Abstract
We present the first large-scale identification of lariats—the transient branched introns that are released as a byproduct of pre-mRNA splicing. The locations of the branchpoints in these introns provide insight into the early steps of splicing. From this data set, we have developed a comprehensive model of 3′ splice-site selection, identified new mechanisms of alternative splicing and mapped the distribution of splicing factors around branchpoints.
Pre-mRNA contains short sequences coding for proteins (exons) interrupted by long, noncoding sequences (introns). The process called splicing connects two exons and releases the intron as a branched RNA (Fig. 1a, lariat). The lariats are produced in an equal quantity to the exon junctions in mRNA, but they degrade rapidly. Almost all we know about splicing in vivo comes from mRNA: hundreds of thousands of splice sites have been mapped from mRNA and genomic alignments, but fewer than 100 lariats have been characterized in vivo. Consequently, the mechanism of 3′ splice site (3′ss) selection is poorly understood. Here, we report what is, to our knowledge, the first large-scale detection of the transcript branchpoints from deep-sequencing data.
Figure 1.
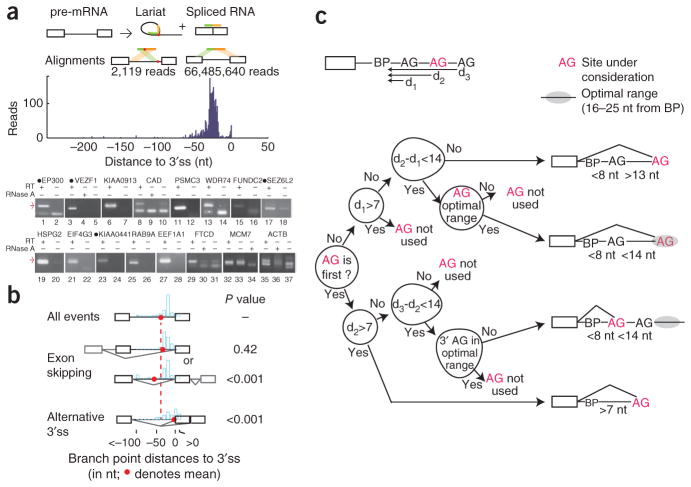
Identifying pre-mRNA lariats in human transcripts. (a) Each splicing event creates a lariat and spliced exon junction. Reverse transcriptase can extend through branched RNA. Inverted and conventional gapped alignments to the genome define branchpoint (red dot) and splice sites. The histogram represents the distribution of branchpoints mapped by distance in nucleotides to the 3′ splice site (3′ss). Predicted branchpoint validation gels are shown below. Black circles preceding the gene names indicate circularized introns. Red arrows represent lariat product sizes predicted from deep sequencing. The RNA dependence of the amplification was verified by the omission of reverse transcriptase (RT) or pretreatment with ribonuclease A (RNase A). (b) The mean locations and distribution of branchpoints for exons separated into three functional classes: all, skipped and alternative 3′ss exons. P values comparing mean branchpoint distances for alternative events to all events were calculated by resampling. (c) A decision tree describing AG selection to an accuracy of 95.6% using features identified in the literature. Upstream and downstream AGs (black font) are negative cases, while the observed 3′ss (in red) are positives. The most concise decision tree reveals four scenarios of AG selection (diagrammed to the right). Distances between the branchpoint and the upstream AG, used AG and downstream AG are represented as d1, d2 and d3, respectively.
Reverse transcriptase approaching the branchpoint by reading through the 5′ splice site (5′ss) occasionally traverses the branched nucleotide to copy the region of intronic RNA immediately upstream of the branchpoint1. This read contains two juxtaposed intronic segments that align in an inverted order, defining the 5′ss and the branchpoint (Fig. 1a). Reasoning that this same read-through phenomenon occurs during the construction of deep-sequencing libraries, we screened 1.2 billion total RNA reads for these unconventional, inverted alignments2. We found 2,118 reads that corresponded to 861 lariats from 759 U2-dependent introns and a single read from a U12-dependent intron (Fig. 1a). An additional 3% (70 lariats) corresponded to events with no transcript support. Although nine of these reads suggested splicing events that occurred deep within an intron, the lariat more often appeared to have arisen from a known site splicing to a nearby cryptic site (61 lariats). Overall, the data suggest that splicing occurs with high fidelity. None of the recovered lariats corresponded to recursive splicing or trans-splicing events that could be corroborated by transcript data or, in the case of recursive splicing, by nearby composite splice sites.
