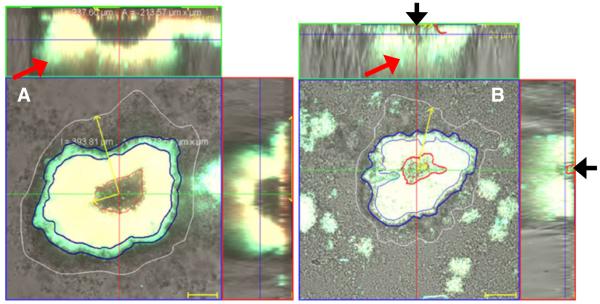
At two different influent nutrient concentrations, biofilm were observed to form mushroom- or tulip-shaped microcolonies and clusters protruding away from the surface (Fig. 1). Quantitative GFP and DsRed expression levels as a function of position across a microcolony were generated for biofilms cultivated in a continuous flow cell system, at influent glucose concentrations of 20 or 200 mg/L, respectively (Fig. 2). Biofilm architecture and the localization of plasmid transfer correlated well with the variation in nutrient loading. Distinct microcolonies obtained average thicknesses of 90–100 μm at 200 mg/L of glucose after 8 days of cultivation with medium, whereas biofilms reached only 50–60 μm at 20 mg/L glucose. Table 2 summarizes values of “angle of inner cone”, thickness and volume for populations of transconjugants and recipients, and efficiencies of plasmid transfer in P. putida mature biofilms cultivated at 20 and 200 mg/L glucose, respectively. Further, by combining the nondestructive in situ CLSM monitoring techniques with COMSTAT software, a wide range of variables up to 30 can be calculated that describe biofilm 3-D architectures. Table 3 summarizes values for two such variables, “roughness” and “surface area to volume ratio”, of P. putida biofilms accumulated with 20 and 200 mg/L glucose as carbon source. Three independent experiments and nine image stacks for each condition were performed to acquire mean values. Clear differences on roughness and surface area to volume ratio were observed between these biofilms. It was found in this study that “roughness” and “surface area to volume ratio” increased with decreasing glucose concentrations.
Fig. 1.
In situ monitoring of plasmid transfer in a biofilm cultivated in a continuous flow cell system irrigated with 200 mg/L (a) or 20 mg/L (b) concentrations of glucose, respectively. Spatial distribution of green transconjugants (P. putida KT2442 with TOL-gfpmut3b plasmid, green, regions between blue or red boundaries, marked as white or red arrows on y–z plane), donors (P. putida TUM-PP12 with TOL-gfpmut3b plasmid, yellow, region between red and blue boundaries), recipients (P. putida KT2442, region between white and blue boundaries, colorless of the reflection image) in this view, and segregants (P. putida TUM-PP12 (miniTn5Putdsred), red). Red arrows point to regions of transconjugants on x–z plane and black arrow points to cells bedded inside the inner cone. The micrograph is displayed in an orthogonal view to show the biofilm structure in three sets of 2-D sections; xy, xz, and yz. Bar size 20 μm
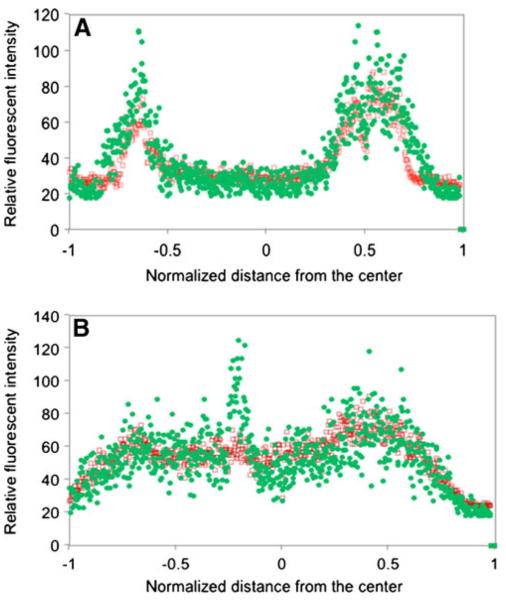
Fig. 2.
Quantitative analysis of GFP and DsRed expression levels across microcolonies cultivated in a continuous flow cell system irrigated with the glucose concentrations of a 200 or b 20 mg/L, respectively. Biofilms of donor cells (P. putida TUM-PP12 with TOL-gfpmut3b plasmid) were formed in flow cells for 4 days, and then recipient cells (P. putida KT2442) were introduced into the flow chambers and continuously cultivated for an additional 4 days. Data are relative fluorescent intensity profiles along the green lines in Fig. 1. The x–y planes were chosen at a depth close to the top of the internal water voids. The profiles were plotted as a function of the normalized distance from the center of the microcolonies
Table 2.
