Abstract
During the past decade, cancer drug development has shifted from a focus on cytotoxic chemotherapies to drugs that target specific molecular alterations in tumors. Although these drugs dramatically shrink tumors, the responses are temporary. Research is now focused on overcoming drug resistance, a frequent cause of treatment failure. Here we reflect on analogous challenges faced by researchers in infectious diseases. We compare and contrast the resistance mechanisms arising in cancer and infectious diseases and discuss how approaches for overcoming viral and bacterial infections, such as HIV and tuberculosis, are instructive for developing a more rational approach for cancer therapy. In particular, maximizing the effect of the initial treatment response, which often requires synergistic combination therapy, is foremost among these approaches. A remaining challenge in both fields is identifying drugs that eliminate drug-tolerant “persister” cells (infectious disease) or tumor-initiating/stem cells (cancer) to prevent late relapse and shorten treatment duration.
The modern era of antimicrobial therapy is ~60 years old. It has produced agents that target bacterial cell wall biosynthesis (e.g., penicillins, cephalosporins, vancomycin, Isoniazid), protein synthesis (e.g., aminoglycosides, tetracyclines, chloramphenicol, oxazolidinones, macrolides), RNA synthesis (e.g., rifampin), and DNA metabolism (e.g., sulfonamides, quinolones). Despite this diversity of targets, resistance remains a universal accompaniment to antimicrobial therapy. Microbes use remarkably diverse strategies to overcome selective pressure, and much is known about the mechanisms of antimicrobial resistance. Although antimicrobial resistance remains a major problem on a population level, the emergence of drug resistance in an individual patient with a chronic infection can be prevented by the administration of a highly effective combination therapy regimen, which either cures the patient or prevents death from previously lethal infections.
The history of targeted cancer therapy is much shorter than that of infectious diseases, but already it is replete with a similarly diverse range of resistance mechanisms. However, effective combinations leading to cures have not yet emerged. Oncology has a track record of prior success in developing curative combination chemotherapy for pediatric leukemia, germ cell tumors, and lymphoma, but this progress required decades of empirically mixing and matching available agents. There is optimism that this timeline can be shortened with targeted cancer drugs because our understanding of cancer biology today is markedly more advanced.
Here we compare and contrast examples of drug resistance in infectious diseases and cancer, with the hope that lessons learned in one field may inform the other. We acknowledge that this is a forced comparison. There are fundamental differences in the principles underlying the search for drugs that target a foreign invader (i.e., in infections) versus mutant cells that emerge from the host (i.e., in cancer), particularly with regard to anticipated toxicities. Yet, current drug-targeting strategies in both fields share the goal of exploiting the unique dependencies of each disease, such as tumor-specific mutations in cancers or microbe/virus-specific targets in infectious agents. Another challenge in comparing these disciplines are the different definitions of treatment success. Resolution of the illness in the patient is central to both, but the infectious diseases field must also consider the impact of drug resistance on public health. Overtreatment with broad spectrum antibiotics cures most patients but hastens the emergence and spread of multi-drug-resistant strains, which can impact the health of currently uninfected individuals.
Rather than divide the discussion into separate sections on infectious diseases and cancer, we consider both fields together, beginning with mutational and nonmutational mechanisms of resistance (Table 1). We follow with a review of successful combination drug strategies in infectious diseases. We provide insights into why they worked and highlight a few instances when monotherapy is surprisingly effective in both disciplines. We conclude with the argument that molecular diagnostics, which already play a critical role in defining drug-sensitive subsets of cancer patients, could also transform current infectious diseases treatment. To learn more about the mechanistic details of and treatment options for drug resistance in HIV, tuberculosis (TB), and malaria, see Review by Goldberg et al. on page 1271 of this issue.
Table 1.
Mechanisms of Resistance
| Mutational | Nonmutational |
|---|---|
| mutation of drug target | drug-tolerant persister cells |
| amplification of drug target | tumor-initiating cells/cancer stem cells |
| bypass of drug targeted pathway | signaling pathway feedback |
| drug inactivation | lineage switching |
Mutational Mechanisms of Resistance
Mutation of the Drug Target
A common resistance mechanism shared across antimicrobial and anticancer agents is mutation in the gene that encodes the drug target (Figure 1A). HIV serves as an illustrative example of this resistance mechanism. The goal of HIV therapy is long-term suppression of viral replication with combinations of antiretroviral agents targeting viral reverse transcriptase (RT), protease, or integrase enzymes. Loss of viral suppression is often associated with emergence of HIV-1 variants, which express drug-resistant alleles of the viral RT, protease, or integrase gene due to mutations (Blanco et al., 2011; Zolopa et al., 1999). Mutations in drug targets are also common in antibacterial resistance. For example, rifampicin binds and inhibits the β subunit of the bacterial RNA polymerase enzyme complex (Campbell et al., 2001). β subunit mutations that impair drug binding confer rifampicin resistance. Resistance to β-lactam antibiotics can also occur through drug target mutation. β-lactam antibiotics, such as penicillins and cephalosporins, inhibit bacterial peptidoglycan biosynthesis by binding and inhibiting transpeptidases (i.e., penicillin-binding proteins or PBPs), enzymes that crosslink the peptidoglycan peptide side chains. PBP mutations that diminish the affinity for β-lactam confer β-lactam resistance in a wide variety of gram-positive pathogens, including Staphylococcus aureus, Streptococcus pneumoniae, and Enterococcus (Zapun et al., 2008).
