Abstract
In cells, there exists a delicate balance between accumulation of charged metal cations and abundant anionic complexes such as phosphate. When phosphate metabolism is disrupted, cell-wide spread disturbances in metal homeostasis may ensue. The best example is a yeast pho80 mutant that hyperaccumulates phosphate and as result, also hyperaccumulates metal cations from the environment and shows exquisite sensitive to toxicity from metals such as manganese. In this study, we sought to identify genes that when over-expressed would suppress the manganese toxicity of pho80 mutants. Two classes of suppressors were isolated, including the histone chaperones SPT16 and HPC2, and RAD23, a well-conserved protein involved in DNA repair and proteosomal degradation. The histone chaperone gene HPC2 reversed the elevated manganese and phosphate of pho80 mutants by specifically repressing PHO84, encoding a metal-phosphate transporter. RAD23 also reduced manganese toxicity by lowering manganese levels, but RAD23 did not alter phosphate nor repress PHO84. We observed that the RAD23-reversal of manganese toxicity reflected its role in protein quality control, not DNA repair. Our studies are consistent with a model in which Rad23p partners with the deglycosylating enzyme Png1p to reduce manganese toxicity through proteosomal degradation of glycosylated substrate(s).
Keywords: Manganese, Phosphate, Yeasts, Metals
1. Introduction
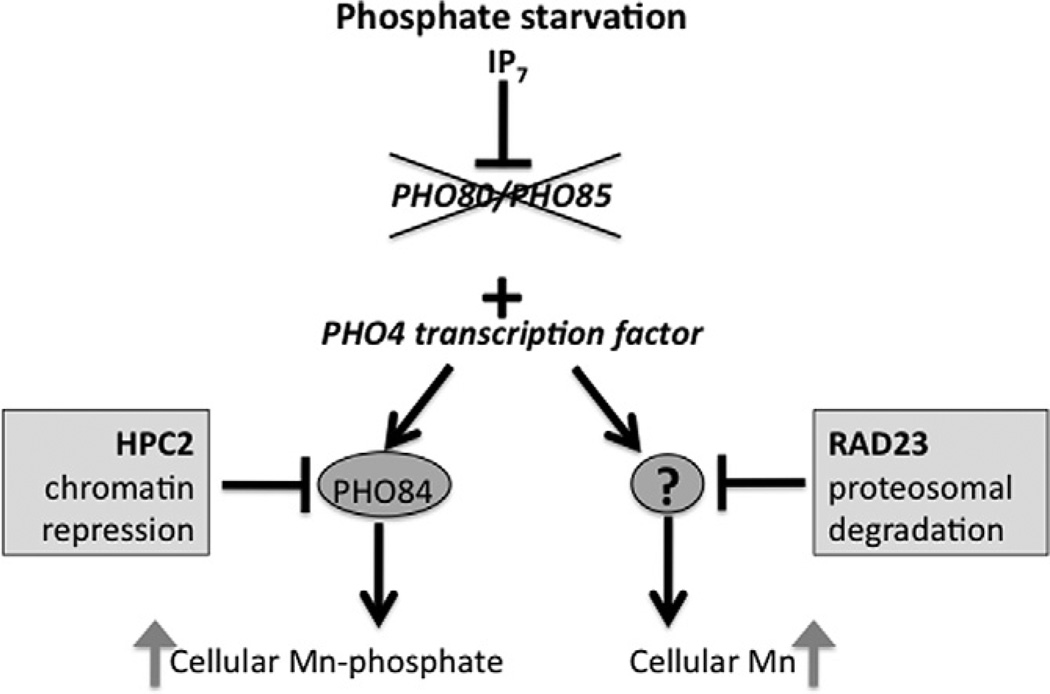
Phosphate is regulated in yeast cells through the Pho85p–Pho80p cyclin dependent kinase–cyclin complex [1]. When phosphate is abundant, this complex phosphorylates the Pho4p transcription factor and prevents the factor from activating phosphate metabolism genes. Conversely, when phosphate is limiting, IP7 inositol pyrophosphate inhibits the Pho85p–Pho80p kinase complex, and the dephosphorylated Pho4p factor is free to enter the nucleus and activate the transcription of a wide assortment of genes that mediate the uptake, storage and metabolism of phosphate (summarized in cartoon of Fig. 4) [2–5].
Fig. 4.
