Abstract
In the modern world people frequently interact with retrieval systems to satisfy their information needs. Humanly understandable well-formed phrases represent a crucial interface between humans and the web, and the ability to index and search with such phrases is beneficial for human-web interactions. In this paper we consider the problem of identifying humanly understandable, well formed, and high quality biomedical phrases in MEDLINE documents. The main approaches used previously for detecting such phrases are syntactic, statistical, and a hybrid approach combining these two. In this paper we propose a supervised learning approach for identifying high quality phrases. First we obtain a set of known well-formed useful phrases from an existing source and label these phrases as positive. We then extract from MEDLINE a large set of multiword strings that do not contain stop words or punctuation. We believe this unlabeled set contains many well-formed phrases. Our goal is to identify these additional high quality phrases. We examine various feature combinations and several machine learning strategies designed to solve this problem. A proper choice of machine learning methods and features identifies in the large collection strings that are likely to be high quality phrases. We evaluate our approach by making human judgments on multiword strings extracted from MEDLINE using our methods. We find that over 85% of such extracted phrase candidates are humanly judged to be of high quality.
Keywords: machine learning, imbalanced data, biomedical phrases, statistical phrase identification, Unified Medical Language System, abbreviation full forms
1. Introduction
Biological concepts are frequently expressed in terms of phrases. Not surprisingly then, studies indicate that a significant fraction of queries in PubMed® are multiword queries which are meaningful phrases, rather than simple collections of terms. This suggests that users, in many cases, have a phrase in mind when they create a query [1]. Therefore identifying high quality phrases can be beneficial for both document indexing and information retrieval.
We are interested in detecting syntactically well-formed high-quality meaningful biological phrases, i.e., given a sequence of tokens in a sentence, our goal is to assess whether or not that expression exemplifies a syntactically well-formed high-quality meaningful biomedical phrase. Central venous pressure, placenta abruption, familial Mediterranean fever are examples of such phrases. In contrast, central nervous is not a syntactically well-formed phrase. One would like to detect central nervous system as a phrase. Moreover, syntactically well-formed phrases are not always high-quality. For example, the phrase different statistical methods is syntactically well-formed, but the phrase statistical methods presents a better choice as a meaningful phrase. We will refer to such high-quality meaningful biological phrases as good phrases. We do not restrict the phrases to terminology or idiomatic expressions, nor do we restrict their length.
The ideal phrase will be useful, meaningful, and aesthetically pleasing. A frequently used phrase is clearly useful. Meaningful means the phrase is comprehensible and understandable without any additional context. Many phrases in a document are useful and valuable in the context where they appear, but would leave obvious questions without that context. Finally, aesthetically pleasing acknowledges that these are human judgments without a final objective criterion.
We limit ourselves to phrases without prepositions or other stop words. By avoiding prepositions we lose the ability to identify all good quality phrases. However, this limitation leaves many good biomedical phrases to discover. In the 2011 edition of UMLS® only 24.9% of the unique phrases containing only alphanumeric characters include stop words. Only 10.1% of these UMLS phrases which appear in MEDLINE include stopwords. Similarly only 16% of the abbreviation full forms we find in MEDLINE® contain stopwords. The list of UMLS phrases with stop words which appear in MEDLINE includes phrases we are sorry to lose, such as “quality of life” and “head and neck.” However there are also many phrases that are not helpful without additional context: “associated with,” “use of,” and “followed by.” Clearly the vast majority of meaningful biomedical phrases used in MEDLINE include neither stopwords nor prepositions. Reliable ways of identify such phrases are useful.
Previous studies have addressed certain aspects of this problem. Some have concentrated on identifying noun phrases [2–4]. Chen [2] designed a system to acquire noun phrases from running text by using a probabilistic chunker to decide phrase boundaries and linguistic knowledge to extract the noun phrases. Bennett et al. [4] simply used a finite set of rules composed of different sequences of part-of-speech tags to detect noun phrases in MEDLINE. Other studies have looked at identifying specific phrases, such as domain-specific multiword terminology [5, 6] or multiword expressions [7], which are idiomatic, fixed or partially separable expressions. Many of these approaches restrict the lengths of phrases they detect.
Several methods have been used in an attempt to extract useful phrases from a document collection. Statistical approaches detect sequences of words that occur contiguously in a corpus more frequently than expected by chance [8–10]. Syntactic approaches are based on the assumption that a sequence of words satisfying certain syntactic relations or a specified structure form a phrase [4].
A comparison of several term recognition algorithms by Zhang et al.[11] identified the C-value algorithm by Frantzi et al. [6] as the best on a biological corpus. The C-value method combines both statistical and syntactic approaches into a hybrid method. Some of our statistical features resemble the statistical values used to calculate the C-value. Our part-of-speech features provide similar information to their part-of-speech filters. Our method uses machine learning to combine this information in a very flexible manner.