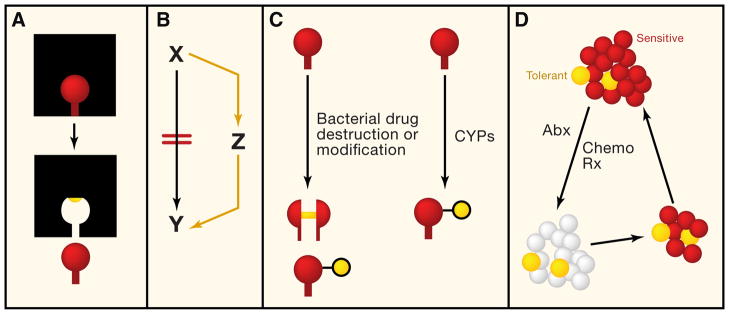
Figure 1. Mechanisms of Resistance to Antimicrobials and Targeted Anticancer Agents.
(A) Resistance via target mutation. This mechanism has been well described in both antimicrobial resistance and tumor cell resistance. The red drug binds tightly to its target (black square). A mutational event leads to alteration in the binding site for the drug (yellow circle), leading to loss of drug binding. This mechanism governs resistance to β-lactam antimicrobials (and other antimicrobial classes) as well as resistance to kinase inhibitors.
(B) Resistance via bypass pathways. Treatment with antimicrobial or anticancer agents (red lines) leads to a block in the pathway converting X to Y. Conceptually, Y can be a metabolite or a phenotypic state (e.g., cell proliferation). Resistance to the effect of the drug can be mediated by upregulation of a parallel pathway that allows Y to be restored. This mechanism of resistance has been documented in anticancer therapy, for example, amplification of MET to bypass a drug-induced block in EGFR signaling.
(C) Resistance by drug destruction or modification. Bacterial enzymes, such as β-lactamases or aminoglycoside-modifying enzymes, mediate antimicrobial resistance by drug destruction or modification. This mechanism has not been described for resistance to anticancer agents, although modification of anticancer agents through CYPs can affect efficacy (although this effect is not mediated by the tumor cell).
(D) Intrinsic, nonmutational, resistance. Here the population of cells (either tumor or microbial) are genotypically identical. The red cells are drug sensitive and are rapidly killed by antimicrobial or anticancer therapy, but the yellow cells are poorly killed by antimicrobial or cancer therapy (tolerant). These tolerant cells are called microbial persisters or cancer stem cells. Therapy with antimicrobials or anticancer agents leads to substantial killing, but the persister cells are able to resist treatment and can repopulate the infection or tumor with drug-sensitive cells, causing disease relapse.
Drug target mutations are also common with many anticancer agents, particularly in the growing class of kinase inhibitors that target oncogenic driver mutations. This mechanism was first demonstrated in patients with chronic myeloid leukemia (CML) who developed resistance to the ABL kinase inhibitor imatinib. Now this mechanism has been observed with nearly every kinase inhibitor tested to date. In the case of CML, mutations in the kinase domain of BCR-ABL impair imatinib binding, although preserving the catalytic activity of the enzyme (ATP hydrolysis) that is required for oncogenicity (Gorre et al., 2001). Some mutations confer resistance by blocking interactions between the drug and target through steric hindrance. Other mutations restrict the flexibility of the enzyme to conformations that are unsuitable for drug binding (Burgess et al., 2005; Gorre et al., 2001; Shah et al., 2002). Similar mechanisms account for resistance to epidermal growth factor receptor (EGFR) kinase inhibitors and ALK kinase inhibitors in lung cancer, KIT kinase inhibitors in gastrointestinal stromal tumors (GIST), platelet-derived growth factor receptor (PDGFR) kinase inhibitors in hypereosinohilic syndrome, and BRAF kinase inhibitors in melanoma (Antonescu et al., 2005; Choi et al., 2010; Cools et al., 2003; Pao et al., 2005; Poulikakos et al., 2011). In CML and lung cancer, drug-resistance mutations can be detected in some patients prior to treatment with the kinase inhibitor and can impact prognosis by shortening the time to disease progression. The drug-resistant allele is generally present in a small minority of cells, but in a few cases, drug-resistant clones have undergone substantial expansion in the absence of drug. This observation raises the question of whether some resistance mutations also confer a tumor fitness advantage (Shah et al., 2002; Bean et al., 2007; Godin-Heymann et al., 2007; Maheswaran et al., 2008; Skaggs et al., 2006). This contrasts with HIV infections, in which virions bearing drug-resistant RT mutations generally have reduced viral fitness (Martinez-Picado and Martínez, 2008).
