Abstract
Low-density lipoprotein receptor–related proteins 5 and 6 (LRP5/6) mediate canonical Wnt–β-catenin signaling by forming a complex with the co-receptor Frizzled, which binds to Wnt proteins. Dickkopf (DKK)–related proteins inhibit the Wnt signaling pathway by directly binding to the ectodomains of LRP5/6. However, the mechanism for DKK-mediated antagonism has not been fully understood as of yet. Crystal structures of the LRP6 ectodomain in complex with DKK1, along with mutagenesis studies, provide considerable insights into the molecular basis for DKK-mediated inhibition and Wnt signaling through LRP5/6.
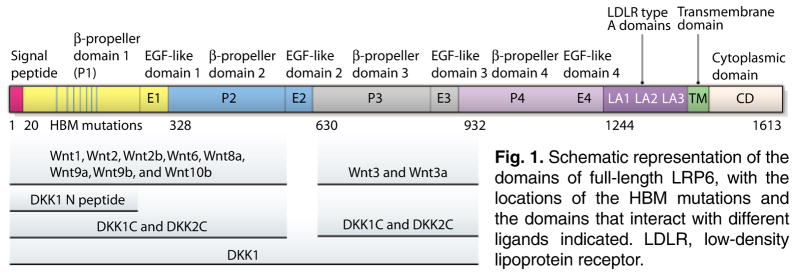
The Wnt signaling pathway is important in many biological processes, including embryonic development, organogenesis, tissue homeostasis, stem cell biology, and lipid and glucose metabolism. Dysregulation of Wnt signaling can lead to many pathophysiological conditions, including degenerative diseases and cancers (1–4). The canonical Wnt signaling pathway is activated when Wnt proteins bind to the receptor complex formed by Frizzled and low-density lipoprotein receptor–related protein 5 or 6 (LRP5/6). The full-length LRP5/6 can be divided into the ectodomain (ECD), transmembrane domain, and cytoplasmic domain. The LRP5/6 ECD contains four YWTD (Tyr-Trp-Thr-Asp) β-propeller domains, each of which is coupled to an epidermal growth factor (EGF)–like domain. These four β-propeller (P) and EGF-like (E) domains are followed by three low-densitylipoproteinreceptortypeA (LA) domains. Various mutagenesis studies have indicated that different Wnt molecules bind to different regions of LRP5/6 ECDs (Fig. 1) (5–8).
Fig. 1.
Schematic representation of the domains of full-length LRP6, with the locations of the HBM mutations and the domains that interact with different ligands indicated. LDLR, low-density lipoprotein receptor.
Wnt signaling is regulated by various antagonists that include the Dickkopf (DKK)–related proteins and the Wnt modulator in surface ectoderm (WISE) and sclerostin (SOST) families of proteins (3). Except for DKK3, the other three members of the DKK family are effective antagonists of canonical Wnt–β-catenin signaling that directly bind to LRP 5/6 with high affinities (7, 9, 10). The DKK molecules contain two conserved cysteine-rich domains (N and C for their relative position in the protein) that are connected by a linker domain (11). The C domains of DKK1 and DKK2 alone can inhibit canonical Wnt–β-catenin signaling (12, 13). The overall structure of DKK2C, as determined by nuclear magnetic resonance (NMR), is composed of six β-sheet regions that are stabilized by five disulfide bonds formed by 10 highly conserved cysteine residues (14). DKK2C is a relatively flat molecule, and each of the two sides is a potential binding surface. Mutagenesis studies have revealed that one side of DKK2C binds to LRP5/6, and the other side binds to Kremen (15), a molecule that modulates DKK-mediated Wnt antagonism (16). WISE and SOST proteins also inhibit Wnt–β-catenin signaling by directly binding to LRP5/6 (17, 18).