Statistical and syntactic information is undoubtedly fundamental knowledge one can obtain about a string of tokens. However, it is not evident how to combine that information optimally. In this work we propose using a machine learning approach that learns from statistical and syntactic features utilizing existing sets of high-quality phrases. One such source of high-quality phrases is UMLS, which is a curated source and contains meaningful and well-formed content. Another source of good phrases is the list of full forms (defining phrases) coming from abbreviation-definition pairs extracted from MEDLINE using an abbreviation-definition identification algorithm [12].
We formulate our problem as follows. Suppose we have a large collection of multiword strings that are unlabeled. Suppose further that we have an additional smaller set of phrases which are known to be high-quality meaningful biomedical phrases. We believe that there are more of these high-quality phrases in our large unlabeled collection. Our goal is to detect and retrieve these unknown high-quality phrases from the unlabeled set. We treat the labeled high-quality phrases as the positive class, and the unlabeled data as the negative class. We use the term negative class in spite of the fact that phrases in the negative class are not necessarily negative, they simply do not distinguish possible positive from true negatives. We consider both cost-sensitive SVM (CS-SVM) and a wide-margin classifier with the modified Huber loss function (Huber) as our learning algorithms. We find that Huber is slightly superior in performance to CS-SVM with an optimal cost factor. The result of our processing is a large set of strings (over 700,000) of which by our evaluation at least 85% are high quality phrases.
The paper is organized as follows. Under Methods we describe the learning algorithms and the data sets and define the features used to represent phrases. Under Results we describe our experiments and the evaluation measures used and present the numerical results. This is followed by an Application of our method. Lastly come the Discussion and Conclusion.
2. Material and methods
2.1. Data Sources and Preparation
First, MEDLINE was processed as follows. We processed the titles and abstracts of the MEDLINE records and extracted all contiguous multi-token strings that contained neither punctuation marks nor stop words. This set includes all multi-token substrings of longer strings. The resulting strings were then normalized (lowercased, redundant white space was removed) and duplicates were removed yielding a set of 280,737,434 strings which we call M.
We considered two sources to acquire high quality meaningful multiword phrases for training: UMLS (http://www.nlm.nih.gov/research/umls/) and a list of full forms (abbreviation defining phrases) coming from abbreviation-definition pairs extracted from MEDLINE [12]. UMLS is a curated source and contains meaningful, well-formed content. The list of abbreviation-definition pairs is automatically extracted from MEDLINE using AB3P, an abbreviation-definition identification algorithm, which achieves an F-measure of 90% on several manually evaluated datasets and compares favorably to other existing abbreviation-definition identification algorithms.
From the UMLS Metathesaurus® file we take the subset of English strings. These are normalized and duplicates are dropped. The overlap of this set with M consists of 297,005 phrases which we denote by U. Similarly, we normalize the set of full forms, drop duplicates and find the overlap with M to consist of 733,410 phrases which we denote by F.
For each phrase in U, we randomly select up to 5 MEDLINE sentences containing it. If there are five or fewer sentences containing a phrase we take all of them, but if there are more than five we randomly select five. We denote the resulting set of 1,198,849 MEDLINE sentences by SU. Just as we processed MEDLINE text to obtain M, we process SU to extract all contiguous multiword normalized strings. Of this set 5,789,943 strings are not present in U. We refer to this subset as MU. Starting with F we carry out exactly the same procedure to obtain SF and MF. Selecting up to 5 MEDLINE sentences containing each phrase in F, we obtain the set SF and subsequently the set MF. MF consists of 12,044,070 strings and by construction has no overlap with F. For machine learning we consider U to be the positive class and MU the negative class. Similarly for F and MF. These numbers have been summarized in Table 1.
Table 1.
Sizes of the phrase and string sets.
| M, unique MEDLINE strings | 280,737,434 |
| U, the part of UMLS contained in M | 297,005 |
| SU, sentences | 1,198,849 |
| MU, unique MEDLINE strings in set SU other than UMLS phrases | 5,789,943 |
| F, full forms contained in M | 733,410 |
| SF, sentences | 2,732,911 |
| MF, unique MEDLINE strings in set SF other than full forms | 12,044,070 |
2.2. Description of Features
In order to apply machine learning we must define features to represent a phrase (or multi-token string). We first define basic statistical features based on properties of a phrase and its appearances in MEDLINE text. We then discretize these values.
2.2.1. Basic Statistical Features
Given a phrase ph that is composed of n words, i.e., ph = w1w2…wn, we extract a set of 11 associated numeric values . These values are defined as:
f1: Number of occurrences of the phrase ph throughout MEDLINE;
Rationale: Phrase frequency reflects the usefulness of the phrase. Good phrases tend to be frequently used.
f2: Number of occurrences of w2…wn not following w1 in documents with ph;
Rationale: Is w1 a critical part of the phrase ph or is it a modifier of w2…wn, the true essential phrase? We look at the documents that contain ph. If w2…wn occurs frequently in these documents without following w1, then w1 is likely to be an optional word and is not a fundamental part of the phrase. Documents without ph are ignored because we are not directly concerned with whether w2…wn itself is a meaningful phrase.