Gene Amplification of the Drug Target or a Bypass Pathway
Drug resistance can also occur through amplification of the drug target gene in the absence of mutation (Figure 1B). A prime example of this mechanism in cancer is the androgen receptor (AR). The AR gene is amplified in ~30% of prostate cancers that have acquired resistance to standard androgen deprivation therapy with drugs (e.g., leuprolide) that lower testosterone production and AR antagonists (e.g., bicalutamide or flutamide) that block ligand binding (Scher and Sawyers, 2005). BCR-ABL gene amplification can also drive resistance to ABL kinase inhibitors in CML, although this mechanism is less common than mutations in the kinase domain (Gorre et al., 2001). Amplification of the dihydrofolate reductase (DHFR) gene confers resistance to the chemotherapeutic agent methotrexate in cancer cell lines (Schimke et al., 1978), but this mechanism has not emerged as a common resistance mechanism in patients. However, amplification of the bacterial DHFR gene can cause resistance to the antibiotic trimethoprim (Steen and Sköld, 1985).
Approximately 20% of patients with EGFR mutant lung cancer develop resistance to EGFR inhibitors by amplification of another receptor tyrosine kinase, MET (Engelman et al., 2007). This mechanism has been termed “oncogene bypass” because the primary drug target remains unaltered and continues to be inhibited by drug. Resistance occurs because MET activates downstream components of the EGFR signaling pathway, bypassing the need for EGFR (Figure 1B). A conceptually similar mechanism has been documented with thymidine auxotrophs of E. coli and Enterococcus. These strains can become resistant to sulfonamides because the inhibition of thymidine biosynthesis by the antibiotic can be bypassed by the acquisition of thymidine from the environment (Maskell et al., 1978) (Figure 1B).
Drug Inactivation
The most common mechanism of antibacterial resistance is drug destruction by bacterial enzymes (Figure 1C). β-lactamases, which cleave the amide bond of the β-lactam ring, confer resistance to the antibiotic through drug destruction. Progressive chemical modification of the β-lactam nucleus to prevent destruction by β-lactamases has yielded compounds that are active against β-lactamase-producing organisms. However, progressively broader spectrum β-lactamases, including some capable of hydrolyzing all β-lactams and carbapenems, have consequently become widespread. These broad spectrum β-lactamases are an escalating concern and threaten the utility of this class of antibiotics (Cornaglia et al., 2011).
Resistance through antibiotic modification is not limited to the β-lactam class of antibiotics. Aminoglycoside antibiotics inhibit protein synthesis by binding to the 30S subunit of the bacterial ribosome. The most common mechanism of resistance to these drugs is through bacterial acquisition of enzymes that covalently modify the aminoglycoside by phosphorylation, acetylation, or adenylation. The modified aminoglycoside no longer binds its target on the ribosome.
In contrast to antibacterials, drug inactivation has not emerged as a major cause of resistance to anticancer agents. Cytochrome P450 enzymes (CYPs) play a critical role in the metabolism of many drugs through oxidation reactions that can inactivate the compound or lead to its rapid elimination (Figure 1C). Although this metabolic degradation was problematic for many classes of drugs in the past, it is less relevant today because most drug candidates are routinely counterscreened early in development against panels of CYPs. Compounds that score as potent CYP substrates are typically eliminated or chemically modified to “dial out” the CYP activity while preserving the desired anti-cancer function.
Nonetheless, there are examples of drugs in which CYP modification may affect therapeutic response. The antiestrogen tamoxifen, which is widely used in the treatment of estrogen receptor-positive breast cancer, is metabolized by CYP2D6 to its primary active metabolite endoxifen. Some genetic variants of CYP2D6 confer reduced levels of enzyme activity, and there is evidence that women with these variants may not respond as well to tamoxifen because they generate lower levels of endoxifen. Furthermore, CYP2D6 is inhibited by selective serotonin reuptake inhibitors (SSRIs). SSRIs are commonly prescribed to ameliorate “hot flashes,” but they may counteract the clinical benefit of tamoxifen by blocking production of its primary metabolite (Borges et al., 2006).
Resistance to the antimycobacterial drugs isoniazid and pyrazinamide, both important agents in the treatment of TB, also occurs because of the failure to generate the active metabolite of the drug. Both antibiotics are prodrugs that must be activated by the TB bacterial enzymes KatG (for isoniazid) and PncA (for pyrazinamide). Resistance to both drugs often occurs through mutations in their respective activator enzymes (Altamirano et al., 1994; Scorpio and Zhang, 1996; Zhang et al., 1992).
Although conceptually similar, these mechanisms differ from the example with tamoxifen/CYP2D6 because resistance to isoniazid and pyrazinamide is cell autonomous. We are not aware of examples of cancer drug inactivation mediated specifically by tumor cells. This mechanism has not been considered by most cancer scientists due to challenges in measuring concentrations of the drug and potential metabolites in tumor biopsies. However, advances in mass spectrometry technology should enable such measurements on a more routine basis. Drug destruction should be evaluated as a potential cause of tumor-mediated resistance in cases in which other mechanisms have been excluded.