The effects of phosphate dysregulation and the HPC2 and RAD23 suppressors on manganese accumulation in S. cerevisiae Shown is a cartoon depicting the phosphate regulation pathway in S. cerevisiae. In phosphate starved cells, IP7 inositol pyrophosphate inhibits the Pho80p/Pho85p kinase pair which allows for activation of the Pho4p transcription factor [2–5]. Pho4p is also constitutively active in pho80Δ or pho85 Δ null strains (depicted by X). Pho4p activates expression of the PHO84 gene encoding the metal-phosphate transporter through chromatin remodeling effects, and as a result, cells hyperaccumulate manganese and phosphate. The Pho84p-increase in manganese toxicity can be reversed by over-expression of the histone chaperone HPC2 that represses PHO84 transcription, presumably by nucleosome deposition on the PHO84 gene promoter. The Pho4p transcription factor also activates a secondary pathway for increasing manganese accumulation and manganese toxicity that is currently of unknown nature (represented by “?”) but works independent of PHO84 and without obvious changes in phosphate [11]. This secondary pathway can be suppressed by over-expression of RAD23 through a pathway involving the proteosomal degradation of glycosylated substrates.
One of many targets of Pho4p regulation is PHO84 which encodes the cell surface high affinity transporter for phosphate [4–7]. Compared to other gene targets of Pho4p, PHO84 exhibits a lower threshold for induction and is rapidly activated with relatively small changes in intracellular phosphate [8]. Interestingly the substrate for the Pho84p phosphate transporter is a phosphate–divalent metal ion complex, such as manganese–phosphate [9,10]. In this manner, Pho84p not only controls cellular phosphate but can also influence the uptake and toxicity of divalent metals. We have previously shown that cells lacking PHO84 are resistant to toxicity from manganese, zinc and cobalt [10]. Moreover, activation of PHO84 gene expression, through a mutation in the aforementioned Pho80–Pho85 kinase, leads to elevated uptake of manganese from the environment [11]. However, this elevated manganese uptake by Pho84p was by itself not sufficient to explain all the manganese toxicity in pho80 (or pho85) mutants. There are other targets of the Pho4p transcription factor that cause increased sensitivity to metals when phosphate regulation is lost [11]. The nature of this Pho84-independent manganese toxicity is not understood.
In an attempt to better understand the changes in metal sensitivity associated with loss of phosphate control, we conducted a dosage suppressor screen for genes that alleviate the manganese sensitivity of a pho80Δ mutant. This screen led to the identification of two histone chaperones HPC2 and SPT16, as well as RAD23, involved in nucleotide excision repair and proteosome function. The histone chaperones were found to suppress metal toxicity by repressing transcription of the metal-phosphate transporter gene, PHO84. By comparison RAD23 reversed manganese toxicity without changes in PHO84 gene activity. Our mechanistic studies implicate a role for RAD23 in proteosomal degradation of a certain glycosylated protein that controls manganese accumulation and manganese toxicity.
2. Materials and methods
2.1. Strains and growth conditions
All the strains in this study are isogenic to BY4741 (Mata, leu2Δ0, met15Δ0, ura3Δ0, his3Δ1). Commercially available deletion strains described throughout (Open Biosystems) were verified using DNA sequencing. PHO84 was disrupted in BY4741 and in the pho80Δ::KanMX4 strain to create LR122 (pho84Δ::LEU2) and LR154 (pho80Δ::KanMX4 pho84Δ::HIS3) using the previously published pLJ246 [10] and pLJ089 [12] pho84 deletion plasmids. LR237 (pho80Δ::LEU2), LR359 (rpn10Δ::KanMX4 pho80Δ::LEU2) and LR329 (png1Δ::KanMX4 pho80Δ::LEU2) were constructed using the pLR001 deletion plasmid [11] in wild type BY4741, or the respective isogenic single gene deletion strains from the Open Biosystems collection. RAD4 was disrupted in BY4741 using the LR01 deletion plasmid (described below) in pho80Δ::KanMX4 to generate the LR222 (pho80Δ::KanMX4 rad4Δ::LEU2) strain.
Cells were maintained by growth at 30 °C in either enriched YPD (1% bacto-yeast extract, 2% bacto-peptone, 2% dextrose), or SC (synthetic complete) media with 2% agarose added for solid media [13]. For metal toxicity tests, 105, 104, 5 × 102 and ~25 cells were spotted onto solid YPD medium supplemented with defined metal concentrations and allowed to grow at 30 °C for 48 h.