Effect of substrate concentrations on biofilm struc- tures of P. putida KT2442 using cluster mapping
| Substrate glucose concentration (mg/L) | 200 | 20 |
|---|---|---|
| Angle of cone (θ), degree | 56.89 a | 44.69 b |
| Outer layer thickness L (μm) | 21.82±3.98 (4.46±1.23) | 12.54±2.55 (5.21±0.87) |
| Donors (transconjugants) | ||
| Inner layer thickness L (μm) | – | 5.34±1.58 (2.12±0.98) |
| Donors (transconjugants) | ||
| Volume of transconjugants VT (μm3) | 1.27E+04 | 7.37E+03 |
| Volume of donors VD (μm3) | 1.12E+05 | 3.15E+04 |
| Transfer frequencies (VT/VD) | (1.13E-01) a | (2.34E-01) b |
Values with different letters differ signifi- cantly (P<0.05) within row
Table 3.
Effect of substrate concentrations on P. putida KT2442 bio- film structure analysis using COMSTAT
| Substrate glucose concentration (mg/L) |
Roughness coefficient |
Surface area to volume ratio (μm2·μm−3) |
|---|---|---|
| 20 | 1.78±0.09 | 7.37±0.73 |
| 200 | 0.74±0.06 | 2.57±0.38 |
Roughness coefficient=, where Lfi is the ith individual thickness measurement, is the mean thickness, and N is the number of thickness measurements
Surface area to volume ratio=surface area/biovolume
In order to validate the enumeration of bacterial populations by non-invasive CLSM, different approaches, CFU and FACS, were performed to enumerate T, D, R, and S. Plasmid conjugative transfer frequency was expressed as the ratio of transconjugant to donor (T/D) cells. Table 4 lists the distinct concentrations of transconjugants and transfer frequencies determined by these three counting approaches. Compared with CLSM direct counts, similar counts of transconjugants on the order of 106 were detected by the use of flow cytometry analysis approach.
Table 4.
Enumeration of conjugative transfer frequency (T/D) of TOL-gfpmut3b plasmid from P. putida TUM-PP12 to P. putida KT2442 by CFU, FACS, and CLSM
| Methods | Transconjugants (cells/cm2) | Transfer frequency (T/D) | Transfer frequency standard deviation |
|---|---|---|---|
| CFUa | (9.73×104)a | 0.82×10−1 | 2.16×10−2 |
| FACSb | (1.26×106)b | 3.30×10−1 | 4.23×10−2 |
| CLSMc | (4.07×106)c | 1.24×10−1 | 3.06×10−2 |
Values with different letters differ significantly (P<0.05) within column
CFU colony forming unit. Enumeration of cells by plating assorted destructive biofilms on plates
FACS fluorescent activated cell sorting. Flow cytometry enumeration of cells by using the defined gates. Four populations of D, T, R, and S in biofilms can be isolated by FACS
CLSM confocal laser scanning microscope. Estimation of D and R directly by using CLSM microscopy combined with quantification method of image analysis
Non-invasive estimation of plasmid conjugational transfer frequencies in biofilms as a function of antibiotic selection pressure
The effect of antibiotic exposure on cell survival in planktonic cultures was examined under the range of concentrations for kanamycin (0.1–20 μg/ml) and imipenem (0.05– 10 μg/ml). Small and negligible effects on cell growth rates were observed in recipient (P. putida KT2442) and donor (P. putida TUM-PP12 with TOL-gfpmut3b plasmid) cultures exposed to sublethal concentration of 2.5 μg/ml kanamycin or 0.10 μg/ml imipenem for up to 9 h (data not shown). These results may not be extrapolated to biofilms exposed to antibiotics, since these cells embedded deep within a thick biofilm experience totally different conditions of ecological factors affecting host metabolism versus those cells spatially closer to the bulk fluid.