Nonmutational Resistance Mechanisms
Drug-Tolerant “Persister” Cells
In the earliest days of antimicrobials, it was observed that the killing of a microbial population was rarely complete. A small subpopulation of cells survived but did not have a drug-resistance mutation. When expanded, these cells reverted to antimicrobial sensitivity (Figure 1D). This phenomenon, termed bacterial persistence, is observed in a wide variety of bacterial taxa, including M. tuberculosis, E. coli, and others (Connolly et al., 2007; Lewis, 2010). It is widely suspected that persister cell populations are responsible for the slow sterilization of many chronic infections and consequent requirement for prolonged antibiotic therapy. There is great interest in identifying molecular determinants of persistence because knowledge of such pathways could lead to more effective antimicrobials that quickly eliminate this reservoir of cells. Such drugs could reduce the chance of late relapse and shorten duration of treatment. In the case of TB, a curative regimen administered over several weeks rather than 6–9 months could have profound consequences on treatment compliance, with obvious public health implications.
We currently have little insight into the molecular basis of persistence. Recent evidence suggests that toxin-antitoxin (TA) modules may be molecular determinants of persistence in some bacteria, presumably by the induction of growth arrest in a subpopulation of cells through toxin-mediated mRNA cleavage (Maisonneuve et al., 2011; Moyed and Bertrand, 1983; Wolfson et al., 1990). But, many questions remain, including the exact mechanisms of TA-mediated persistence, the generalizability of this mechanism to chronic infections, and the therapeutic benefit of targeting TA modules. Similarly, recent evidence from the M. marinum system suggests that M. tuberculosis drug tolerance in vivo may be mediated by host-inducible drug efflux pumps (Adams et al., 2011). Although the exact contribution of drug efflux to drug tolerance in vivo remains to be determined, these findings suggest a druggable target that would increase antimicrobial killing by presently available antimyco-bacterials.
Tumor-Initiating/Cancer Stem Cells
The phenomenon of bacterial persistence shares conceptual similarities with evidence that late relapses of some cancers may be due to tumor-initiating or cancer stem cells that are not eliminated by most treatments (Figure 1D). The existence of cancer stem cells is a topic of considerable controversy (Rossi et al., 2008), but this is not germane to our discussion here. What is relevant is the observation that, even following effective treatment regimens, residual cancer cells persist and are responsible for late relapse. Acute myeloid leukemia (AML) serves as an illustrative example, in which initial treatment with induction chemotherapy is quite effective in inducing remission, but relapse is inevitable unless additional rounds of consolidation chemotherapy are administered.
Several preclinical studies have documented that these residual cells do not have mutations in the drug target or other factors that could explain resistance. Like residual persister cells after antibiotic treatment, these residual cancer cells, when expanded in the absence of drug, give rise to drug-sensitive populations of cancer cells (Sharma et al., 2010). These cancer persister cells tend to express cell-surface antigens typically present on tissue stem cells, raising the possibility that stem cells are inherently drug resistant.
One potential explanation for their resistance is expression of drug efflux pump proteins, such as MDR (multidrug resistance), which are naturally expressed at higher levels in stem cells. However, cancer cells can persist following treatment with drugs that are not MDR substrates. Another potential mechanism is survival signaling mediated through growth factors expressed by adjacent cells in the tumor microenvironment or metastatic niche (Guise, 2010). For example, the CXCR4 receptor plays a critical role in anchoring hematopoietic stem cells (HSCs) and CXCR4-positive AML cells to niches in the bone marrow microenvironment through interaction with the stromal factor SDF-1. CXCR4 inhibitors mobilize HSCs into circulation (DiPersio et al., 2009) and enhance the efficacy and duration of response to induction chemotherapy (Nervi et al., 2009). Similar to antibacterial persister drugs, compounds that selectively eliminate tumor-initiating cells could complement existing cancer therapies (Gupta et al., 2009; Reya et al., 2001).
Feedback due to Signaling Pathway Inhibition
When cancer cells are exposed to drugs that block signaling pathways, such as kinase inhibitors, many tumor cells compensate by activating other signaling pathways. The mechanism of these feedback responses is complex and related to concepts of network robustness and redundancy that have emerged from systems biology research (Lander, 2011). In the context of cancer, inhibition of one signaling pathway initiates compensatory feedback responses that can lead to the activation of another (Carver et al., 2011; Chandarlapaty et al., 2011; O’Reilly et al., 2006; Pratilas et al., 2009). These observations predict that monotherapy with some inhibitors, particularly those targeting the PI3K signaling pathway, is unlikely to be effective unless paired with inhibitors of the compensatory pathway. Interestingly, there is currently no evidence for feedback as a mechanism of resistance to anti-infectious agents, likely because these drugs tend to target essential steps in the replication of the organism (e.g., cell wall synthesis, viral replication, etc.) rather than signaling pathways. However, similar mechanisms of resistance may arise with antimicrobials that target virulence pathways that are not essential to cell viability (Clatworthy et al., 2007).
Lineage Switching
Studies of acquired resistance to EGFR inhibitors in lung cancer reveal that recurrent tumors in some patients undergo a lineage switch from non-small cell to small cell carcinoma (Sequist et al., 2011). Clonality studies document that both tumors arose from a common EGFR mutant tumor rather than from two independent cancers, indicating that the tumor has shifted to a different cellular differentiation pathway. Switching to the small cell lineage (by an unknown mechanism that presumably involves cellular reprogramming) relieves cells of their dependence on EGFR, which was critical for survival of the non-small cell tumor lineage. A somewhat analogous phenomenon has been observed with antibiotics, in which resistance to drugs targeting the bacterial cell wall can occur through the cell wall-deficient bacteria, called L-forms (Allan et al., 2009).