2.2. Plasmids and genomic library screening
To isolate multi-copy suppressors of pho80 manganese toxicity, the LR801 pho80Δ::KanMX4 strain was transformed with a genomic URA3/2µ library derived from pRS202 [14]. Approximately 100,000 colonies were tested for growth on SC medium lacking uracil and supplemented with 40 mM MnCl2. Plasmids conferring manganese resistance were isolated and sequenced, and through a series of sub-cloning experiments, the manganese-resistance loci identified. In total, the complementing PHO80 gene was isolated four times, MNR2 and RAD23 isolated four and five times respectively; HPC2 was isolated seven times and a single isolate of SPT16 was obtained. All subsequent analyses of SPT16 involved the original library isolate pS3 that represents a Sau3A/Sau3A genomic DNA fragment from chromosome VIII sequences 98003 to 104554. The p3LΔEcoRI plasmid for expressing HPC2 (spanning chromosome II sequences 653413–657032) is a derivative of the original genomic library clone in which an EcoRI/EcoRI fragment outside of the HPC2 containing region was deleted. The p1FΔKpnI plasmid for expressing RAD23 was derived from the original genomic library clone by deleting a Kpn1/KpnI fragment outside of the HPC2 containing region. The RAD4 disruption plasmid, LR01, was generated by PCR amplification of upstream (−610 to −98) and downstream (2665–3465) sequences introducing BamHI and NotI or PstI and BamHI sites, respectively. The RAD4 PCR products were digested with the indicated enzymes and ligated in a trimolecular reaction into pRS305 (LEU2) [15] digested with SalI and NotI. Transformation with pLR01 digested with BamHI resulted in the deletion of chromosomal RAD4 from to −98 to +2665.
2.3. Biochemical assays
For measurements of phosphate and for atomic absorption spectroscopy (AAS) analysis of manganese, cells were seeded at an OD600 = 0.05 in YPD medium and grown for 18 h at 30 °C. In the case of metals analysis, the growth media was supplemented with either 70 µM (pho80Δ single mutants) or 400 µM (pho80Δ pho84Δ double mutants) MnCl2 to monitor metal accumulation under manganese toxicity conditions. The pho80Δ pho84Δ is far more resistant to manganese than the single pho80Δ mutant, hence the need for higher levels of the metal. Whole cell manganese analysis by AAS was carried out on a PerkinElmer Life Sciences AAnalyst 600 graphite furnace atomic absorption spectrometer according to the manufacturer’s specifications, as described [10]. Inorganic phosphate was assayed using molybdate reactivity as described [12]. Polyphosphate was detected using polyacrylamide gel electrophoresis and toluidine blue staining as described [12].
For quantitative PCR, triplicate cultures of WT and pho80 mutants were grown in YPD + 75 µM MnCl2 to an OD600 = 1.0. Manganese was added to cultures to maintain selective advantage of 2µ plasmids in pho80Δ cells. Cells were harvested, RNA extracted using the hot phenol method [16] and analysis of PHO84 expression by quantitative PCR carried out precisely as previously done using ACT1 as control [11].