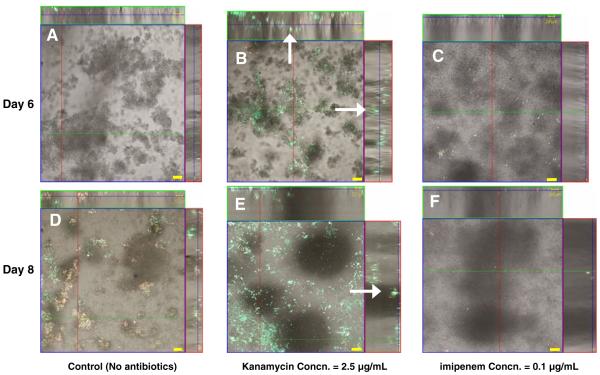
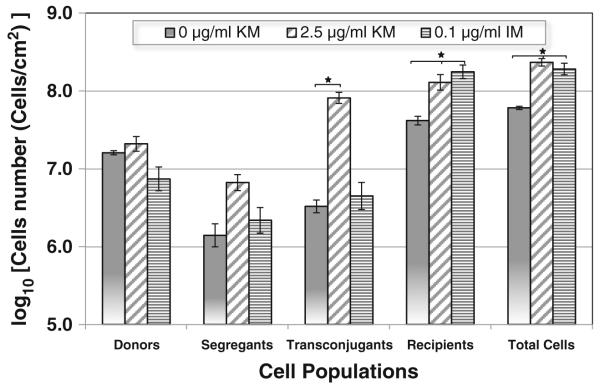
Transfer efficiency of a plasmid containing the kanamycin resistance gene, kanR+, were determined locally within a biofilm exposed to (a) no antibiotics (control), (b) a sublethal dose of kanamycin (2.5 μg/ml), or (c) a sublethal dose of imipenem (0.1 μg/ml). Using the biofilm architecture analysis software, COMSTAT (Christensen et al. 1998; Heydorn et al. 2000) and our cluster mapping analysis, biofilm cell populations (donor, segregants, recipients, and transconjugants) and the overall architecture of biofilms (controls; no antibiotic) are compared (Fig. 3) to biofilms subjected to sublethal doses of kanamycin and imipenem, respectively. By day 8, biofilms formed under either antibiotic selection pressure were ~110 μm in average thickness compared to the 50–60 μm thick biofilm formed with no antibiotic exposure. Bacterial total cell counts were also significantly higher in biofilms exposed to either antibiotic versus controls. Biofilm-averaged concentrations of transconjugant, segregant, and recipient populations within the total cell population were determined for all three biofilms (Fig. 4). Sub-inhibitory concentrations of kanamycin, not only enhanced formation of biofilm but also significantly increased the total number of transconjugant cells. However, under imipenem exposure, biofilm cell population and total cell numbers were also enhanced compared to the no-antibiotic controls (equaling the biofilm formation enhancement observed in kanamycin exposure), yet transconjugant populations were not increased. Table 5 lists conjugational transfer frequencies of plasmid under the presence of sub-inhibitory concentrations of kanamycin and imipenem reported in the different expression methods. Results indicate that the transfer efficiency for a plasmid expressing kanR+ was significantly higher when exposed to kanamycin versus when exposed to imipenem (Table 5).
Fig. 3.
In situ monitoring of plasmid transfer in a biofilm cultivated in a continuous flow cell system cultivated on FAB medium containing concentrations of 0 (control, no antibiotics, a and d), 0.5 μg/ml kanamycin (b and e) or 0.1 μg/ml imipenem (c and f), respectively. Spatial distribution of green transconjugants (P. putida KT2442 with TOL-gfpmut3b plasmid, green, marked as white arrow), donor (P. putida TUM-PP12 with TOL-gfpmut3b plasmid, yellow) and segregants (P. putida TUM-PP12 (miniTn5Putdsred), red). Recipients (P. putida KT2442) are colorless cells (of reflection images) in this view. The micrograph is displayed in an orthogonal view to show the biofilm structure in three sets of 2-D sections; xy, xz, and yz. A–c Images monitored on day 6. c–e Matured biofilms formed on day 8. Bar size 20 μm
Fig. 4.
Direct counts of the cell populations for plasmid transfer from donor (P. putida TUM-PP12 with TOL-gfpmut3b plasmid) to the established biofilms of recipients (P. putida KT2442) on day 8. Biofilms were challenged with either kanamycin (KM) or imipenem (IM) at the concentrations shown after inoculation with donor cells. Each point in the graph is the mean of three different positions around the middle of the flow cell. Error bar represent standard deviation. *P<0.05 as compared to control (without antibiotic treatment)
Table 5.
Effect of kanamycin (KM) and imipenem (IM) antibiotics on plasmid transfer efficiency of plasmid TOL-gfpmut3b from P. putida TUM-PP12 to P. putida KT2442
| Antibiotics concentration (μg/ml) |
T/R | T/D·R |
|---|---|---|
| 0 | (8.55±2.72)×10−2 | (5.24±1.48)×10−9 |
| 2.5 (KM) | (6.62±2.71)×10−1 | (3.03±1.17)×10−8 |
| 0.1 (IM) | (2.66±1.05)×10−2 | (3.74±1.75)×10−9 |
Conjugation efficiency determined non-invasively via CLSM versus destructive biofilm sampling followed by both flow cytometry and PCR analysis
The same procedure was used to cultivate recipient biofilms, incubate with donor cells, but then biofilms were destructively removed from the substratum and suspended in PBS for flow cytometry enumeration and sorting. Thus, spatial resolution of the various populations was lost by destructive sampling. Figure 5 shows the dot plot of green and red fluorescence for one sample biofilm. Four regions on this dot plot were defined by the different FACS gates; thus, four populations were detected according to their size scatter and relative green and red fluorescence. Pure biofilm cultures of recipient (P. putida KT2442) and donor (P. putida TUM-PP12 with TOL-gfpmut3b plasmid) strains were used as the negative and positive controls respectively to define the gate for the population of noncolor recipients (data not shown). When biofilms were exposed to sub-inhibitory concentration of kanamycin 2.5 μg/ml, the percentage of transconjugants was the fivefold higher than control, without the stress of antibiotic. Thus, the plasmid transfer frequencies (T/R) were detected in the order of 10−1. In contrast, without antibiotic, the transfer frequencies were kept the order of 10−2 in control biofilms, which is tenfold lower.