f3: Number of occurrences of w1…wn−1 not preceding wn in documents that contain ph;
Rationale: Is wn a critical part of the phrase ph ? We look at the documents that contain ph. If w1…wn−1 occurs frequently in these documents not followed by wn, then wn is likely to be an optional word and is not a fundamental part of the phrase. Documents without ph are ignored because we are not directly concerned with whether w1…wn−1 itself is a meaningful phrase.
f4: Number of occurrences of strings of the form xw1…wn throughout MEDLINE;
Rationale: This feature represents the tendency of ph to be preceded by another term. Can it stand alone or does it need another word to begin the phrase? For example, `fibrosis patients' is not a good stand alone phrase, as it most frequently appears as `cystic fibrosis patients'.
f5: Number of occurrences of phrases of the form w1…wnx throughout MEDLINE;
Rationale: This feature represents the tendency of ph to be followed by another term. Again can it stand alone or does it need another word to end the phrase? For example, `central nervous' is not a stand alone phrase, it most frequently appears as `central nervous system'.
f6: Bayesian weight between w1 and w2,
Rationale: Given that w2 is the second word in the phrase ph, how likely is w1 the first word? The more likely w1 appears before w2, the more likely w1 is an important part of the phrase.
f7: Mutual information between w1 and w2;
Rationale: Mutual information measures the mutual dependency between w1 and w2. The more w1 and w2 appear together, the more likely w1 is an important part of the phrase.
f8: Bayesian weight between wn−1 and wn,
Rationale: Given that wn−1 is the next to last word in the phrase ph, how likely is wn to be the last word? The more wn appears after wn−1, the more likely wn is an important part of the phrase.
f9: Mutual information between wn−1 and wn;
Rationale: Mutual information measures the mutual dependency between wn−1 and wn. The more wn−1 and wn appear together, the more likely wn is an important part of the phrase.
f10: Number of different multiword phrases beginning with w1 in MEDLINE;
Rationale: The more different phrases begin with w1, the more likely w1 is a generic modifier and is not an essential part of the phrase.
f11: Number of different multiword phrases ending with wn in MEDLINE.
Rationale: The more different phrases end with wn, the more likely wn is a generic word and is not an essential part of the phrase.
We also normalize fi values for 1< i ≤11 by dividing each fi by f1 and denote it as
| (1) |
Therefore we have a total of 21 basic statistical feature values (11 fi values and 10 normalized ) to be used for machine learning.
2.2.2. Discretization
While one can use numerical values as features to train a classifier, we discretize these numeric values into categorical values in order to get more robust behavior of classification algorithms. Given the set of phrases, every feature variable defines a range of values that the feature assumes. At discretization, that range of values is partitioned into a small number of bins, and all values that fall into one bin are represented using a single nominal feature. Thus, for each numerical feature, discretization reduces the space of feature values to a much smaller set of categorical values. The advantage to this is the machine learning can then independently weight these different discrete features.
Several discretization methods are proposed in the literature, including entropy-based methods that are among the most commonly used discretization techniques. Such methods work well for obtaining an optimal result for a single numerical feature. But when one has a number of numerical features to discretize, such methods can make no guarantee that the final set of features is optimal for learning on all features. Thus we use a uniform discretization approach and examine different numbers of bins in search of good machine learning results.
2.2.3. Syntactic Features
In addition to statistical features we include features based on part-of-speech tags for a phrase ph. We use the MedPost tagger [13]. To obtain the tags for a given phrase ph, we randomly select a sentence from SU (or SF) containing the phrase ph, tag the sentence, and consider the tags t−1t1t2…tn−1tntn+1 where t−1 is the tag of the word preceding word w1 in phrase ph, t1 is the tag of word w1 in phrase ph, and so is the tag of the word preceding word on. We construct the features
These features emphasize the left and right ends of the phrase and include parts-of-speech in the middle without marking their position. A phrase can have up to n+2 features, if the interior words have unique parts of speech. If the phrase begins at the beginning of the sentence, then a feature (Lend,1) replaces the (t−1,1) feature. Similarly, if the phrase ends at the end of the sentence, then a (Rend,4) feature replaces the (tn+1,4) feature. The resulting features are included with the discretized features we discussed in the previous section to represent the phrase.
2.3. Learning Algorithms
The set MU is a large collection of unlabeled multiword strings to compare with the set U of high-quality phrases. Our goal is to use machine learning to identify additional high-quality phrases in MU based on the high quality data in U. Similarly for F and MF.
A naïve approach to this problem would simply take the known high-quality phrases as the positive class and the rest of the collection as the negative class (unlabeled documents) and apply support vector machine learning to learn the difference and rank the negative class based on the resulting scores. It is reasonable to expect that the top of this ranking would be enriched for the positive class. But previous studies [14–17] have shown that due to the imbalanced nature of the problem an appropriate choice of methods can improve over this naïve approach.
The issue with imbalanced learning is that the dramatic difference in class size compromises the performance of some classification techniques. The large prevalence of negative documents dominates the decision process and harms classification performance. Several approaches have been proposed to deal with the problem including sampling methods and cost-sensitive learning methods and are described in [18–20]. These studies show that there is no clear advantage of one approach versus another. In fact, cost-sensitive methods and sampling methods are related in the sense that altering the class distribution of training data is equivalent to altering misclassification cost. Based on these studies we chose to examine cost-sensitive learning in which the cost for misclassifying elements of the positive set is increased.