There appear to be differences between introns in terms of lariat abundance and the number of branchpoints used. It is known that branchpoints are functional elements whose disruption can result in a loss of splicing activity. (For example, numerous hereditary disease alleles in branch-point motifs result in splicing defects3; see Supplementary Fig. 1 and Supplementary Table 1). Whereas many introns appeared to splice via a single, presumably nonredundant branchpoint, other introns splice via multiple, presumably redundant sites (for example, the most extreme case listed in Supplementary Table 2, MCM7, utilizes 11 sites; Supplementary Fig. 1 shows the distribution of site multiplicity across introns). Analysis of sequence conservation supports this notion of two classes of introns. If an intron splices via a single branchpoint, we found its branchpoint region to be 17 times more conserved than those in introns that use alternate branchpoints (P value = 10−22 by t-test; see Online Methods). Short introns were sampled at a higher rate than long introns, even after correction for transcript abundance (Supplementary Fig. 2). For pre-mRNAs that undergo exon skipping, we detected expected levels of ‘branch-site skipping’ (lariats that encompass the skipped exon) consistent with an early-commitment model in which splice sites pair together before the first catalytic step of splicing (Supplementary Fig. 2)4.
Within annotated introns, 80% of the branchpoints mapped between 18 and 35 nucleotides (nt) upstream of the 3′ss (Fig. 1a). Of the 15 lariats validated in a nested RT-PCR-and-sequencing strategy, 11 mapped to within 2 nt of the predicted branchpoints (Fig. 1a, Supplementary Table 2). About 3% of all branchpoints appeared to map to the last intronic position (that is, the 3′ss), suggesting circularization. Some of these circles could be validated by RT-PCR in human cell lines, and, in both cases that we tested, circularization also occurred in the orthologous intron in mouse, as determined using brain RNA samples (Supplementary Fig. 3). However, these products did not seem to arise directly through splicing. The RNA appeared to have been spliced via a conventional lariat and then been circularized through a 3′-to-5′ linkage, possibly through a third nucleophilic attack or a debranching-and-ligation event. Unlike in branched RNA, the nucleotide in the circle corresponding to the last intronic position was not associated with the elevated substitution rate (40-fold) typical of reverse transcription through a 2′-to-5′ linkage (Supplementary Fig. 4, P < 10−12) and of some examples of circles spliced via conventionally located branchpoints during validation (for example, Fig. 1a, lane 23). Overall, these data suggest that there is an optimal branch-point location relative to the 3′ss. Branchpoints that map to the class of skipped exons, whose recognition is conditional, are located on average 15 nt upstream of this optimal distance (Fig. 1b). Alternative 3′ss exons have a bimodal distribution of branchpoints. Almost half (44%) of these branchpoints appear to enforce the usage of the distal 3′ss by mapping downstream of, on or within a few nucleotides of the proximal site. This analysis suggests that branchpoint location can determine splice site selection.
The existence of this data set of branchpoints provides an opportunity to test existing models of splice site recognition. For example, the widely accepted model of AG selection proposes a downstream scanning mechanism that starts from the branchpoint and selects the first AG for the 3′ss5. For 80% of the branchpoints, the first downstream AG is indeed the annotated 3′ss (P < 0.001, by permutation test). This phenomenon has been used to predict branchpoints by the ‘AG exclusion zone’ that often exists between the branchpoint and the 3′ss6. Other criteria have been proposed to modify this scan (such as secondary structure, context around the AG, distance to neighboring AGs and an optimal distance between the branchpoint and AG)7–10. To determine the relative importance of these influences, we used the branchpoint data to fit the process of AG selection to a decision tree. A decision tree optimized by the ID3 algorithm finds the most concise and effective hierarchy of decisions to explain the process of branch-point selection11. Because this framework is hierarchical, like the underlying scanning process, an optimal decision tree could provide insight about the dominance of these mechanisms and the order in which they are applied in vivo. For example, certain criteria previously reported to affect AG selection (such as secondary structure and AG context) did not contribute to the accuracy of our implementation of this classifier. Instead, almost all 3′ss decisions made after lariat formation could be explained by distances of the AGs to the branch-point and to each other. Optimizing the decision tree by testing a range of constraints returned four scenarios of 3′ss recognition that used the following types of spatial information: a preference for the first AG, a minimal distance (8 nt) and an optimal distance (16–25 nt) from the branchpoint (Fig. 1c). Separating the data into a testing set and training set using these optimized parameters returned a correct prediction rate of 95.6% for AG dinucleotides located downstream of identified branchpoints. This approach organized the conclusions of numerous biochemical case studies into a general unifying mechanism that is, in general, simpler and more complete than the models proposed in the separate studies.