Some of the gaps in our understanding of Wnt signal transduction include the method by which Wnt interacts with its receptors and how Wnt antagonists, particularly those that directly bind to LRP5/6, inhibit Wnt signaling. Given that some of the human high bone mass (HBM) mutations have been mapped to the ECD of LRP5, the elucidation of molecular mechanisms for the interactions of Wnt and DKK or SOST with LRP5/6 could have important practical implications. Earlier mutagenesis studies suggested that β-propeller–EGF-like domains 3 and 4 (P3E3P4E4) of LRP5 and 6 are not required for Wnt1 signaling but are essential for DKK1-mediated inhibition of Wnt1 signaling (7, 8). However, it was puzzling that DKK1 could still bind to LRP5 without the P3E3P4E4 domain (8). In addition, DKK1 could bind to β-propeller–EGF-like domains 1 and 2 (P1E1P2E2) of LRP6 (5, 19). The issue was further compounded by the observations that DKK-mediated Wnt antagonism is inhibited by HBM mutations in the β-propeller domain 1 (P1) of LRP5 and a monoclonal antibody that binds to this region (20–23). Attempts were also made to probe the DKK interaction surface on β-propeller domain 3 (P3) of LRP5, based on a deduced structure modeled on the YWTD repeat domain of the low-density lipoprotein receptor, which revealed that several amino acids, particularly Glu721 in LRP5 and Glu708 in LRP6, are important for DKK binding (8). However, the lack of assessment of direct binding of purified recombinant proteins and atomic structures of LRP5/6 ECDs by themselves and in complex with DKK prevented reconciliation of the apparent paradoxes and understanding Wnt signaling through LRP5/6 and its inhibition by DKK or SOST at a mechanistic level.
Four recent publications reported the crystal structures of LRP6 P1E1 in the presence of an antibody, a DKK peptide or a SOST peptide (24); LRP6 P1E1P2E2 (25); LRP6 P3E3P4E4 (25–27); and the LRP6 P3E3P4E4-DKK1C complex (25, 26). These publications have enhanced our understanding of the molecular basis of the Wnt signaling pathway and of DKK- and SOST-mediated inhibition of Wnt signaling through LRP6.
The backbone conformations of the four LRP6 β-propeller domains, each of which consists of six β propellers, are similar to each other and to those of other YWTD repeat domains (24–27). However, the amino acid compositions and surface properties vary. The top cavities of the β propellers in P1, P2, and P3, but not those in P4, possess hydrophobic patches formed by several conserved hydrophobic residues, indicating that LRP6 P4 may not serve as the primary site for ligand binding. Overall, the orientations of the two propellers in P1E1P2E2 and P3E3P4E4 of LRP6 are spatially restricted due to the interactions involving the propellers and the EGF-like domains in between [EGF-like domain 1 (E1) in P1E1P2E2 and EGF-like domain (E3) in P3E3P4E4]. A negative-stain EM (EM) study reveals that the sequence between E2 and P3 serves as a hinge between P1E1P2E2 and P3E3P4E4 in LRP6; thus, the four tandem PE repeats in the LRP6 ECD form two rigid domains with limited flexibility in the hinge area (25). Nevertheless, in another negative-stain EM study, Chen et al. reported a skewed planar horseshoe-like model of the LRP6 ECD, with three additional LA domains in the central hinge area, which contact four β-propeller domains and stabilize the overall compact horseshoe shape (27).