3. Results and discussion
3.1. Isolation of manganese resistant suppressor genes
We screened a multi-copy yeast genomic library for genes that would reverse the manganese sensitivity of pho80Δ mutants. This screen resulted in the identification of five genes that when overexpressed, permitted growth of pho80Δ mutants on high manganese. These genes included the complementing PHO80 locus, the vacuolar magnesiumand cobalt transporter MNR2 that also confers resistance to manganese [17], the DNA repair and ubiquitin receptor protein RAD23 [18], and two genes involved in chromatin remodeling, namely SPT16 and HPC2. Roles for SPT16, HPC2 and RAD23 in modulating manganese toxicity had not been previously documented and these genes were selected for further study.
3.2. The chromatin remodeling factors and metal toxicity in pho80 mutants
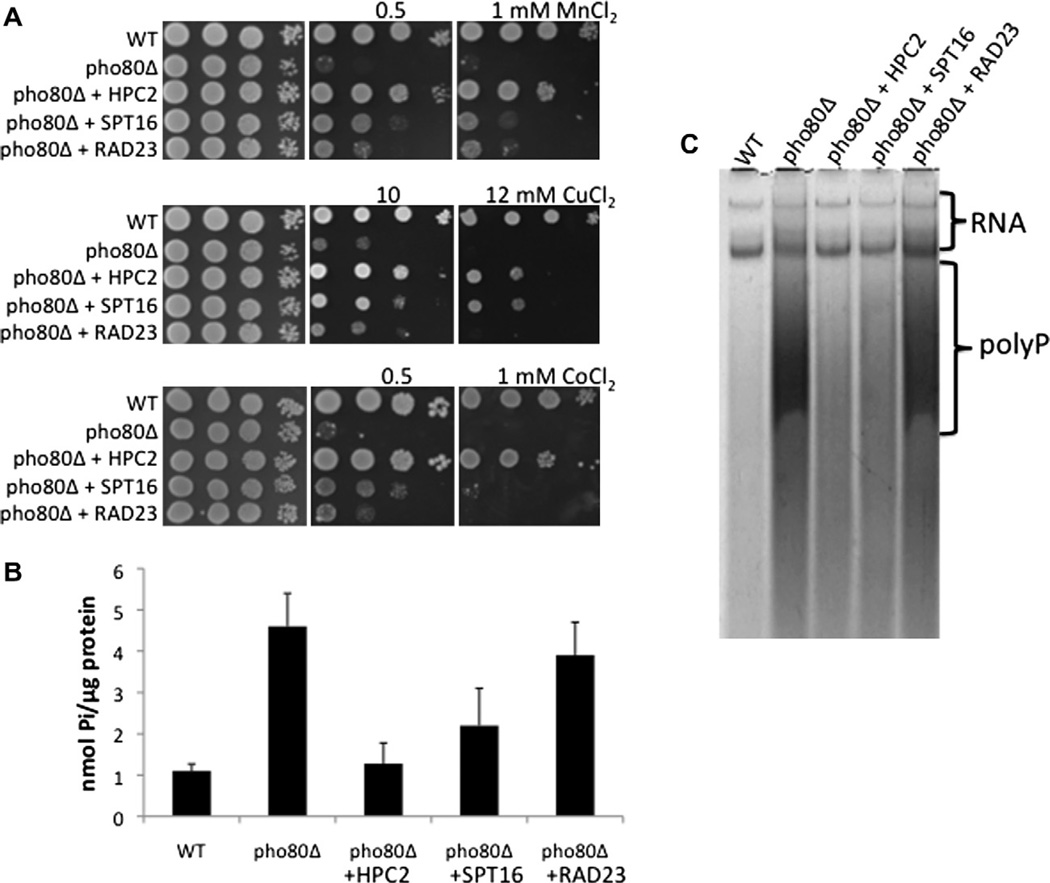
Spt16p and Hpc2p are among a class of proteins known as histone chaperones that facilitate the assembly and disassembly of nucleosomes. Spt16p is a component of the FACT nucleosome remodeling complex that acts on H2A/H2B histones [19], while Hpc2p is part of the HIR complex that operates on histones H3/H4 [20]. As seen in Fig. 1A, over-expression of HPC2 and SPT16 not only reduced manganese toxicity in pho80Δ mutants, but also suppressed toxicity from cobalt and copper. The effects of HPC2 and SPT16 were not limited to metals. Mutants of pho80Δ are known to hyperaccumulate phosphate [6,12] and over-expression of either HPC2 or SPT16 helped reduce levels of orthophosphate (Fig. 1B) and polyphosphate (Fig. 1C) in the pho80Δ mutant. In all these assays, the effects of over-expressed HPC2 were more pronounced than that of SPT16 and we therefore focused on HPC2 for further analysis.
Fig. 1.
Effects of over-expressing HPC2, SPT15 and RAD23 on metal toxicity and phosphate accumulation of pho80Δ mutants. (A) Metal toxicity was measured by spotting 1 × 105, 1 × 104, 5 × 102 and 25 cells of the indicated strains onto YPD medium supplemented with the designated concentrations of metal salts and by allowing growth for 2 days at 30 °C. (B) Whole cell orthophosphate analysis of cell lysates was carried out using the molybdate detection method. Values represent the averages of two independent cultures from independent experimental trials; error bars represent standard deviation. (C) Polyphosphate analysis of cells was conducted by polyacrylamide gel electrophoresis and toluidine blue staining as described in Section 2. The positions of RNA and large polyphosphate molecules are shown. Strains used: WT, BY4741; pho80Δ, LR801. Multi-copy yeast plasmids harboring genomic copies of the designated genes were as follows: HPC2, p3LΔEcoRI; SPT16, pS3; RAD23, p1FΔKpnI.