Zhang and Iyengar [16] considered the problem of recommender systems which use historical data on customer preferences, purchases and other available data to predict items that might be of interest to a customer. By their nature recommender systems deal with imbalanced data. They showed a wide margin classifier with a quadratic loss function to be very effective for this purpose. It may be a better method than varying costs because it requires no searching for the optimal cost relationship between positive and negative examples.
In this study we apply both cost-sensitive SVM and a wide-margin classifier with modified Huber loss function with quadratic properties designed to take advantage of the insight of Zhang and Iyengar [16]. Both algorithms address the problems with imbalanced and noisy data sets.
Here we write the standard equations for an SVM following Zhang [21]. Given training data {(xi, yi)} where yi is 1 or −1 depending on whether the data point xi is classified as positive (C+ or negative (C−), an SVM seeks that weight vector which minimizes
| (2) |
where the loss function is defined by
| (3) |
The SVM classifier can be sensitive to a large class imbalance, resulting in a drop in classification performance. It is susceptible to generating a classifier that has an estimation bias towards the majority class, resulting in a large number of false negatives. Cost sensitive learning has been proposed to deal with that problem. The cost-sensitive version of SVM modifies (3) to become and now we can choose
| (4) |
and now we can choose r+ and r− to magnify the losses appropriately. Generally we take r− to be 1, and r+ to be some factor larger than 1. Choosing r+ to be greater than r− helps overcome the dominance of negative points in the decision process. Generally, the same algorithms used to minimize (2) can be used to minimize (4).
Zhang and Iyengar [16] proposed a wide margin classifier with the quadratic loss function h(z)2 as effective for imbalanced and noisy training sets. We use a variation of quadratic loss function, the modified Huber loss function [21], which is quadratic where this is important and has the form
| (5) |
We refer to this as the Huber method as opposed to SVM. We compare it with CS-SVM. We also used the multivariate Bernoulli naïve Bayesian classifier [22, 23] as a baseline with which we compare results.
We believe that MU and MF contain many syntactically well-formed and meaningful phrases, and our goal is to identify them. In order to do that, we set up machine learning experiments to learn the difference between the phrases in set U and elements in set MU. Similarly, we learn the difference between the phrases in set F and elements in set MF.
We perform 3-fold cross validation by training the method on two-thirds of U ∪MU and scoring the remaining one-third of the phrases. When all three folds are completed the whole list of phrases in U ∪MU have been scored. Similarly, we train and score phrases in F ∪MF. We then rank the phrases in sets MU and MF and expect the top of these respective rankings will be enriched for high-quality phrases.
We evaluate performance of the learning using the Mean Average Precision measure (MAP) [24]. Average precision is the average of the precisions at each rank that contains a true positive element, i.e. a high-quality phrase from the positive class U. What we report are the MAPs or mean of the average precisions coming from the three rounds of cross validation. We believe that better classification between phrases in U and MU leads to better ranking of phrases in set MU. Since we are only interested in phrases in MU that look like phrases in U, the more successfully we can learn to separate the phrases in MU that don't look like U from U the more useful the resulting ranking of MU will be. On the other hand we expect that there will be many phrases in MU that look so close to U they cannot be separated by the learning and this puts an upper bound on how good the learning can be based on counting only U as the positive set. The same remarks apply also to F and MF.
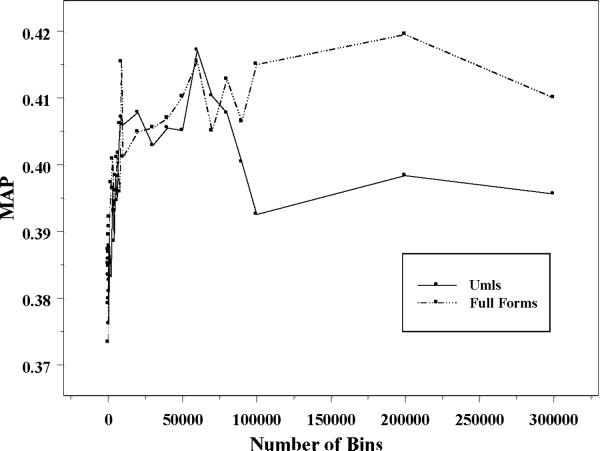
Before finalizing our approach we had to determine the number of bins in the uniform discretization approach to obtain the best learning performance. Figure 1 shows the MAP values for the classification problems U ∪MU and F ∪MF based on 3-fold cross-validation applying Huber learning with different numbers of uniform bins. We conclude that 60,000 bins is a reasonable choice. For F ∪MF, one could consider more than 60,000 bins, but we doubt the improvement from 60,000 is significant. Therefore, we chose to use 60,000 uniform bins for discretization of all numerical features for both problems and used the resulting features for all learning algorithms tested.
Figure 1.