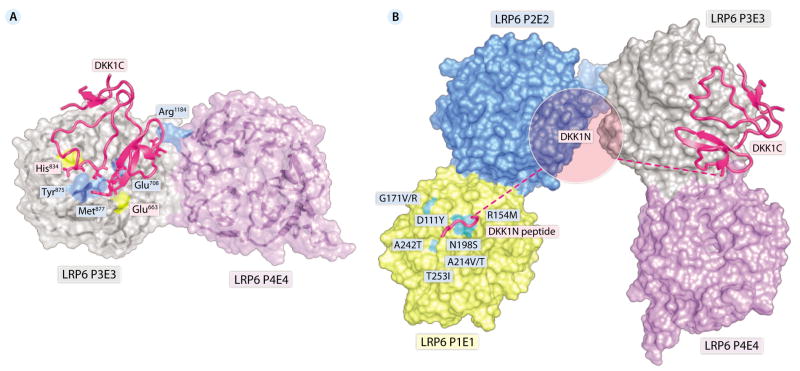
The structure of the DKK1C-LRP6 P3E3P4E4 complex (25, 26) confirms a previous prediction that DKK1 binds to the top surface of LRP5/6 P3 (8, 14). However, two LRP6 P3 domains form contacts with both sides of DKK1C through the top-surface hydrophobic cavities of the β propellers in the crystals. Experiments including mutagenesis and stoichiometry measurements (24–26) suggest that DKK and LRP5/6 form a 1:1 complex and validate that the LRP P3 binding surface on DKK1C described previously (14, 15) is directly related to DKK1C-mediated inhibition of Wnt signaling. The affinity of DKK1C for the Kremen-binding surface, although weak, raises the possibility that DKK2C, particularly when overexpressed, may bind two LRP5/6 molecules simultaneously, leading to LRP5/6 dimerization and, hence, weak activation, as reported previously (28, 29). The overall topology of bound DKK1C is similar to the NMR structure of unbound DKK2C, whereas some local areas undergo substantial conformational changes. In particular, the long loop region in DKK1C, which is unstructured in the NMR structure of unbound DKK2C, becomes partially helical in structure in the cocrystal complex with LRP6 P3E3. This helical structure stabilizes the binding of LRP6 P3 to the DKK1C cavity, which contains residues critical for Kremen binding (15).
Consistent with the previous studies (5), Chen et al. showed Wnt3a binds to the top face of LRP6 P3E3P4E4 (27). Because the Wnt3a binding surface on LRP6 P3E3P4E4 partially overlaps with DKK1C binding surface, it is likely that DKK1C antagonism comes from its direct competition with Wnt3a for binding to LRP6 P3P4 (Fig. 2A). Ahn et al. (26) also reached a similar conclusion. Although the majority of the regions that interact with DKK1C are located in LRP6 P3, a potential salt bridge between Glu185 in DKK1C and Arg1184 in LRP6 P4 may also stabilize the complex and further contribute to DKK1-mediated inhibition of Wnt3a signaling, because Arg1184 may also be involved in the Wnt3a-LRP6 interaction (27).
Fig. 2.
(A) Molecular mechanism of inhibition of Wnt signaling by DKK1C. The binding site of DKK1C partially overlaps with that of Wnt3a on LRP6 P3P4. DKK1C is shown in cartoon representation, and LRP6 P3E3P4E4 is shown in surface representation. Residues of LRP6 P3E3P4 involved in both DKK1C and Wnt3a binding are shown in blue; residues of LRP6 P3P4 that interact only with Wnt3a are shown in yellow. Residues of DKK1C that directly compete with Wnt3a for binding on LRP6 P3E3P4E4 are shown in stick representation (including Glu185, His204, Phe205, Trp206, Leu231, and Ile233; for simplicity, the labels of these residues are not shown). The atomic coordinates are from Protein Data Bank accession no. 3S8V. The Wnt3a binding surface is adopted from Chen et al. (27). (B) Proposed mechanism of the DKK1-LRP6 ECD interaction. All four β propellers of LRP6 ECD are shown in surface representation. DKK1 (including the N-terminal peptide and DKK1C) is shown in cartoon representation. Selected HBM mutations are indicated.
The most unexpected finding of these four studies is the identification of a previously unknown LRP5/6 interaction motif [Asn-X-Ile/Val (NXI/V), where X is any amino acid] at the N-terminal end of DKK1 (24). This motif is present in all of the LRP5/6-binding Wnt inhibitors including DKK1, DKK2, DKK4, WISE, and SOST. The complex structures with the DKK1 or SOST peptide containing this motif revealed the binding of the peptides to the top center of P1. Ahn et al. also observed that DKK1 N-terminal domain (DKK1N), which contains this motif, bound to LRP6 P1E1P2E2 (26). This leads to a model (Fig. 2B) that DKK1 binds to LRP5/6 in a bipartite manner, with DKK1C binding to P3E3P4E4 and the NXI/V motif binding to P1E1. The model explains why DKK1 can inhibit Wnt proteins that bind to both P1P2 (Wnt9b) and P3P4 (Wnt3a) (5). The model also helps to explain the molecular mechanism by which the HBM mutations in LRP5 result in reduced DKK- and SOST-mediated inhibition of Wnt signaling (20, 23, 30). These mutated residues, which are highly conserved in LRP5 and LRP6, occur on the top surface of LRP5 P1, where peptide binding occurs (Fig. 2B).