Fig. 5.
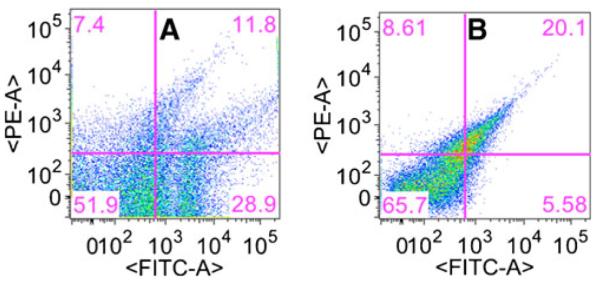
Detection of the conjugational gene transfer by flow cytometry equipped with florescent activated cell sorting (FACS). Dot plots of green fluorescence (GFP-H) versus red fluorescence (PE-H) under the presence of kanamycin at concentrations of 2.5 (a) and 0 (b)μg/ml. Regions defining four populations (D, T, R, and S) were set according to size scatter and relative green and red fluorescence. Red region plasmid segregational lost cells, green region transconjugant cells, yellow region dual labeling donor cells, none-fluorescent region recipient cells. Biofilms of recipient cells (P. putida KT2442) were formed in flow cells for 3 days, and then donor cells (P. putida TUM-PP12 with TOL-gfpmut3b plasmid, yellow) were introduced into the flow chambers and continuously cultivated for additional 5 days
The populations of assorted cells collected from FACS flow cytometry were utilized as PCR templates for characterization of conjugative transfer genes. Specific size bands based on the different primers designs have been detected by agarose gel electrophoresis as shown in Fig. 6. The electrophoresis gel results shown here confirmed the events of GFP-based plasmid transfer from donors to recipients, as well as the possibility of any false reading donors as transconjugants.
Fig. 6.
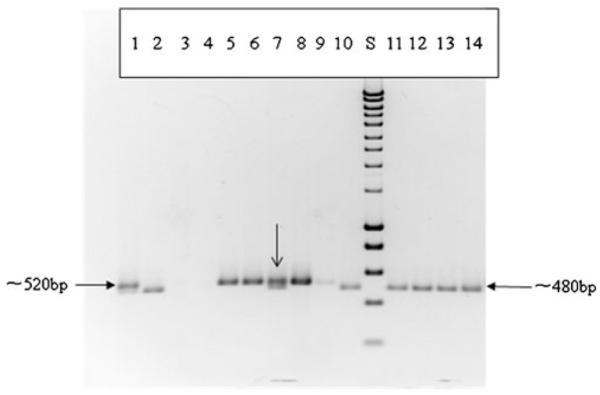
Colony PCR analyses of transconjugants, donors, segregants, and recipient cells obtained by plating sorted cells from flow cytometry equipped with FACS machine on LB plates. a1, 2 Positive control, using donor cells (dual labeling) and segregational lost cells (DsRed) as DNA templates; 3, 4 negative control, using recipient cells (no color) and diH2O as DNA templates; 5, 6, 7, 8, 9 using Transconjugants as DNA templates; 10, 11, 12, 13, 14 using segregational lost cells as DNA templates; S standard DNA HyperLadder I from Bioline. Both DsRed and GFP specific primers used for all PCR reactions. Arrow points to a false reading of a donor cell as a transconjugant cell
Discussion
In this study, a non-invasive microscopic analysis coupled with an in situ quantification method, the viability mapping image analyses tool, was applied to quantify plasmid transfer in heterogeneous biofilms locally. The occurrence and growth of transconjugants was quantified non-invasively by CLSM observation of donor, recipients, and transconjugants cells. Conventional methods for detecting plasmid gene expression, transfer, and stability have been carried out using mostly destructive sampling and selective plating techniques (Oliver 1995). However, selective plating is based on bacterial replication and thus may underestimate the overall extent of the HGT because only 1 % of the total number of bacteria determined by direct counts is frequently culturable (Torsvik et al. 1990; Amann et al. 1995). Destructive samples of biofilms provide gene transfer estimates that are spatially averaged population value that cannot provide direct estimates of local transfer kinetics. However, numerous studies have revealed that the real spatial structure of biofilms is heterogeneous, with numerous pores and channels surrounding dense clusters of biofilm-bound cells (Wuertz et al. 2001; Haagensen et al. 2002, Molin and Tolker-Nielsen 2003). Only a few studies have been carried out to examine the survival and transfer of plasmid DNA in biofilms (Beaudoin et al. 1998; Bathe et al. 2004; Maeda et al. 2006), and those provide very limited information on the efficiency of transfer on a local scale.