Huber Classifications with Different Number of Uniform Bins
3. Results
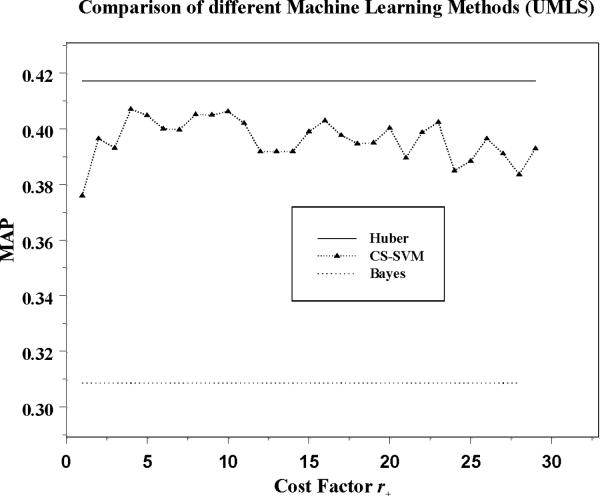
Here we provide results of applying our machine learning methods to U ∪MU and F ∪MF. Figure 2 and Figure 3 represent the MAP values for U ∪MU and F ∪MF based on 3-fold cross-validation. We present results for three different classifiers: Naïve Bayes which we use as a baseline, Huber, and CS-SVM with a range of cost factors. We observe that the Huber and CS-SVM classifier performs much better than Naïve Bayes. CS-SVM with the cost factor of 1 (standard SVM) is not optimal. As we increase the cost factor, the performance of CS-SVM improves and reaches a best performance with an optimal cost factor. However, the Huber performs better than the CS-SVM with the optimal cost factor. Furthermore the Huber does not require a search for the optimal cost factor as CS-SVM requires. Therefore, we conclude that the Huber classifier may represent the best approach for this problem.
Figure 2.
Huber, CS-SVM and Bayes classifiers applied to U ∪MU
Figure 3.
Huber, CS-SVM and Bayes classifiers applied to F ∪FU
The MAP scores represent how well the positive labeled points are ranked ahead of the negative labeled (actually unlabeled) points. As we argue above, it is reasonable to use MAP to compare different classification methods. However our main goal is to use the trained classifiers to rank the unlabeled sets and to achieve a ranking that places the unlabeled yet high quality phrases high in this ranking. In order to evaluate the quality of such rankings, the Huber learner was used to produce rankings for both and MU and MF. We then extracted four different sets of 100 multiword strings:
Set 1: 100 candidate phrases randomly selected from the subset of MU strings, whose Huber scores are above the median score of the set U phrases. There are 224,249 such strings in MU.
Set 2: 100 candidate phrases randomly selected from the subset of MF strings, whose Huber scores are above the median score of the set F phrases. There are 520,848 such strings in MF.
Set 3: 100 phrases randomly selected from set F.
Set 4: 100 randomly selected multiword MEDLINE strings from set M.
These four sets of phrases were individually judged by all four authors of the paper. The phrases from the different sources were randomly arranged and the judging was performed without knowledge of their source. Judges were instructed not to mark a phrase containing a token used as a verb or a phrase ending in a token used as an adverb or adjective as a good phrase. Beyond this they were to use their judgment based on whether the phrase was aesthetically pleasing and meaningful without additional context. A string was considered to be a high quality phrase if it was rated high quality by at least three judges. The results are in Table 2.
Table 2.
Quality assessments for four humanly judged sets. Sets 1 and 2 each contain 100 phrases randomly selected from the subset of MU and MF phrases, whose Huber scores are above the median score of the set U and set F phrases, respectively. Sets 3 and 4 each contain 100 phrases randomly selected from sets F and M, respectively.
| Set 1 | Set 2 | Set 3 | Set 4 | |
|---|---|---|---|---|
| Fraction of high-quality phrases | 85% | 91% | 92% | 35% |
We found that 85% of the phrases in the set MU that scored above the median score of the set U are well-formed high quality meaningful phrases. This is a dramatic improvement over randomly selecting a phrase from set M, which yielded only 35% high quality phrases. Similarly, we found that 91% of the phrases in the set MF that scored above the median score of the set F are well-formed high quality meaningful phrases. Set 3 was included as a check on the quality of F and showed that while it is not perfect, it is indeed high quality.
Based upon the size of M and the above humanly judged sets, we can estimate a lower bound on the number of the high quality biomedical phrases in MEDLINE. We expect at least 9.2 million high quality well-formed phrases based upon the set 1 of quality comparable to U. Likewise, we expect at least 11 million high quality well-formed phrases estimated from the set 2 of quality comparable to F. Of course there are likely to be many more phrases that have some level of acceptability to a human, but which do not score high on either of the scales we are using (at or above the median for either U or F based learning) and this includes half of U and F themselves. Thus we see these estimates as quite conservative lower bounds on the number of high quality phrases.