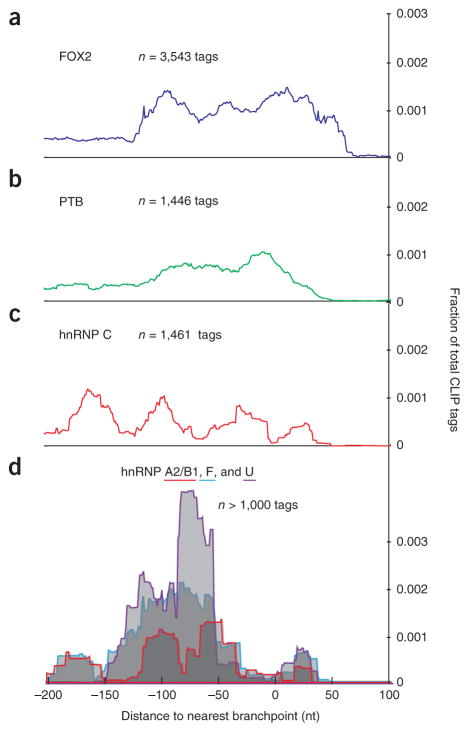
Finally, the availability of a large set of branchpoints allowed for the description of the architecture of RNA-protein interactions around the 3′ termini of introns. Overlaying genome-wide maps of splicing- factor binding reveals nonuniform distributions around the database of branchpoints12–15 (Fig. 2). In general, hnRNP proteins avoid the branchpoint (Fig. 2b,c: PTB and hnRNP C in HeLa cells; Fig. 2d: hnRNP A2/B1, hnRNP F and hnRNP U in 293 cells). HnRNP C has been shown to bind pre-mRNA with a periodicity of 165 nt (ref. 12). Overlaying these data suggests that the binding is phased in two registers around the branchpoint (that is, hnRNP C appears to contact the RNA either immediately upstream or downstream of the branchpoint). RNA maps illustrate the importance of position to splicing-factor function. To date, this type of analysis has classified binding-site function relative to splice sites; however, this is an incomplete division. With the availability of branchpoints, pre-mRNAs can be fully resolved into regions delimited by the main functional landmarks of splicing: the splice sites and the branchpoints.
Figure 2.
CLIP-tag density of splicing factors relative to the closest branchpoint. (a–d) Published alignments of cross-linking and immunoprecipitation (CLIP) tags were overlapped with lariat introns from four separate studies of human splicing factors: FOX2 in human embryonic stem cells14 (a); PTB13 (b) and hnRNP C12 in HeLa cells (c); and hnRNP A2/B1, hnRNP F and hnRNP U in 293 cells15 (d). The genomic coordinates of tag alignments were converted to distance from branchpoints. Tag density (y axis) is plotted along this branchpoint coordinate system (x axis) for three regulators of splicing. The branchpoint is located at x-axis position zero.
ONLINE METHODS
Lariat discovery
Data set
Lariat reads were identified in the Illumina Human Body Map 2.0 total RNA deep-sequencing library (GEO series accession number GSE30611). Reads consist of RNA samples derived from 16 tissues (GEO accession numbers GSM759522–GSM759537).
Hg19 annotated
We discovered branchpoints by searching for reads with inverted alignments. Reads with conventional alignments with fewer than four mismatches were eliminated. Each remaining read was split into all possible head and tail segments, in which each segment was at least 15 nt long. We mapped all head and tail segments to the genome using the Bowtie aligner16. Head and tail segments were required to have exactly one valid alignment and zero mismatches. Reads with segments that mapped in the expected order or did not map intronically were filtered. The remaining inverted reads were mapped to splice sites. In cases where the tail began at the first nucleotide of the intron and the head mapped within 500 nt upstream of a 3′ss, we determined that the read spans the lariat 2′–5′ linkage. In cases where there was alignment ambiguity, we allowed up to two mutations and assumed the alignment in which the tail mapped to the first nucleotide of the intron. The last nucleotide of the head was determined to be the branchpoint. Through this screen, 2,066 lariat reads were discovered (lariat_0–lariat_2065 in BED track). Reads that suggested branchpoints supported by spliced EST evidence were also reported. Nine lariat reads were discovered through this screen (lariat_2110–lariat_2118).