Overcoming Resistance
Although our knowledge is still incomplete, progress in deciphering mechanisms of resistance to targeted cancer therapy is already guiding solutions to overcome it. The recognition that point mutations in the BCR-ABL kinase domain confer resistance to the first-generation inhibitor for the ABL kinase inhibitor imatinib led rapidly to efforts to find other ABL inhibitors effective against the mutant BCR-ABL alleles. Within 5 years, two next-generation ABL inhibitors, dasatinib and nilotinib, were approved for the treatment of imatinib-resistant CML (Kantarjian et al., 2007; Talpaz et al., 2006). Dasatinib is notable because its activity against imatinib-resistant BCR-ABL mutants is explained by its ability to bind multiple distinct conformations of BCR-ABL, thereby restricting the potential for escape mutants (Burgess et al., 2005; Shah et al., 2002). A second example has emerged from the discovery that prostate cancers resistant to standard androgen deprivation therapy remain dependent on AR signaling (Chen et al., 2004). Two new drugs, abiraterone and MDV3100, impair AR signaling in this drug-resistant setting and are effective in men with metastatic prostate cancer resistant to standard hormone and chemotherapy (de Bono et al., 2011; Scher et al., 2010; Tran et al., 2009).
These examples underscore the importance of understanding resistance mechanisms and the rapid progress that can be made by leveraging these insights. But they also represent partial solutions because sequential treatment with these different targeted agents is not curative. This current scenario in cancer contrasts strikingly with the current treatment of two previously lethal infections, TB and HIV: combinations of antibiotics cure TB, whereas combinations of antivirals can indefinitely suppress the HIV virus. Physicians treating these diseases in the past faced a situation remarkably similar to that faced by oncologists today. Successful combination regimens arose only after attempts to treat these infections with monotherapy failed.
HIV: Importance of Maximal Suppression of Viral Load at Treatment Initiation
HIV cannot be cured with current antiretroviral therapy, but mortality is dramatically reduced through combination antiretro-viral drug therapy. The first major success in HIV therapy came from nucleoside analogs, such as zidovudine (AZT). These analogs inhibit viral replication by causing chain termination when RT incorporates the analogs into the viral cDNA. Although testing of serum viral loads was not available at that time, the clinical benefit of AZT was evident after 12 weeks in patients with AIDS. However, the efficacy of the therapy was short lived: CD4 T cell counts returned to the levels of placebo-treated patients in 24 weeks, indicating a significant but transient clinical benefit (Fischl et al., 1987). Similar results were observed with the nucleoside RT inhibitor lamivudine (3TC): nearly 100% of patients had lamivudine resistance by week 12 of monotherapy (Schuurman et al., 1995). Monotherapy with the protease inhibitor saquinavir also revealed substantial short-term clinical benefit, which was lost over the course of the 24 week observation period (Schapiro et al., 1996). This failure of HIV antiviral monotherapy is schematically depicted in Figure 2A.
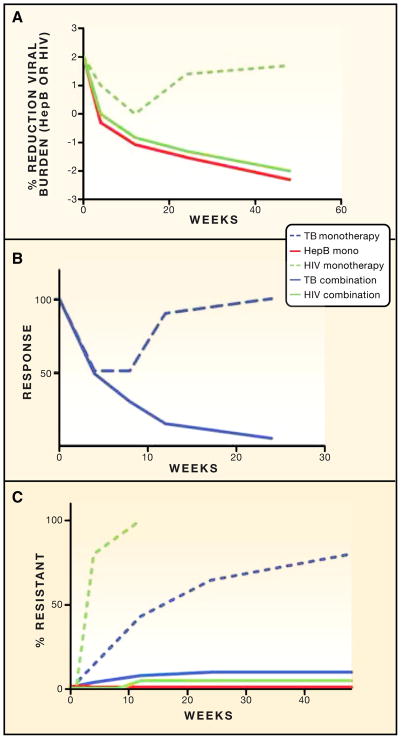
Figure 2. Relative Efficacy of Mono- versus Combination Therapy for Three Chronic Infections and Effect on Resistance.
(A) The three curves schematically indicate the reduction in viral burden during monotherapy of HIV (dashed green line), combination therapy for HIV (solid green line), and monotherapy for hepatitis B (solid red line). Monotherapy of HIV produces a transient reduction in viral load, which becomes more dramatic and sustained with combination therapy. In contrast, monotherapy for hepatitis B (with entecavir or tenofovir) produces sustained virologic suppression. See text for specific references.
(B) Monotherapy and combination therapy for tuberculosis. The y axis schematically represents clinical response (reduction in bacterial load, clinical improvement, radiographic improvement). Monotherapy and combination therapy have similar efficacy early in treatment, but the benefit of monotherapy is not sustained.