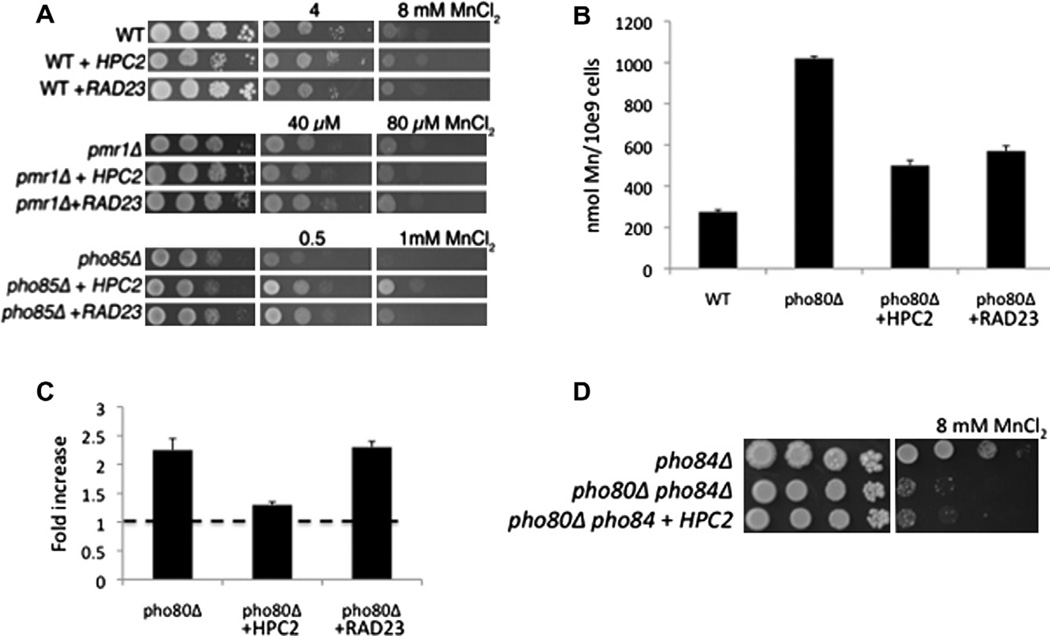
We tested whether the HPC2-resistance to manganese was unique to cells with disrupted phosphate control or was independent of phosphate. As seen in Fig. 2A, over-expression of HPC2 did not increase manganese resistance in a wild type strain, nor in a pmr1 mutant that is highly sensitive to manganese toxicity without changes in phosphate [21,22]. By comparison, HPC2 did increase manganese resistance of a pho85 mutant that like pho80 strains, cannot control phosphate uptake (Fig. 2A bottom) [12,22]. Hence, the effects of HPC2 on manganese are specific to strains with disruptions in phosphate metabolism.
Fig. 2.
HPC2 and RAD23 lower manganese accumulation in pho80 mutants by distinct methods. (A) The indicated strains were tested for manganese toxicity as in Fig. 1A. The different levels of manganese salts represent the differing sensitivities of the parental strains to the metal where WT strains are most resistant and pmr1Δ mutants are most sensitive to manganese toxicity. (B) The indicated strains were tested for whole cell manganese by AAS as described in Section 2. Values are averages of duplicate measurements from two independent cultures and error bars indicate standard deviation. (C) Real time PCR analysis of PHO84 mRNA in the indicated strains was carried out as described in Section 2 and previously published [11]. Fold change represents the increase in PHO84 mRNA over the WT control. Dotted line shows value of 1.0 assigned to WT control. Values are averages of duplicate measurements from three independent cultures; error bars indicate standard deviation. (D) The indicated strains were tested for manganese toxicity as in Fig. 1A. The pho84Δ mutation reverses much of the manganese toxicity of a pho80Δ strain; hence the higher level of manganese used in toxicity testing. Strains used: (A) WT, BY4741; pmr1Δ and pho85Δ are KanMX4 deletion derivatives of BY4741; (B and C) as described in Fig. 1; (D) pho84Δ, LR122; pho80Δ pho84Δ, LR154. The indicated plasmids for expressing HPC2 and RAD23 are as described in Fig. 1.
Manganese toxicity of pho80 mutants correlates with increased uptake of manganese from the growth medium [11]. As seen in Fig. 2B, pho80 mutants accumulate ≈4-fold higher levels of manganese than WT cells exposed to manganese. Over-expression of HPC2 substantially reversed this high accumulation of manganese (Fig. 2B), indicating that HPC2 reverses manganese toxicity by inhibiting manganese uptake. We tested whether HPC2 was working through PHO84, encoding the metal-phosphate transporter [10,11]. By quantitative PCR, PHO84 mRNA levels increased more than 2-fold in pho80Δ mutants (Fig. 2C). Notably, HPC2 overexpression decreased PHO84 mRNA to near-control levels. To determine if this lowering of PHO84 mRNA was by itself responsible for HPC2 suppression of manganese toxicity, we created a pho80 pho84 double mutant. As seen in Fig. 2D, over-expressed HPC2 could not suppress manganese toxicity in this strain lacking Pho94p (Fig. 2D). Together these results indicate that the HPC2 is reversing metal toxicity of pho80 mutants specifically by repressing transcription of the PHO84 metal-phosphate transporter, presumably through chromatin remodeling effects.