Unannotated transcripts with Illumina transcript support
To discover lariats forming in unannotated transcripts, we built a library of potential spliced products inferred from the inverted reads. For each inverted read that did not map to an annotated 5′ss, we constructed a potential upstream exon by taking an 85-nt window upstream of the read tail. We created an array of 200 potential downstream exons by taking 85-nt windows at a distance of 1–200 nt from the end of the read head. These windows were artificially spliced together to create a set of potential spliced products. We aligned the Illumina reads against these spliced products, requiring that each read contain at least 15 nt on either side of the splice junction and not have any mismatches. In cases where a splice product was found and the implied intron contained a 5′ GT and 3′ AG sequence, the inverted read was determined to be a true lariat forming in an unannotated transcript. Through this screen, 44 lariat reads were discovered (lariat_2066–lariat_2109).
Lariats forming deep within introns
From intronic out-of-order reads without transcript support, we filtered a high-confidence set of lariats by requiring that both the heads and tails were never annotated as exons (alternative events), the beginning of the tail had a patser score of at least 6.0 against a 5′ss position–specific weight matrix, and the read had a mutation at the branchpoint17.
Scoring bona fide lariats for matches to the 5′ss position–specific weight matrix, we found that 83% score greater than 6.0. Of inverted gapped intronic alignments that lack transcript support, 0.5% score greater than 6.0—a two-fold excess relative to noninverted gapped intronic alignments (0.22%) or random intronic windows sampled (0.29%). The mutation rate of reverse transcriptase is 40-fold higher when reading through a 2′–5′ (as opposed to 3′–5′) phospho-diester bond (Supplementary Fig. 2). Demanding this mutation at potential lariats increases this 2-fold excess to 13-fold.
We counted the number of splicing events that used an annotated 5′ss without a 3′ss, an annotated 3′ss without a 5′ss or, lastly, an event deep within an intron that used no annotated splice sites. We also counted the number of bona fide lariats that passed the patser score and mutational and intronic filters, and we used that fraction to extrapolate how many true lariats without transcript support we expected to exist in our data.
Identifying hereditary disease mutations in branch-point motifs
We selected the 66 Human Gene Mutation Database (HGMD) disease-causing splicing mutations located 20–35 bases upstream of a 3′ss (62 unique mutation positions) for examination. These mutations plus 7 nt of flanking sequence on either side were used as input for a ClustalW multiple-sequence alignment18. We used a maximal gap-open penalty in order to align the sequences without gaps. The ClustalW output was used to create a sequence logo with the application WebLogo 3 (ref. 19). We counted the number of times a mutation occurred at each position within the sequence logo to create a histogram showing how often a particular position within the sequence was affected by a splicing mutation.
Branchpoint characterization
Conservation
Introns with a minimum of five reads were separated into single- and multiple-branchpoint categories. Fifty-one introns had single branchpoints. Nineteen introns were multiclass, for a total of 71 branchpoints. Conservation of branchpoint motifs was estimated from mammalian phastCon score averaged over a 7-nt window centered on each branch-point. Single-branchpoint introns had significantly greater conservation (0.229, as compared to 0.013 for multiclass introns; P = 10−22 by t-test).
Distance
The branchpoint distance was measured as the distance between the last nucleotide of the read head to the first downstream annotated 3′ss.
Mutational profile
Circular introns have a significantly different mutational profile than conventionally located branchpoints in lariat introns. The χ2 test was used to compare the proportion of reads with an unambiguous base substitution at the transition site when this site occurred at position 0 as opposed to other locations.
Comparison of branchpoint distance between alternative and constitutive splicing events
mRNA exon-junction data were studied using the TopHat program20. We created a junction file consisting of all possible constitutive and exon skipping events within each annotated transcript. We aligned the Illumina reads using this junction file to determine how many reads spanned each splice junction. We calculated overall rates of alternative splicing and intersected these data with our lariat branchpoints. In Figure 1b, the average branchpoint-3′ss distance was measured for 2,066 reads. Resampling was used to compare the average distance of a series of 1,000 samples from different subsets of alternative events from the listed categories to the measured value.
Building a decision tree to model 3′ss selection
The number of AG dinucleotides between the branchpoint and the AG of the 3′ss was counted for the 2,066 annotated lariats. To determine the significance of our finding that 80% of the 3′ss were the first AG downstream of the splice site, 1,000 permutation trials simulated branchpoints (maintaining average 3′ss–branch-point distance) in introns to scan for the first AG.