(C) Effect of combination therapy on emergence of resistance. The y axis represents the % of patients with resistant bacteria or viruses according to week of treatment. HIV and TB resistance emerges rapidly during mono-therapy, leading to the loss of therapeutic effect depicted in panel A (HIV) and panel B (TB). Combination therapy for HIV and TB suppresses the emergence of resistant organisms and allows the sustained therapeutic benefit depicted in (A) and (B). In contrast, monotherapy for hepatitis B with entecavir or tenofovir is not associated with the emergence of resistant viruses, allowing sustained therapeutic benefit with monotherapy.
In all of these examples (and other attempts of monotherapy), the cause of clinical failure was the emergence of virions with mutations that confer resistance to the administered agent (Richman et al., 1994; Schuurman et al., 1995; Zolopa et al., 1999) (Figure 2C). The biologic basis for this rapid emergence of resistance is the massive pool of HIV virions in an infected host, estimated to be 1010 new virions per day (Perelson et al., 1996). This pool generates a large number of possible mutant virions, which have a selective growth advantage during monotherapy.
These results rapidly led to the testing of combination regimens, first with dual-nucleoside analogs (Eron et al., 1995) and then with triple-drug regimens, including two nucleosides and a protease inhibitor, such as saquinavir or indinavir. Trials comparing AZT/3TC and AZT/3TC/indinavir revealed that the three-drug combination suppressed HIV-1 viral load more potently than the two-drug combination and substantially lowered the number of HIV-related deaths (Gulick et al., 1997; Hammer et al., 1997). These results have been replicated multiple times with multiple different combination therapy regimens and have led to a highly evolved standard of care for HIV, infection consisting of three-drug combination therapy now known as HAART (highly active antiretroviral therapy). The key to this success is the more rapid and sustained decline in HIV burden achieved through HAART, thereby preventing the emergence of resistant virions (Figures 2A and 2C).
TB: Importance of Preventing Outgrowth of Drug-Resistant Subclones
TB is remarkable for its insidious onset and slow progression, which, in the preantibiotic era, was usually fatal. The first anti-TB drugs were paraminosalicylic acid (PAS) and streptomycin. Both drugs were initially used as monotherapy with significant clinical improvement over 1–2 months of therapy. This success was quickly followed by loss of clinical efficacy due to the emergence of streptomycin- or PAS-resistant TB strains (British Medical Research Council, 1947, 1948, 1950; Lehmann, 1946). Next, a series of trials were performed comparing streptomycin and PAS monotherapy to streptomycin plus PAS combination therapy. Ultimately, the combination therapy proved superior over monotherapy. All three treatment strategies showed similar improvement at 6 months in terms of the resolution of fever, radiographic improvement, and weight gain, but the combination therapy was ultimately superior because it prevented the emergence of streptomycin resistance (Figures 2B and 2C) (British Medical Research Council, 1950).
Similar results emerged from trials with the current standard-of-care drug isoniazid (INH). INH monotherapy was initially as effective as streptomycin/PAS combination therapy but ultimately failed due to the emergence of INH-resistant strains (British Medical Research Council, 1950, 1952). These insights led to the current, curative three- to four-drug, 6–12 month regimens that are now the standard of care (Fox et al., 1999). The key concept underlying the success of antimicrobial TB therapy is that combination therapy is necessary to prevent the emergence of resistance. In contrast, the success of HIV combination therapy is partially based on a more rapid, synergistic suppression of the infectious agent and partially due to the suppression of resistance (Figure 2).
Exceptions: Examples of Successful Monotherapy
Acute Bacterial Infections
Antibiotic monotherapy for most acute bacterial infections is curative in the absence of pre-existing antimicrobial resistance. Despite substantial evidence for synergistic killing by antibiotic combinations in vitro, definitive evidence supporting the clinical benefit of combination therapy for acute-onset infections, such as bacterial pneumonia or urinary tract infection, is generally lacking (Del Favero et al., 2001). It is not known why antibiotic monotherapy is so effective for most acute infections, but we speculate that the magnitude and kinetics of decline in bacterial burden are sufficiently steep to prevent the emergence of resistance and therefore obviate the need for combination therapy. In contrast, more slowly progressive bacterial infections, such as enterococcal endocarditis, require multiple antibiotics for elimination. It is also likely that the host immune system plays a role in eliminating minimal residual disease because patients with reduced neutrophil counts (e.g., from chemotherapy treatment) often require more prolonged antibiotic therapy and generally recover quickly once the neutropenia resolves.
Hepatitis B
Hepatitis B is a chronic viral infection of hepatocytes that eventually can lead to cirrhosis and hepatocellular carcinoma. Nucleoside analogs, such as entecavir, lamivudine, and tenofovir (the latter two are also used as antiretroviral therapy for HIV), have similar activities against the hepatitis B DNA virus due to the RT activity of the viral polymerase (which copies the pregenomic RNA into DNA). Monotherapy with each of these drugs suppresses viral burden substantially but, surprisingly, is not accompanied by rapid emergence of drug-resistant hepatitis B virions. The incidence of lamivudine-resistant hepatitis B is 20% after 1 year of therapy, and almost no resistance is observed with the newer agents tenofovir or entecavir, with 1 and 5 years of follow up, respectively (Chang et al., 2006; Lok and McMahon, 2009; Marcellin et al., 2008). Notably, the decline in viral burden with hepatitis B monotherapy occurs over months, at a pace comparable to monotherapy for HIV or TB (Figure 2A), indicating that the clinical efficacy of hepatitis B monotherapy is based on the fact that drug-resistant virions rarely emerge. The biologic basis for this serendipitous lack of resistance in hepatitis B is unknown because the same drugs targeting a similar enzyme (i.e., RT) rapidly generate resistant HIV when used as monotherapy (Figure 2C).