Activation of Pho4-target genes in yeast is known to involve chromatin remodeling. As a histone chaperone, Hpc2p could conceivably repress (or activate) the chromatin of any number of Pho4-targets, but the studies of Fig. 2D demonstrate that repression of PHO84 is the only target relevant to the manganese resistance observed. Compared to other Pho4-target genes, the chromatin of PHO84 in particular has a particularly low threshold for activation by low phosphate [23]. Rapid activation of PHO84 involves histone eviction through the remodelers Snf2 and Ino80 and the histone acetyltransferase Gcn5 [23] as well as the binding of topoisomerase II to the gene promoter [24]. However, nothing has been reported for factors that mediate repression of PHO84 chromatin when phosphate is abundant, other than the histone deactylases Hda1p and Hda2p [25]. Based on our findings here with Hpc2p, the H3/H4 histone chaperones of the HIR complex are likely candidates for repressing PHO84.
It is noteworthy that the histone chaperone, Spt16 is also a multicopy suppressor of pho80 metal toxicity and phosphate accumulation (Fig. 1). Spt16 is a member of the FACT complex that is best known for its role in histone eviction during elongation, but can also re-assemble nucleosomes after the wake of RNA polymerase II [19]. Based on a synthetic lethality of spt16 and hpc2 mutations, it was proposed that Spt16 and Hpc2 play parallel roles in nucleosome deposition [26]. Like Hpc2p, Spt16p may facilitate repression of PHO84 chromatin when phosphate is abundant.
3.3. The effects of RAD23 on manganese toxicity in pho80 mutants
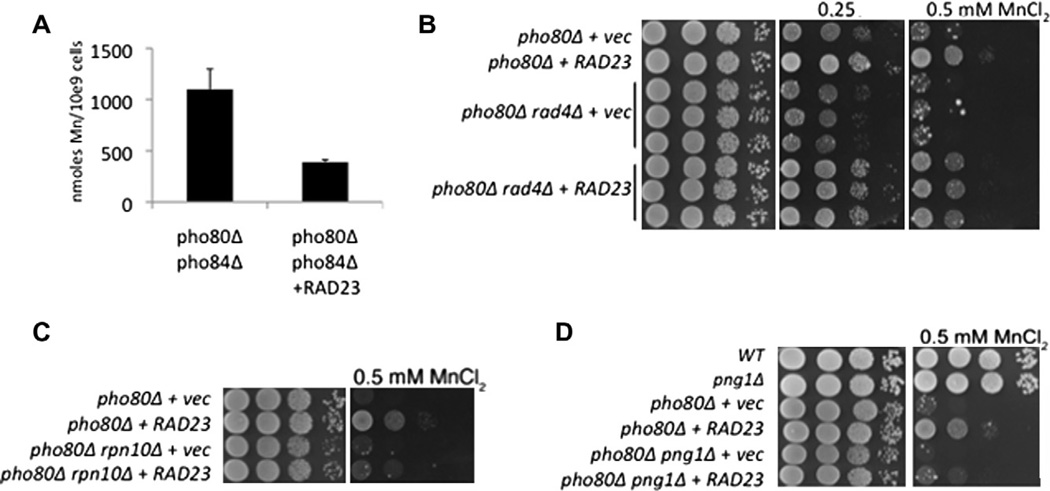
Manganese toxicity in pho80Δ mutants is also reversed by overexpression of RAD23. As with HPC2, the suppression of manganese toxicity by RAD23 was specific to cells with disrupted phosphate control (Fig. 2A) and correlated with a reduction in cellular manganese levels (Fig. 2B). However, unlike the histone chaperones HPC2 and SPT16, RAD23 seemed relatively specific to manganese. RAD23 afforded no increased resistance to other metals including copper and cobalt (Fig. 1A) and did not significantly alter phosphate levels (Fig. 1B, C). These results would suggest that RAD23 does not work to repress the metal-phosphate transporter Pho84p. Consistent with this, over-expression of RAD23 did not reduce mRNA levels of the PHO84 gene (Fig. 2B). Moreover, RAD23 still lowered manganese accumulation in a strain lacking pho84 (Fig. 3A). RAD23 works independent of Pho84p to reverse manganese toxicity.
Fig. 3.
RAD23 suppresses manganese toxicity through a mechanism involving the proteosome, not DNA repair. (A) AAS measurements of whole cell manganese in the indicated strains was determined as in Fig. 2B. (B–D) Manganese resistance in the indicated strains was monitored as in Fig. 1A. In part B, results from three independent transformants of the pho80Δ rad4Δ mutant are shown. Strains utilized: pho80Δ pho84Δ, LR154; pho80Δ, LR801; pho80Δ rad4Δ, LR222; pho80Δ rpn10Δ, LR359; pho80Δ png1Δ, LR329. Vec = strains transformed with empty 2µ URA3 pRS326 plasmid [15]. The RAD23 expressing plasmid is as described in Fig. 1.