AG-selection analysis was determined using the C5.0 software tool11. Introns with exactly one discovered branchpoint and one used 3′ss were considered in this analysis. The used AG, upstream AG (if extant), and downstream AG were included in the data set. First, the data were organized into a decision tree using classifiers from the literature, including the branchpoint-AG distance, the distance to surrounding AGs, the nucleotide upstream of the AG and the presence of secondary structure (as indicated by the Gibbs free energy, determined using RNA fold)21. In this initial run, the only informative classifiers were the presence or lack of an upstream AG, the branchpoint-AG distance and the distance to surrounding AGs. Next, a range of constraints for each of the distance classifiers was applied to the data set, and C5.0 was run on each of combination of constraints. The classifier sets with error rates lower than those of the literature classifier sets were run again on the data set, this time using half of the data set as training data and the other half as testing data. The highest-predictive-scoring classifier sets were subjected to ten-fold cross-validation trials. These trials were completed 1,000 times. The classifier set with the highest average predictive accuracy was used to create the decision tree.
RNA-protein interactions
Published cross-linking and immunoprecipitation (CLIP) data for FOX214, PTB13, hnRNP C12 and a panel of other hnRNP proteins15 were mapped around the branchpoints. The FOX2, PTB, and hnRNP A1, hnRNP A2/B1, hnRNP F, hnRNP M and hnRNP U data sets were smoothed using the center coordinate and adding 15 nt to either side. The raw hnRNP C CLIP reads were aligned using Bowtie, and the last nucleotide was used as the binding point (as described in this study). We smoothed the hnRNP C data by adding 15 nt to either side of the binding point. Of this set of proteins, we included CLIP-tag density plots for proteins with at least 1,000 CLIP-tag– lariat-intron overlaps.
Lariat recovery
The number of reads spanning each annotated splice junction was determined using TopHat. Lariats and splice junctions were both binned by intron size. The recovery rate of a lariat read was calculated by counting the number of detected lariat reads and dividing it by the number of detected splice-junction reads within each intron size bin. The error bars were calculated by resampling the lariat read data 1,000 times and using the 95% confidence interval.
Experimental methods
We performed nested RT-PCR to validate the branch-point predictions of lariats in total RNA from HEK293 cells. All sequences (primer, PCR), experimental or computational protocols and statistical tests are available at http://fairbrother.biomed.brown.edu/data/Lariat/.
Supplementary Material
Acknowledgments
We would like to thank R. Freiman and members of the Fairbrother lab for useful comments, suggestions and assistance; G. Schroth and Illumina for sharing body-map sequencing data; and G. Yeo for sharing RNA binding data before publication. The lab was supported by US federal funding sources R01GM095612–01 and NSF1020552 (both to A.J.T., W.G.F., J.S.S.) and by Brown University through the use of the OSCAR cluster (administered by the Center for Computation and Visualization) and the genomics core facility (8P30GM103410).
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
A.J.T. and W.G.F conceived and planned the project. A.J.T., A.M.D., J.S.S. and M.E.F collected data. A.J.T., A.M.D., J.S.S. and M.E.F performed analysis. A.J.T., A.M.D., J.S.S. and W.G.F. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Vogel J, Hess WR, Borner T. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkinson H, et al. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stenson PD, et al. Hum Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 4.Legrain P, Seraphin B, Rosbash M. Mol Cell Biol. 1988;8:3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith CW, Porro EB, Patton JG, Nadal-Ginard B. Nature. 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- 6.Gooding C, et al. Genome Biol. 2006;7:R1. doi: 10.1186/gb-2006-7-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S, Anderson K, Moore MJ. Proc Natl Acad Sci USA. 2000;97:593–598. doi: 10.1073/pnas.97.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chua K, Reed R. Mol Cell Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer M, Plass M, Perez-Valle J, Eyras E, Vilardell J. Mol Cell. 2011;43:1033–1039. doi: 10.1016/j.molcel.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 10.Smith CWJ, Chu TT, Nadalginard B. Mol Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinlan R. Mach Learn. 1986;1:81–106. [Google Scholar]
- 12.König J, et al. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue Y, et al. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeo GW, et al. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huelga SC, et al. Cell Reports. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertz GZ, Stormo GD. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 18.Chenna R, et al. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crooks GE, Hon G, Chandonia JM, Brenner SE. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C, Pachter L, Salzberg SL. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding Y, Chan CY, Lawrence CE. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.