Besides shedding light into Wnt/LRP signaling and the inhibition mechanism for DKK and SOST, these four studies also provide solid molecular models for developing small molecules that regulate Wnt signaling and target relevant diseases. However, some important questions remain. Because Wnt1 signals independently of LRP5/6 P3E3 (7, 8), it presumably binds to P1E1P2E2. This idea is supported by data provided by Gong et al. (6), which also suggest that Wnt8 may additionally bind to P1E1P2E2. The question is whether P1 or P2 (or both) is the binding site for these Wnt proteins. Based on the Wnt3a binding surface mapping studies, Wnt1 may not bind to P1 because the HBM mutations do not affect Wnt signaling. This would also explain why HBM mutations only occur in LRP5 P1 and why DKK1 without its C-terminal cysteine-rich domain fails to inhibit Wnt1 activity (13). The second question is whether P2E2 contributes to DKK or SOST binding. The data from Bourhis et al. (24) suggest that this domain may not contribute substantially, but this conclusion is in apparent conflict with the observation that DKK1C can inhibit signaling by Wnt1 and Wnt8, which are supposed to bind to P1E1P2E2 (12, 13). The conclusion is also inconsistent with the observation that the binding of SOST to LRP5 is inhibited by mutations in both Asp111 and Asp418, which are located at the top centers of the β propellers of P1 and P2, respectively, and are equivalent to Glu708 in LRP6 P3 (8). Indeed, DKK1C binds to both LRP6 P1E1P2E2 and P3E3P4E4 fragments (25). Finally, what is the role of the N terminus of DKK (apart from the NXI/V motif) in LRP5/6 interaction and Wnt inhibition? Does this domain merely function as a linker, or does it have a role in providing steric hindrance to interfere with the binding of Wnt proteins to P1P2? Does this domain directly interact with LRP5/6 P2? Clearly, more studies are needed to clarify these questions.
References
- 1.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 3.MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moon RT. Wnt/β-catenin pathway. Sci STKE 2005. 2005:cm1. doi: 10.1126/stke.2712005cm1. [DOI] [PubMed] [Google Scholar]
- 5.Bourhis E, Tam C, Franke Y, Bazan JF, Ernst J, Hwang J, Costa M, Cochran AG, Hannoush RN. Reconstitution of a frizzled8.Wnt3a.LRP6 signaling complex reveals multiple Wnt and Dkk1 binding sites on LRP6. J Biol Chem. 2010;285:9172–9179. doi: 10.1074/jbc.M109.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong Y, Bourhis E, Chiu C, Stawicki S, DeAlmeida VI, Liu BY, Phamluong K, Cao TC, Carano RA, Ernst JA, Solloway M, Rubin-feld B, Hannoush RN, Wu Y, Polakis P, Costa M. Wnt isoform-specific interactions with coreceptor specify inhibition or potentiation of signaling by LRP6 antibodies. PLoS ONE. 2010;5:e12682. doi: 10.1371/journal.pone.0012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Wang Y, Li X, Zhang J, Mao J, Li Z, Zheng J, Li L, Harris S, Wu D. The LRP5 high-bone-mass G171V mutation disrupts LRP5 interaction with Mesd. Mol Cell Biol. 2004;24:4677–4684. doi: 10.1128/MCB.24.11.4677-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/ Arrow. Nat Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- 10.Semënov MV, Tamai K, Brott BK, Kühl M, Sokol S, He X. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr Biol. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- 11.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 12.Brott BK, Sokol SY. Regulation of Wnt/LRP signaling by distinct domains of Dickkopf proteins. Mol Cell Biol. 2002;22:6100–6110. doi: 10.1128/MCB.22.17.6100-6110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Mao J, Sun L, Liu W, Wu D. Second cysteine-rich domain of Dickkopf-2 activates canonical Wnt signaling pathway via LRP-6 independently of dishevelled. J Biol Chem. 2002;277:5977–5981. doi: 10.1074/jbc.M111131200. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Wang K, Shao Y, Huang J, Li X, Shan J, Wu D, Zheng JJ. Structural insight into the mechanisms of Wnt signaling antagonism by Dkk. J Biol Chem. 2008;283:23364–23370. doi: 10.1074/jbc.M802375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Zhang Y, Li X, Chen L, Wang H, Wu J, Zheng J, Wu D. Characterization of the Kremen-binding site on Dkk1 and elucidation of the role of Kremen in Dkk-mediated Wnt antagonism. J Biol Chem. 2008;283:23371–23375. doi: 10.1074/jbc.M802376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, Glinka A, Niehrs C. Kremen proteins are Dickkopf receptors that regulate Wnt/β-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280:19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 18.Semënov M, Tamai K, He X. SOST is a ligand for LRP5/LRP6 and a Wnt signaling inhibitor. J Biol Chem. 2005;280:26770–26775. doi: 10.1074/jbc.M504308200. [DOI] [PubMed] [Google Scholar]
- 19.Liu CC, Pearson C, Bu G. Cooperative folding and ligand-binding properties of LRP6 β-propeller domains. J Biol Chem. 2009;284:15299–15307. doi: 10.1074/jbc.M807285200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Mol Cell Biol. 2005;25:4946–4955. doi: 10.1128/MCB.25.12.4946-4955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhat BM, Allen KM, Liu W, Graham J, Morales A, Anisowicz A, Lam HS, McCauley C, Coleburn V, Cain M, Fortier E, Bhat RA, Bex FJ, Yaworsky PJ. Structure-based mutation analysis shows the importance of LRP5 β-propeller 1 in modulating Dkk1-mediated inhibition of Wnt signaling. Gene. 2007;391:103–112. doi: 10.1016/j.gene.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Binnerts ME, Tomasevic N, Bright JM, Leung J, Ahn VE, Kim KA, Zhan X, Liu S, Yonkovich S, Williams J, Zhou M, Gros D, Dixon M, Korver W, Weis WI, Abo A. The first propeller domain of LRP6 regulates sensitivity to DKK1. Mol Biol Cell. 2009;20:3552–3560. doi: 10.1091/mbc.E08-12-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 24.Bourhis E, Wang W, Tam C, Hwang J, Zhang Y, Spittler D, Huang OW, Gong Y, Estevez A, Zilberleyb I, Rouge L, Chiu C, Wu Y, Costa M, Hannoush RN, Franke Y, Cochran AG. Wnt antagonists bind through a short peptide to the first β-propeller domain of LRP5/6. Structure. 2011;19:1433–1442. doi: 10.1016/j.str.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Cheng Z, Biechele T, Wei Z, Morrone S, Moon RT, Wang L, Xu W. Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat Struct Mol Biol. 2011;18:1204–1210. doi: 10.1038/nsmb.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn VE, Chu ML, Choi HJ, Tran D, Abo A, Weis WI. Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev Cell. 2011;21:862–873. doi: 10.1016/j.devcel.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Bubeck D, MacDonald BT, Liang WX, Mao JH, Malinauskas T, Llorca O, Aricescu AR, Siebold C, He X, Jones EY. Structural and functional studies of LRP6 ectodomain reveal a platform for Wnt signaling. Dev Cell. 2011;21:848–861. doi: 10.1016/j.devcel.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brott BK, Sokol SY. Regulation of Wnt/LRP signaling by distinct domains of Dickkopf proteins. Mol Cell Biol. 2002;22:6100–6110. doi: 10.1128/MCB.22.17.6100-6110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Mao J, Sun L, Liu W, Wu D. Second cysteine-rich domain of Dickkopf-2 activates canonical Wnt signaling pathway via LRP-6 independently of dishevelled. J Biol Chem. 2002;277:5977–5981. doi: 10.1074/jbc.M111131200. [DOI] [PubMed] [Google Scholar]
- 30.Semenov MV, He X. LRP5 mutations linked to high bone mass diseases cause reduced LRP5 binding and inhibition by SOST. J Biol Chem. 2006;281:38276–38284. doi: 10.1126/scisignal.2003028. [DOI] [PubMed] [Google Scholar]