Our study validates a novel non-invasive quantification method that can provide fundamental information on plasmid transfer within developing biofilm systems. The non-invasive microscopic assessments of plasmid transfer were verified using destructive sampling quantification (flow cytometry and CFU) and subsequent PCR characterization. Our results show (Table 4), although experimental biases may be introduced through the procedure of collecting destructive biofilm samples, that both flow cytometry FACS and CLSM analysis techniques provided compatible results for the concentration of transconjugants. However, the number of transconjugants detected by CLSM direct counts was 40-fold higher than plates counts (CFU). These direct plate count underestimations may be due to the so-called “viable-but-non-culturable” status of many sampled cells (Oliver 1995; Ohlsen et al. 2003). Transfer frequencies were on the order of 10−1 when measured by quantitative CLSM microscopy, CFU counts, and flow cytometry. Transfer frequency measured by CFU is 33 % lower than measured by CLSM, but 166 % higher when FACS method was applied. These results (Table 4) indicate that FACS analysis incorporates more error into the estimates of transfer efficiency. A major drawback of the FACS protocol is that “gates” defining bacterial populations are set manually according to size scatter and relative fluorescence, which renders flow cytometry less valuable for plasmid transfer studies within biofilms since both the sizes and fluorescent levels of transconjugants differs significantly during long-term biofilm formation. Current requirements by flow cytometry of manually defining regions can easily fail to include fluorescent cells into the correct gates (pointed arrow shown in Fig. 6). However, as a novel method to determine the occurrence of plasmid transfer, flow cytometry equipped with FACS takes the advantage of rapid enumeration and sortation of single bacteria cells for up to 25,000 cells per second. Therefore, this method was used in this study as a supplement approach to valid the non-invasive quantification method, but not use independently to measure the conjugational plasmid transfer efficiencies.
To investigate gene transfer in heterogeneous biofilms, an in situ quantification method, the viability mapping image analyses tool, was utilized to demonstrate the general structure of a single cluster or microcolony as a function of cluster geometry. From Fig. 1, we observed clusters stabilized by their extracellular polymer matrix and separated from each other by medium-filled channels that allow nutrient access, waste removal and suspended cell movement. After being introduced to a structured biofilm of recipient strains, the donor cells preferentially appeared and established as a subpopulation on the already existing microcolonies of the recipient strain. Few donor cells adhered to the bare substratum. The heterogeneity of the biofilms was evaluated by COMSTAT, an image analysis tool with two major parameters of “roughness coefficient” and “Surface area to volume ratio” (Table 3). The “roughness coefficient” parameter reflects biofilm heterogeneity and measures the differences of local biofilm thicknesses relative to the average thickness. “Surface area to volume ratio” indicates fraction of the biofilm exposed to the nutrient flow (Heydorn et al. 2000). It was found in this study that “roughness” and “surface area to volume ratio” increased significantly with decreasing glucose concentrations. It is most likely that low nutrient concentrations resulted in heterogeneous biofilm structures that were more porous. In contrast, thick, more uniform and compact biofilms were developed under increasing nutrient concentrations.
Our mathematical quantification method, coupled with the ability to map the layers of each population in one microcolony of a mature biofilms, allowed us to quantitatively compare biofilm 3-D structures and plasmid transfer efficiencies within individual microcolonies in biofilms cultivated under 200 or 20 mg/L glucose as limiting carbon nutrient (Table 2). Results showed that significant differences in θ and L′ were observed between mature biofilms formed under the different nutrient concentration of glucose. Low nutrient concentrations resulted in porous biofilm clusters with open channels that enabled easier access and deeper penetration of donor cells throughout a cluster; the more extensive contact of donors with recipients thus led to higher transfer efficiencies. Conversely, lower plasmid transfer rates and efficiencies were observed for thick uniform densely packed biofilm clusters formed under high substrate concentrations, where donor cells were unable to gain access to recipient cell.
Environmental effects on biofilm formation and plasmid transfer are not yet clearly defined. Numerous reports on the effects of nutrient concentrations on plasmid transfer are at best contradictory (Hausner and Wuertz 1999; De Kievit et al. 2001). Reports suggest that plasmid transfer in biofilms is dependent on nutrient concentration varying the structures of the biofilms, but not by alternating the metabolic activities of the host cells (Wimpenny et al. 2000; Hendrickx et al. 2003; Slater et al. 2008). Several studies have reported that instead of developing a uniform distribution throughout a biofilm, a transconjugant cell layer is established on top of recipient microcolonies in a flow chamber system (Sorensen and Jensen 1998). These reports proposed direct correlations between nutrient concentration and transfer efficiency. In these cited studies, the biofilms attained an averaged thickness of 20 μm and exhibited all the characteristic features of differentiated premature conditions, i.e., they were surface-associated microcolonies without the more elaborate structures of mushrooms and water voids; thus cells and nonstable biomass were free to move. At these early stages of biofilm formation, cell replication on the interface was the main process governing accumulation of biomass; thus increasing nutrient concentration can change the density of the biofilms, but not the plasmid transfer efficiency, which requires direct cell–cell contact. However, when biofilms are mature, attachment and movement of swimming bacteria primarily account for population access and plasmid transfer. The spatial architecture of a biofilm, which is highly dependent on nutrient concentration, is considered to be the most important factor affecting plasmid transfer in biofilms.