Rad23p functions in both nucleotide excision DNA repair and the proteasomal pathway for protein degradation [18]. We sought to determine which of these roles was responsible for the RAD23-reversal of manganese toxicity. When DNA is damaged, Rad23p helps to repair the lesion through a complex with Rad4p, the yeast homolog of human Xeroderma pigmentosum group C complementing protein (XPC) [18]. To determine whether this Rad4-dependent DNA repair pathway was relevant to suppression of manganese toxicity, we created a pho80 rad4 double mutant. As seen in Fig. 3B, RAD23 was still capable of suppressing manganese toxicity in the absence of Rad4p. To test whether the proteosomal function of RAD23 was involved, we introduced a mutation in RPN10 encoding a component of the 19S proteasomal lid [27]. As seen in Fig. 3C, loss of RPN10 inhibited the ability of RAD23 to reverse manganese toxicity. Hence RAD23 attenuates the manganese toxicity of pho80 mutants through a mechanism that involves its role in the ubiquitin-proteosome system, not DNA repair.
Rad23p functions as an adaptor protein to transport ubiquitinated misfolded proteins to the proteosome for degradation [18]. Rad23p has numerous substrates, many of which are products of ERAD or endoplasmic reticulum (ER)-associated degradation. One such class of Rad23 substrates are N-glycosylated proteins that were misfolded in the ER. Rad23p recognizes these substrates through a partnership with Png1p, the primary deglycosylating enzyme in the cytosol [28,29]. We observed that RAD23 is not capable of reversing manganese toxicity in a pho80 strain that also lacks Png1p (Fig. 3D). These studies strongly indicate that RAD23 is reversing manganese toxicity by mediating the proteosomal degradation of a glycosylated protein in the secretory pathway.
What is the target of RAD23 that affects manganese toxicity? Because RAD23 over-expression reduces the accumulation of cellular manganese (Figs. 2B and 3A), a membrane transporter for manganese is likely. Moreover, this transporter should be regulated by phosphate and the Pho80/Pho85 pathway because RAD23 only reverses manganese toxicity in strains that have lost phosphate control (Fig. 2A). To date, Pho84p is the only manganese transporter known to be regulated by phosphate in yeast. We previously published that loss of pho80 increases manganese toxicity through two mechanisms: one resulting from increased manganese–phosphate uptake by Pho84p and a second that is independent of Pho84p [11]. Although the identity of this Pho84p-independent manganese transport system is still unknown, our studies indicate that this secondary pathway for manganese is a target for quality control of glycosylated proteins through Rad23p.
3.4. Overview
These findings underscore the complex ways in which loss of phosphate control can impact on manganese uptake in a eukaryotic cell. On one hand, activation of the metal-phosphate transporter Pho84p results in the uncontrolled uptake of toxic levels of manganese from the growth medium. Epigenetic effects on PHO84 must be a major contribution to the metal toxicity because the repression of PHO84 by the histone chaperone Hpc2p was in itself sufficient to restore much of the manganese resistance to pho80Δ cells (Fig. 4). Yet PHO84 is not the entire story: loss of phosphate control also activates a secondary pathway for manganese uptake that is still of unknown nature [11]. We provide evidence here that this secondary pathway represents a phosphate regulated manganese transport system that is subject to quality control by RAD23 proteosomal degradation (Fig. 4).
Acknowledgments
We thank Forrest Spencer for providing the yeast genomic DNA library and Edison Leung and Mark Carroll for analysis of genomic DNA plasmids. This work was supported by the JHU NIEHS center and by NIH grant ES 08996. LR was supported by NIEHS training grant ES 007308.