We also used the Huber algorithm to learn the difference between U and MU, and applied the trained classifier to classify F ∪MF, and vice versa. The average precision is 0.21 in classifying F ∪MF and in the reverse direction 0.19. This suggests a systematic difference between the sets U and F which deserves further investigation.
In Table 3, we measured the contribution of each individual feature by removing it only in the Huber machine learning. One can observe that the part of speech tags are especially important features for this problem.
Table 3.
Contribution of features measured by the effect of removing them one at a time.
| Feature Removed | UMLS | FULL FORM | Feature Removed | UMLS | FULL FORM |
|---|---|---|---|---|---|
| None | 0.417 | 0.415 | |||
| f 1 | 0.417 | 0.405 | |||
| f 2 | 0.409 | 0.406 | 0.414 | 0.413 | |
| f 3 | 0.397 | 0.403 | 0.399 | 0.407 | |
| f 4 | 0.411 | 0.423 | 0.395 | 0.393 | |
| f 5 | 0.407 | 0.396 | 0.401 | 0.292 | |
| f 6 | 0.393 | 0.407 | 0.413 | 0.412 | |
| f 7 | 0.405 | 0.404 | 0.410 | 0.410 | |
| f 8 | 0.397 | 0.405 | 0.407 | 0.405 | |
| f 9 | 0.400 | 0.397 | 0.407 | 0.418 | |
| f 10 | 0.389 | 0.396 | 0.397 | 0.400 | |
| f 11 | 0.364 | 0.376 | 0.392 | 0.409 | |
| POS | 0.343 | 0.319 |
4. Application
Frequently PubMed queries retrieve many more documents than a user can examine. For example, queries such as `alzheimer's disease', `deafness', `autism' and `hypertrophic cardiomyopathy' retrieve 81,326, 31,046, 18,974, and 13,543 documents respectively. Obviously, one cannot manually examine them all. We are currently involved in developing a system that can provide alternative ways of browsing these results.
One way to present retrieved contents is by dividing these documents by underlying theme. Then we must present the phrases that are most central and provide the best representation for each theme. While developing the themes, we treat as synonymous multiword strings that stem the same in any order and strings that occur in the same UMLS concept. For example, the text strings autoimmune disease, autoimmune diseases, autoimmune disorder, autoimmune disorders, autoimmunity disease, autoimmunity diseases, autoimmunity disorders are recognized as variants of the same concept. If one of the best strings to represent a theme is one of several synonyms, we want to present the reader with the variant most likely to be a good biomedical phrase.
We use the method of identifying well-formed biomedical phrases introduced in this paper. We score all the text string variants using our methods and choose the one with the highest score to represent the group of related text strings. Scores are computed using both U ⋃ MU and F ⋃ MF training and averaged. This process ensures that with high probability a good quality phrase is displayed to the user.
As an example, phrases identified by this system to represent documents retrieved with the query `deafness' are:
a1555g mutation; non syndromic deafness; waardenburg's syndrome; hearing disorders, genetics; deafness, congenital; vestibular aqueduct; alport's syndrome; cochlear implants; pure tone; hair cells, auditory
Of these 10 phrases, 6 phrases belong to groups of string variants. In particular, the phrase cochlear implants belongs to a group containing 12 string variants, presented in Table 4. When these string variants are scored the phrase cochlear implants is the highest scoring and is chosen to represent this group of string variants.
Table 4.
The group of string variants is scored using the method of identifying well-formed biomedical phrases presented in this paper. The phrase cochlear implants is chosen to represent this group of string variants.
| 0.189 | auditory prostheses |
| 0.042 | auditory prosthesis |
| 0.739 | cochlear implant |
| 0.326 | cochlear implant procedure |
| 0.319 | cochlear implant procedures |
| 0.511 | cochlear implantation |
| 0.471 | cochlear implantations |
| 0.858 | cochlear implants |
| 0.601 | cochlear prostheses |
| 0.178 | cochlear prosthesis |
| 0.063 | cochlear prosthesis implantation |
| 0.165 | hearing prosthesis |
5. Discussion and Conclusions
We find that given a collection of well-formed and meaningful biomedical phrases such as can be obtained from the UMLS Metathesaurus or the full forms coming from abbreviations in MEDLINE, we can learn to distinguish these phrases from other similarly prepared phrase candidates taken from the same set of sentences at a MAP level of about 42%. When we use such a trained model to score the phrase candidates we find a large number score at or above the median score for the high quality set we used in training and on judging such high scoring phrases we find that over 85% of them are high quality meaningful phrases. We chose to look at those candidate phrases that scored higher than the median of the high quality set because with perfect learning this would imply they are at least as good as half of the phrases in the positive training data we started with. Of course no training is perfect, so we are not surprised when they prove to consist of between 80 and 90% high quality phrases. However, we do believe the results justify the approach and prove that there is significant learning taking place here.