In this work, we studied effects of nutrient concentrations on efficiencies of plasmid transfer in mature biofilms of thickness above 50 μm with the presence of water-filled voids. We found, in mature biofilms comprised of established mushroom-like clusters surrounded by water-filled channels and voids, the difference in medium composition remarkably affected plasmid transfer. High nutrient concentrations resulted in thick and compact biofilm and biofilm clusters that may impede penetration of donor cells from outer to inner regions. Thus, plasmid transfer creates a thin layer of transconjugants on the outer surfaces of the biofilm microcolonies, resulting in lower efficiencies of plasmid transfer. Conversely, biofilms, exposed to low nutrient concentrations, developed clusters that were less dense, seemed more porous and water-filled, thus enabling more donor cell access and deeper penetration of donor cells throughout the cluster leading to a faster spread of genes by conjugative plasmid transfer.
Two studies have shown that sublethal doses of antibiotic can exacerbate biofilm formation. Bagge et al. (2004) reports that sublethal doses of the β-lactam antibiotic, imipenem, not only affected fivefold gene regulation in ~34 genes in P. aeruginosa but also resulted in a 20× increase in alginate matrix synthesis that translated to an increase in biofilm volume and 2 orders of magnitude higher cell numbers. Hoffman et al. (2005) reports similar results for both P. aeruginosa and E. coli biofilms exposed to the aminoglycoside, tobramycin. Here, we asked if exposure to a sublethal level of antibiotic would enhance plasmid conjugative transfer.
Sub-inhibitory concentrations of kanamycin, not only enhanced formation of biofilm, but also significantly increased the total number of transconjugant cells. This increase in plasmid transfer efficiency could be naively rationalized based simply on the increased biofilm cell densities and more rapid biofilm formation. However, under imipenem exposure, biofilm cell population and overall biofilm amount were also enhanced compared to the no-antibiotic controls (equaling the biofilm formation enhancement observed in kanamycin exposure), yet transconjugant populations were not increased. Results indicate that the transfer efficiency for a plasmid expressing kanR+ was significantly higher when exposed to kanamycin versus when exposed to imipenem (Table 5). Conjugation efficiencies determined non-invasively by CLSM were verified with destructive samples of biofilm populations, followed by both flow cytometry assays and PCR analysis. Our results suggest that biofilm bacteria can “sense” antibiotics to which they are resistant and subsequently enhance the spread of that resistance.
Table 5 lists conjugational transfer frequencies of plasmid under the presence of sub-inhibitory concentrations of kanamycin and imipenem reported in the different expression methods. The number of transfer events per donor–recipient encounter is a theoretical approach to express plasmid transfer efficiency that allows true comparisons of transfer efficiencies of different plasmids. However, such a ratio is hard to define owing to the nonhomogeneous 3-D spatial distributions of bacterial populations within biofilms. Rather than the ratio of T/D, two different reporting schemes have been used here to express transfer frequencies: the ratios T/R and T/D·R, since recipient cells were initially formed biofilms on surface and provided a large pool for incoming donor cells to initiate plasmid conjugational transfer. A “paired t test” was performed and resulted in the P value of 4.67e-8 (<<0.05). This result implies that donor to recipient ratio has significant effect on plasmid transfer, at least when frequency was defined as T/R.
The majority of colony-direct PCR products, when the sorted “transconjugants” from FACS used as templates, had resulted in single fragment corresponded to the GFP gene on TOL-gfpmut3b plasmid, but not DsRed-containing sequence on chromosome DNA. However, PCR analysis also revealed that one “transconjugant” group had appeared to have genes coded both for GFP and DsRed, which could result from the false reading of donors as transconjugants. Cells sorted as “segregants” then processed by PCR showed that they only contained genes coded for DsRed and no GFP, which indicated authentic TOL-gfpmut3b plasmid segregation loss from the donor cells. No GFP or DsRed fragments were obtained from the negative controls, indicating that in the absence of templates, no PCR products were produced during the reactions (Fig. 6).
In summary, we have developed non-invasive microscopic analyses to quantify local plasmid conjugational transfer between bacterial populations within a developing biofilm. Frequencies of plasmid conjugational transfer within biofilm populations were affected by limiting substrate loading. Low concentrations of limiting substrate (glucose) produced thinner biofilms comprised of more porous biofilm clusters that allowed greater penetration of plasmid donor cells throughout the clusters with more exposure of recipient population to donor cells, plasmid conjugation efficiencies increased. The opposite was true at high substrate concentrations that produced very dense compact biofilm clusters, with corresponding low plasmid transfer efficiencies. Sublethal exposure of antibiotics (kanamycin, imipenem) exacerbates biofilm formation, producing thicker biofilm volume and increasing cell number populations. More importantly, our studies indicated that plasmid transfer efficiency for a plasmid containing an antibiotic resistance gene (kanR+) was significantly increased when exposed to that specific antibiotic versus an alternative antibiotic stress. These results suggest that biofilm bacteria might “sense” antibiotics to which they are resistant and subsequently enhance the spread of that resistance. We are currently exploring in more depth this effect of sublethal doses of antibiotics on conjugative transfer of plasmids containing antibiotic resistance genes for both different host: recipient bacterial species and different antibiotic: resistant gene combinations.