Chronic Myeloid Leukemia
As discussed earlier, resistance to monotherapy with ABL kinase inhibitors occurs, but the relapse rate is quite low (~4%–5% in the first year) if treatment is initiated in early chronic phase. By comparison, more than half of patients with most other types of tumors (e.g., lung cancer and melanoma) treated with kinase inhibitors relapse after 1 year. Longer follow up of CML patients receiving imatinib monotherapy has shown that the risk of relapse declines after 5 years and may plateau (Hochhaus et al., 2009), suggesting that many patients with CML can be successfully managed indefinitely with monotherapy. The rate of decline in CML disease burden over the initial 3–6 months of treatment, as measured by PCR for BCR-ABL mRNA transcripts in the blood, is a critical determinant of the durability of response. Specifically, CML patients who obtain a three log reduction in tumor burden on imatinib have a low risk of subsequent relapse (Hughes et al., 2010). Early data with the second-generation inhibitors dasatinib and nilotinib indicate that an even greater proportion of CML patients reach this endpoint (Kantarjian et al., 2010; Saglio et al., 2010). Similar to the importance of achieving a rapid and substantial decline in viral load with HIV therapy, the success of CML monotherapy is most likely due to the dramatic reduction in tumor burden. Extrapolating to lung cancer and melanoma, it may be possible to achieve similar long-term success if we focus our efforts on obtaining deeper reductions in tumor burden using more potent agents and relevant combinations.
Precision Diagnostics: Essential for Targeted Cancer Drugs, Missed Opportunity for Antibacterials
The advent of targeted cancer therapies that are active against a subset of tumors with specific mutations mandated the parallel development of diagnostic tests that can determine the presence of these mutations in clinically relevant timeframes. Examples include the assessment of Her2/Neu gene amplification in breast cancer to identify women who would benefit from trastuzumab; EGFR mutation in lung cancer for sensitivity to erlotinib or gefitinib; BRAF mutation in melanoma for response to vemurafenib (PLX4720); and others. This priority represents a move away from broadly targeted cytotoxic therapies in which tumor/patient-specific molecular diagnostics have not been used to make treatment decisions.
A similar logic could easily guide precision diagnostics of suspected infections to determine the presence of a specific pathogen, rapidly define its drug susceptibility profile, and guide therapy. Examples of this strategy that have gained some traction are (1) CCR5 tropic HIV and the use of maraviroc, a CCR5 antagonist entry inhibitor (Gulick et al., 2008); (2) PCR-based determination of rifampin resistance in TB (Boehme et al., 2010); and (3) HIV genotypic resistance testing, in which the susceptibility of a patient’s HIV virus can be predicted based on protease, RT, or integrase mutations present.
Despite these advances, similar diagnostics are either not available or not in widespread use for the most common infectious diseases, largely because of the false comfort in giving broad-spectrum antimicrobials that are relatively nontoxic and safe for any individual patient (Casadevall, 2009). This has led to complacency and blunted the push for specific diagnostics that would allow (1) administration of narrow-spectrum antimicrobials; (2) early recognition of antibiotic resistance; and (iii) differentiation of infections from noninfectious (or viral) diseases with similar clinical presentations. Consequently, we have a situation of antibiotic overuse, which compromises public health by selection for drug-resistant organisms (particularly in hospitals), increases the frequency of life-threatening side effects, such as antibiotic-associated colitis, and results in inefficient and costly heath care expenditures. We note that the problem of resistance as a public health hazard is unique to infectious diseases because the resistant organism is transmissible, and therefore, resistance in one patient can affect the population as a whole. In contrast, although tumor cell resistance is detrimental to the individual patient, it does not harm others who may be afflicted with the same cancer.
Conclusions and Challenges
Except in rare examples, such as CML, there is little doubt that combination therapy will be required for sustained clinical benefit of targeted agents against cancer. The experience with infectious diseases highlights the importance of combinations that achieve rapid, efficient cancer suppression (as in HIV) and prevent the emergence of resistance (as in HIV and TB). Current paradigms of oncology drug development are not aligned in ways that allow these goals to be pursued efficiently. Instead, business and regulatory incentives drive commercial drug developers to seek approval for monotherapy indications before embarking on combination studies. Based on the short duration of response to many recently approved anticancer agents, coupled with the precedent of failed monotherapy in chronic infectious diseases, one might ask whether the obligatory phase of mono-therapy approval should be bypassed. The FDA recently drafted new guidelines for developing two or more investigational drugs in combination to encourage drug makers to adopt a combination rather than monotherapy strategy for registration (FDA, 2010), but it is too early to assess the impact of these changes. Our perspective as academic physician-scientists with experience in drug development is that additional measures are required to incentivize commercial sponsors to move to combination therapy trials early in drug development. One powerful motivation might be patent life extension, as has been successfully implemented for pediatric indications, particularly when the combination requires collaboration between two different drug makers. Activism on the part of physicians and patients may also be essential. Clinical investigators might collectively “demand” early combination studies from sponsors by refusing to conduct single-agent studies until firm commitments to combination trials are in place. Each of these strategies requires extreme coordination of goals among relevant stakeholders. Early efforts in the TB community to streamline the path of new drugs into combination TB regimens could provide a template for similar efforts in anticancer therapy (Critical Path Institute, 2010).