One may ask what motivated the choice of features we use for the learning. If we were only attempting to identify noun phrases syntactic information may be sufficient and perhaps we only would need parts-of-speech. If we defined a useful phrase as a frequently used phrase again the problem would be relatively simple. But when we ask for the most useful phrases to a human we are asking a much more difficult question without a simple or precise answer. This makes the problem more challenging. Certainly we want well-formed phrases so syntax is important. Also we believe the most useful phrases will be used more frequently than less useful phrases, so frequency information is important. But as we illustrated in the Introduction, frequency does not tell one all that is needed. The machine learning methods we apply have the ability to ignore features that are not useful, so in some sense more features increase the chance of good performance. We have given some justifications for the features we have chosen to use in section 2.2. However, the main justification for using these features is that we found they improved MAP as seen in our tests. Even though features f6 and f7 measure similar things we found including both improved overall performance. As seen in Table 3 many of the features seem to make only a small contribution to performance. We believe this is due to two factors. First, there is some redundancy because of the number of different features we use. Second, some of the features are less important. However, even if features individually make only a small contribution, in aggregate they can make a substantial improvement in performance. It may well be that there are more useful features yet to be discovered.
Again one may ask whether the machine learning methods we have applied are the best possible. Our reason for choosing them is quite simple. Support vector machines generally give performance that is excellent on a wide variety of classification problems [25]. Further cost sensitive SVMs and the related Huber based classifiers are especially well suited to the problem of imbalanced data with which we deal here [14–17]. The fact that we obtain nearly equal performance with the two methods and the results are a very large improvement over naïve Bayes is perhaps some justification for our approach.
Since our approach is a machine learning approach and departs from previous methods, it is of interest how our method may compare with prior approaches. For such a comparison we rely on the work of Zhang, Iria, Brewster and Ciravegna [11] who compared five different methods of term recognition on the GENIA corpus and found that the C-value method of Frantzi and Ananiadou [6] performed the best and even better than a voting combination of the different methods. With this in mind we compared the C-value method with our approach. The C-value method first applies one of several possible filters to restrict to a syntactic class and then ranks all the phrases that pass the filter by their C-value. We can make an essentially equivalent computation by combining the C-value as a feature with the POS features and training the Huber classifier with these features. When we do this and compute MAP numbers using the same cross validation used to compute the results in Table 3 we obtain a MAP of 0.238 for UMLS and 0.219 for Full Forms. Thus our machine learning almost doubles the performance obtained by the C-value approach applied to our data with our evaluation approach. Based on these results we conclude that machine learning with many different features can impart a significant advantage over prior methods when a large set of high quality phrases are available for training.
One peculiarity of our approach is that we have two sets of high quality phrases and they are not completely equivalent. We find that when a classifier is trained with the U ⋃ MU data and the learning is applied to the F ⋃ MF problem or vice versa, the classification MAP drops to approximately half the value obtained in the cross validation experiments. This suggests there are significant differences between U and F . One difference we believe may be important is frequency. There are more low frequency phrases in F than in U. In fact the average frequency of a phrase in F is 263 while the average frequency of a phrase in U is 408. Likewise 20% of phrases in F only appear in MEDLINE text once while this is true of 15% of phrases in U. While this is a consistent difference, we believe it is not the full explanation of the difference between these two phrase sources. Further research may clarify the differences.
While high scoring phrases based on the trained Huber classifier tend to be of high quality, results are not perfect. We examined a number of the high scoring phrases judged by us to be of poor quality. While a number of the errors appear to be unique one-of-a-kind errors, one can detect some patterns. For example it can happen that two names appear together repeatedly in a corpus but the combination does not make for a good phrase. For example, “dia niemela” is a juxtaposition of two author last names which in the text appear as “Dia & Niemela”, but our processing ignored the “&” symbol and led to the error of scoring “dia niemela” as a good phrase. Another example is the expression “cell cell” which was scored high but is not very meaningful as a phrase. A further example is “california los angeles” which occurs in text repeatedly as “University of California Los Angeles”. Were prepositions allowed in the phrases we extract, it is possible that this error would have been avoided. Another type of error is when a phrase is truncated on one end. An example is the phrase “fkhr fusion gene” which is truncated on the left and appears in the text as “PAX7-FKHR fusion gene” where the two genes that are fused are included in the phrase. Clearly a more sophisticated processing would have corrected this error. Another example is “heart association functional class” which appears in the text as “New York Heart Association functional class”. Again a more sophisticated processing could have corrected this error. From these examples it is evident that our processing could be improved. However, for our application where the phrases in a synonymy class are predetermined and we are only choosing the best representative of the class such improvements may provide little benefit.
Our application does not require perfect identification of high quality phrases, but it is benefited by the ability to choose with high probability the representative among several semantically equivalent choices that will be most acceptable to a human user of a system. On the other hand one can imagine other applications where this ability may not be useful. For example, a language generation system would not likely find such a discrimination useful because all the phrases we deal with, both the positive and the unlabeled, are snippets from actual written language produced by a human. Thus for language generation there may be little value in discriminating as we are doing here. Finally, if one requires perfection in choosing phrases most meaningful to a human, then our results would still require human review.
Supplementary Material
Highlights
We study extraction of well formed and high quality biomedical phrases from MEDLINE.
Syntactic and statistical features are employed.
Machine learning is applied using sets of known good phrases.