Supplementary Material
Acknowledgments
This work was partially funded by grants to JDB from NSF (CBET-0450253) and NIH (R01 EB007575).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00253-012-4179-9) contains supplementary material, which is available to authorized users.
Contributor Information
Hongyan Ma, Department of Chemical Engineering, University of Washington, Seattle, WA 98195-5061, USA.
James D. Bryers, Department of Chemical Engineering, University of Washington, Seattle, WA 98195-5061, USA; Department of Bioengineering, University of Washington, Seattle, WA 98195-5061, USA
References
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE. Plasmid transfer between strains of Pseudomonas putida, and their survival, within a pilot scale percolating-filter sewage treatment system. FEMS Microbiol Ecol. 1998;18:15–26. [Google Scholar]
- Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, Hoiby N. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and betalactamase and alginate production. Antimicrob Agents Chemother. 2004;48:1175–1187. doi: 10.1128/AAC.48.4.1175-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathe S, Mohan TV, Wuertz S, Hausner M. Bioaugmentation of a sequencing batch biofilm reactor by horizontal gene transfer. Water Sci Technol. 2004;49:337–344. [PubMed] [Google Scholar]
- Beaudoin DL, Bryers JD, Cunningham AB, Peretti SW. Mobilization of broad host range plasmid from Pseudomonas putida to established biofilm of Bacillus azotoformans. I. Experiments. Biotechnol Bioeng. 1998;57:272–279. [PubMed] [Google Scholar]
- Boon N, De Gelder L, Lievens H, Siciliano SD, Top EM, Verstraete W. Bioaugmenting bioreactors for the continuous removal of 3-chloroaniline by a slow release approach. Environ Sci Technol. 2002;36:4698–4704. doi: 10.1021/es020076q. [DOI] [PubMed] [Google Scholar]
- Christensen BB, Sternberg C, Andersen JB, Eberl L, Moller S, Givskov M, Molin S. Establishment of new genetic traits in a microbial biofilm community. Appl Environ Microbiol. 1998;64:2247–2255. doi: 10.1128/aem.64.6.2247-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg C, Bergstrom M, Hermansson M. In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl Environ Microbiol. 1998;64:2670–2675. doi: 10.1128/aem.64.7.2670-2675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol. 2001;67:1865–1873. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RA, Helm RA, Maloy SR. Increasing DNA transfer efficiency by temporary inactivation of host restriction. Biotechniques. 1999;26:892–894. doi: 10.2144/99265st02. 896, 898 passim. [DOI] [PubMed] [Google Scholar]
- Gilbert P, Allison DG, McBain AJ. Biofilms in vitro and in vivo: do singular mechanisms imply cross-resistance? J Appl Microbiol. 2002;92(Suppl):98S–110S. [PubMed] [Google Scholar]
- Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagensen JA, Hansen SK, Johansen T, Molin S. In situ detection of horizontal transfer of mobile genetic elements. FEMS Microbiol Ecol. 2002;42:261–268. doi: 10.1111/j.1574-6941.2002.tb01016.x. [DOI] [PubMed] [Google Scholar]
- Hausner M, Wuertz S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol. 1999;65:3710–3713. doi: 10.1128/aem.65.8.3710-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx L, Hausner M, Wuertz S. Natural genetic transformation in monoculture Acinetobacter sp. strain BD413 biofilms. Appl Environ Microbiol. 2003;69:1721–1727. doi: 10.1128/AEM.69.3.1721-1727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146(Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- Hohnstock AM, Stuart-Keil KG, Kull EE, Madsen EL. Naphthalene and donor cell density influence field conjugation of naphthalene catabolism plasmids. Appl Environ Microbiol. 2000;66:3088–3092. doi: 10.1128/aem.66.7.3088-3092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope CK, Wilson M. Measuring the thickness of an outer layer of viable bacteria in an oral biofilm by viability mapping. J Microbiol Methods. 2003;54:403–410. doi: 10.1016/s0167-7012(03)00085-x. [DOI] [PubMed] [Google Scholar]
- Huang CT, Peretti SW, Bryers JD. Plasmid retention and gene expression in suspended and biofilm cultures of recombinant Escherichia coli DH5alpha (pMJR1750) Biotechnol Bioeng. 1993;41:211–220. doi: 10.1002/bit.260410207. [DOI] [PubMed] [Google Scholar]
- Huang CT, Peretti SW, Bryers JD. Effects of medium carbon-to-nitrogen ratio on biofilm formation and plasmid stability. Biotechnol Bioeng. 1994;44:329–336. doi: 10.1002/bit.260440310. [DOI] [PubMed] [Google Scholar]