Abbreviations
- YPD
yeast extract peptone dextrose medium
- SC
synthetic complete medium
- AAS
atomic absorption spectrometry
- PCR
polymerase chain reaction
- ER
endoplasmic reticulum
References
- 1.Carroll AS, O’Shea EK. Pho85 and signaling environmental conditions. Trends Biochem. Sci. 2002;27:87–93. doi: 10.1016/s0968-0004(01)02040-0. [DOI] [PubMed] [Google Scholar]
- 2.Lee YS, Huang K, Quiocho FA, O’Shea EK. Molecular basis of cyclin–CDK–CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin–CDK–CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wykoff DD, O’Shea EK. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics. 2001;159:1491–1499. doi: 10.1093/genetics/159.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wykoff DD, Rizvi AH, Raser JM, Margolin B, O’Shea EK. Positive feedback regulates switching of phosphate transporters in S. cerevisiae. Mol. Cell. 2007;27:1005–1013. doi: 10.1016/j.molcel.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa N, DeRisi J, Brown PO. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersson J, Pattison J, Kruckeberg AL, Berden JA, Persson BL. Intracellular localization of an active green fluorescent protein-tagged Pho84 phosphate permease in Saccharomyces cerevisiae. FEBS Lett. 1999;462:37–42. doi: 10.1016/s0014-5793(99)01471-4. [DOI] [PubMed] [Google Scholar]
- 8.Springer M, Wykoff DD, Miller N, O’Shea EK. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 2003;1:E28. doi: 10.1371/journal.pbio.0000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fristedt U, Rest MVD, Poolman B, Konings WN, Persson BL. Studies of cytochrome c oxidase-driven H+-coupled phosphate transport catalyzed by the Saccharomyces cerevisiae Pho84 permease in coreconstituted vesicles. Biochemistry. 1999;38:16010–16015. doi: 10.1021/bi991545c. [DOI] [PubMed] [Google Scholar]
- 10.Jensen LT, Ajua-Alemanji M, Culotta VC. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J. Biol. Chem. 2003;278:42036–42040. doi: 10.1074/jbc.M307413200. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeld L, Reddi AR, Leung E, Aranda K, Jensen LT, Culotta VC. The effect of phosphate accumulation on metal ion homeostasis in Saccharomyces cerevisiae. J. Biol. Inorg. Chem. 2010;15:1051–1062. doi: 10.1007/s00775-010-0664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddi AR, Jensen LT, Naranuntarat A, Rosenfeld L, Leung E, Shah R, Culotta VC. The overlapping roles of manganese and Cu/Zn SOD in oxidative stress protection. Free Radic. Biol. Med. 2009;46:154–162. doi: 10.1016/j.freeradbiomed.2008.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman F. Getting started with yeast. Meth. Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 14.Sobering AK, Jung US, Lee KS, Levin DE. Yeast Rpi1 is a putative transcriptional regulator that contributes to preparation for stationary phase. Eukaryot. Cell. 2002;1:56–65. doi: 10.1128/EC.1.1.56-65.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces ceresiviae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces ceresiviae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pisat NP, Pandey A, Macdiarmid CW. MNR2 regulates intracellular magnesium storage in Saccharomyces ceresiviae. Genetics. 2009;183:873–884. doi: 10.1534/genetics.109.106419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dantuma NP, Heinen C, Hoogstraten D. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 2009;8:449–460. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Reinberg D, Sims RJ., 3rd De FACTo nucleosome dynamics. J. Biol. Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 20.Amin AD, Vishnoi N, Prochasson P. A global requirement for the HIR complex in the assembly of chromatin. Biochim. Biophys. Acta. 2011 doi: 10.1016/j.bbagrm.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Durr G, Strayle J, Plemper R, Elbs S, Klee SK, Catty P, Wolf DH, Rudolph HK. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol. Biol. Cell. 1998;9:1149–1162. doi: 10.1091/mbc.9.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNaughton RL, Reddi AR, Clement MH, Sharma A, Barnese K, Rosenfeld L, Gralla EB, Valentine JS, Culotta VC, Hoffman BM. Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy. Proc. Natl. Acad. Sci. USA. 2010;107:15335–15339. doi: 10.1073/pnas.1009648107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wippo CJ, Krstulovic BS, Ertel F, Musladin S, Blaschke D, Sturzl S, Yuan GC, Horz W, Korber P, Barbaric S. Differential cofactor requirements for histone eviction from two nucleosomes at the yeast PHO84 promoter are determined by intrinsic nucleosome stability. Mol. Cell Biol. 2009;29:2960–2981. doi: 10.1128/MCB.01054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sperling AS, Jeong KS, Kitada T, Grunstein M. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc. Natl. Acad. Sci. USA. 2011;108:12693–12698. doi: 10.1073/pnas.1106834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in Saccharomyces ceresiviae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Jimeno-Gonzalez S, Gomez-Herreros F, Alepuz PM, Chavez S. Gene-specific requirement for FACT during transcription is related to the chromatin organization of the transcribed region. Mol. Cell Biol. 2006;26(23):8710–8721. doi: 10.1128/MCB.01129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T, Park H, Lennarz WJ. Cytoplasmic peptide:N-glycanase (PNGase) in eukaryotic cells: occurrence, primary structure, and potential functions. FASEB J. 2002;16:635–641. doi: 10.1096/fj.01-0889rev. [DOI] [PubMed] [Google Scholar]
- 29.Kim I, Ahn J, Liu C, Tanabe K, Apodaca J, Suzuki T, Rao H. The Png1–Rad23 complex regulates glycoprotein turnover. J. Cell Biol. 2006;172:211–219. doi: 10.1083/jcb.200507149. [DOI] [PMC free article] [PubMed] [Google Scholar]