Over 85% of such extracted candidates are humanly judged to be of high quality.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Yeganova L, et al. How to interpret Pubmed queries and why it matters. Journal of the American Society for Information Science. 2009;60:264–274. doi: 10.1002/asi.20979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chen KH, Chen HH. Extracting noun phrases from large-scale texts: a hybrid approach and its automatic evaluation. ACL '94 Proceedings of the 32nd annual meeting on Association for Computational Linguistics.1994. [Google Scholar]
- [3].Evans DA, Zhai C. Noun-Phrase Analysis in Unrestricted Text for Information Retrieval. Proceedings of the 34th Annual Meeting of the Association for Computational Linguistics.1996. pp. 17–24. [Google Scholar]
- [4].Bennett NA, et al. AMIA'99 Annual Symposium. Washington, D.C.: 1999. Extracting Noun Phrases for all of MEDLINE; pp. 671–675. [PMC free article] [PubMed] [Google Scholar]
- [5].Wermter J, Hahn U. Paradigmatic modifiability statistics for the extraction of complex multiword terms. Proceedings of the conference on Human Language Technology and Empirical Methods in Natural Language Processing HLT 05.2005. pp. 843–850. [Google Scholar]
- [6].Frantzi K, Ananiadou S. The C-value/NC-value Method of Automatic Recognition for Multiword Terms. Proceedings of the Second European Conference on Research and Advanced Technology for Digital Libraries.1998. pp. 585–604. [Google Scholar]
- [7].Baldwin T, Kim SN. Handbook of Natural Language Processing. CRC Press; Boca Raton, USA: 2010. [Google Scholar]
- [8].Kim WG, Wilbur WJ. Corpus based statistical screening for phrase identification. Journal of the American Medical Informatics Association. 2000;7:499–511. doi: 10.1136/jamia.2000.0070499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Murphy R. Phrase detection and the associative memory neural network. presented at the Proceedings of the International Joint Conference on Neural Networks 2003; 2003. Conference. [Google Scholar]
- [10].Kim HR, Chan P. Identifying Variable-Length Meaningful Phrases with Correlation Functions. presented at the IEEE International Conference on Tools with Artificial Intelligence 2004 Conference. [Google Scholar]
- [11].Ziqi Zhang JI. Christopher Brewster and Fabio Ciravegna A Comparative Evaluation of Term Recognition Algorithms. Proceedings of the Sixth International Conference on Language Resources and Evaluation 2008. [Google Scholar]
- [12].Sohn S, et al. Abbreviation definition identification based on automatic precision estimates. BMC Bioinformatics. 2008;9:402. doi: 10.1186/1471-2105-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Smith L, et al. MedPost: A part of speech tagger for biomedical text. Bioinformatics. 2004;20:2320–2321. doi: 10.1093/bioinformatics/bth227. [DOI] [PubMed] [Google Scholar]
- [14].Lewis DD, Yang Y. RCV1: A New Benchmark Collection for Text Categorization Research. Journal of Machine Learning Research. 2004;5:361–397. [Google Scholar]
- [15].Abkani R, Kwek S. Applying Support Vector Machines to Imballanced Datasets. ECML. 2004 [Google Scholar]
- [16].Zhang T, Iyengar VS. Recommender Systems Using Linear Classifiers. Journal of Machine Learning Research. 2002;2:313–334. [Google Scholar]
- [17].Yeganova L, et al. Text Mining for Leveraging Positively Labeled Data. presented at the BioNLP 2011; 2011. Conference. [Google Scholar]
- [18].Chawla NV, Bowyer KW. SMOTE: Synthetic Minority Over-sampling Technique. Journal of Artificial Intelligence Research. 2002;16:321–357. [Google Scholar]
- [19].Maloof M. Learning when data sets are imbalanced and when costs are unequal and unknown. Proc. of the ICML-2003Workshop:Learning with Imbalanced Data Sets II. 2003:73–80. [Google Scholar]
- [20].Weiss G, McCarthy K. Cost-Sensitive Learning vs. Sampling: Which is Best for Handling Unbalanced Classes with Unequal Error Costs?. Proceedings of the 2007 International Conference on Data Mining.2007. [Google Scholar]
- [21].Zhang T. Solving large scale linear prediction problems using stochastic gradient descent algorithms. Twenty-first International Conference on Machine learning. 2004:918–922. [Google Scholar]
- [22].Langley P. Elements of Machine Learning. Morgan Kaufmann Publishers, Inc.; San Francisco: 1996. [Google Scholar]
- [23].Wilbur WJ, Kim W. The ineffectiveness of within-document term frequency in text classification. Information Retrieval. 2009;12:509–525. doi: 10.1007/s10791-008-9069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Baeza-Yates R, Ribeiro-Neto B. Modern Information Retrieval. Addison-Wesley Longman Ltd.; Harlow, England: 1999. [Google Scholar]
- [25].Yang Y, Liu X. A re-evaluation of text categorization methods. 22nd Annual ACM Conference on Research and Development in Information Retrieval; Berkeley, CA. 1999. pp. 42–49. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.