- Ingerslev F, Torang L, Loke ML, Halling-Sorensen B, Nyholm N. Primary biodegradation of veterinary antibiotics in aerobic and anaerobic surface water simulation systems. Chemosphere. 2001;44:865–872. doi: 10.1016/s0045-6535(00)00479-3. [DOI] [PubMed] [Google Scholar]
- Klevens RM, Edwards JR, Richards CL, Jr, Horan TC, Gaynes RP, Pollock DA, Cardo DM. Estimating health care-associated infections and deaths in US hospitals, 2002. Public Health Rep. 2007;122:160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsen L, Sternberg C, Molin S. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol. 2004;6:726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- Ma H, Bryers JD. Non-invasive method to quantify local bacterial concentrations in a mixed culture biofilm. J Ind Microbiol Biotechnol. 2010;37:1081–1089. doi: 10.1007/s10295-010-0756-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Ito M, Ando T, Ishimoto Y, Fujisawa Y, Takahashi H, Matsuda A, Sawamura A, Kato S. Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli. FEMS Microbiol Lett. 2006;255:115–120. doi: 10.1111/j.1574-6968.2005.00072.x. [DOI] [PubMed] [Google Scholar]
- Molin S, Tolker-Nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14:255–261. doi: 10.1016/s0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- Nancharaiah YV, Wattiau P, Wuertz S, Bathe S, Mohan SV, Wilderer PA, Hausner M. Dual labeling of Pseudomonas putida with fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid. Appl Environ Microbiol. 2003;69:4846–4852. doi: 10.1128/AEM.69.8.4846-4852.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsen K, Ternes T, Werner G, Wallner U, Loffler D, Ziebuhr W, Witte W, Hacker J. Impact of antibiotics on conjugational resistance gene transfer in Staphylococcus aureus in sewage. Environ Microbiol. 2003;5:711–716. doi: 10.1046/j.1462-2920.2003.00459.x. [DOI] [PubMed] [Google Scholar]
- Oliver JD. The viable but non-culturable state in the human pathogen Vibrio vulnificus. FEMS Microbiol Lett. 1995;133:203–208. doi: 10.1111/j.1574-6968.1995.tb07885.x. [DOI] [PubMed] [Google Scholar]
- Pinedo CA, Smets BF. Conjugal TOL transfer from Pseudomonas putida to Pseudomonas aeruginosa: effects of restriction proficiency, toxicant exposure, cell density ratios, and conjugation detection method on observed transfer efficiencies. Appl Environ Microbiol. 2005;71:51–57. doi: 10.1128/AEM.71.1.51-57.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravatn R, Zehnder A, Jvan der Meer JR. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl Environ Microbiol. 1998;64:2126–2132. doi: 10.1128/aem.64.6.2126-2132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. First State-Specific Healthcare-Associated Infections Summary Data Report. CDC Public Health Report. 2009.
- Slater FR, Bruce KD, Ellis RJ, Lilley AK, Turner SL. Heterogeneous selection in a spatially structured environment affects fitness tradeoffs of plasmid carriage in pseudomonads. Appl Environ Microbiol. 2008;74:3189–3197. doi: 10.1128/AEM.02383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater FR, Bruce KD, Ellis RJ, Lilley AK, Turner SL. Determining the effects of a spatially heterogeneous selection pressure on bacterial population structure at the sub-millimeter scale. Microb Ecol. 2010;60:873–884. doi: 10.1007/s00248-010-9687-5. [DOI] [PubMed] [Google Scholar]
- Sorensen SJ, Jensen LE. Transfer of plasmid RP4 in the spermosphere and rhizosphere of barley seedling. Antonie Van Leeuwenhoek. 1998;73:69–77. doi: 10.1023/a:1000661115753. [DOI] [PubMed] [Google Scholar]
- Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- Top EM, De Rore H, Collard JM, Gellens V, Slobodkina G, Verstraete W, Mergeay M. Retromobilization of heavy metal resistance genes in unpolluted and heavy metal polluted soil. FEMS Microbiol Ecol. 1995;18:191–203. [Google Scholar]
- Torsvik V, Goksoyr J, Daae FL. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimpenny J, Manz W, Szewzyk U. Heterogeneity in biofilms. FEMS Microbiol Rev. 2000;24:661–671. doi: 10.1111/j.1574-6976.2000.tb00565.x. [DOI] [PubMed] [Google Scholar]
- Wuertz S, Hendrickx L, Kuehn M, Rodenacker K, Hausner M. In situ quantification of gene transfer in biofilms. Methods Enzymol. 2001;336:129–143. doi: 10.1016/s0076-6879(01)36585-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.