Another variable that could delay development of combination cancer therapy regimens is the paucity of data on coadministration of two or more targeted agents. In contrast to infectious diseases, in which most drugs are specific for the pathogen, targeted cancer agents typically impact normal and tumor cells, which could lead to unacceptable side effects. The fact that most targeted cancer therapies have highly favorable toxicity profiles relative to cytotoxic chemotherapy is cause for optimism. However, the current focus on monotherapy could complicate the investigation of tolerable combinations because dose and schedule are selected exclusively based on tolerability of monotherapy. Combination studies of targeted therapies typically begin by evaluating tolerability of each monotherapy regimen given simultaneously. If additive toxicities are observed (as might be expected), the combination might be abandoned prematurely. If early development decisions were, instead, driven by strategies that plan for combination therapy rather than monotherapy, clinical safety and efficacy could be assessed more quickly.
Another challenge is the need for better technologies to quantify disease burden over a wide range, as is possible for viral load assessment in HIV and hepatitis B. The rapid approval of ABL kinase inhibitors in CML was enabled, in part, by the availability of highly sensitive and quantitative assays of tumor burden (cytogenetics, PCR) that detect disease well below the threshold of traditional clinical response. Similar assays for solid tumors do not exist today, but proof of concept for quantitative assessment of circulating tumor DNA levels in blood has been established for several types of tumors (Leary et al., 2010). Once in place, these assays might also serve as early endpoints for drug approval, as they did for HIV, hepatitis B, and CML.
Curative regimens must also overcome the problem of subclinical reservoirs of persistent disease. Experimental strategies to define the molecular basis for persistence of cancer cells or microbes deserve more focused attention because the insights gained have the potential to eliminate drug-tolerant cells and thereby avoid prolonged treatment regimens, which are inevitably compromised by chronic toxicity and noncompliance. Recent data in cancer suggest that the immune system could be harnessed to eliminate minimal residual disease. Stimulation of host T lymphocytes by the anti-CTLA4 antibody ipilimumab induces tumor regression and prolongs survival in patients with metastatic melanoma (Robert et al., 2011). Remarkably, some patients remain in remission for years after therapy ceases and may be cured. This long-term success could be explained by the elimination of drug-tolerant persister cells by immune effector cells or by ongoing antitumor immunity that prevents expansion of tumor cells that persist indefinitely. There is clinical precedent for both of these mechanisms from allogeneic marrow transplantation data in CML in which depletion or infusion of donor T lymphocytes can profoundly impact treatment response (Mackinnon, 1997).
Finally, with more and more evidence supporting the potential value of genome-based medicine, the escalating problem of antimicrobial resistance should motivate the infectious diseases community to strive for more precise diagnostics that would allow more specific or limited use of antimicrobial agents. The advent of these technologies in targeted cancer therapy provides a template that the infectious diseases community should leverage.
Acknowledgments
The authors apologize to those whose work could not be cited due to space limitations. Work in the Sawyers laboratory is supported by the Howard Hughes Medical Institute and grants from NCI and Stand Up to Cancer. Work in the Glickman laboratory is supported by the Geoffrey Beene Cancer Research Center, the Starr Cancer Consortium, and NIAID.
References
- Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell. 2011;145:39–53. doi: 10.1016/j.cell.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan EJ, Hoischen C, Gumpert J. Bacterial L-forms. Adv Appl Microbiol. 2009;68:1–39. doi: 10.1016/S0065-2164(09)01201-5. [DOI] [PubMed] [Google Scholar]
- Altamirano M, Marostenmaki J, Wong A, FitzGerald M, Black WA, Smith JA. Mutations in the catalase-peroxidase gene from isoniazid-resistant Mycobacterium tuberculosis isolates. J Infect Dis. 1994;169:1162–1165. doi: 10.1093/infdis/169.5.1162. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Besmer P, Guo T, Arkun K, Hom G, Koryotowski B, Leversha MA, Jeffrey PD, Desantis D, Singer S, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11:4182–4190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. HIV-1 integrase inhibitor resistance and its clinical implications. J Infect Dis. 2011;203:1204–1214. doi: 10.1093/infdis/jir025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, Jin Y, Storniolo AM, Nikoloff DM, Wu L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- British Medical Research Council. EFFECT of streptomycin upon pulmonary tuberculosis; preliminary report of a cooperative study of 223 patients by the Army, Navy and Veterans Administration. Am Rev Tuberc. 1947;56:485–507. [PubMed] [Google Scholar]
- British Medical Research Council. Streptomycin treatment of pulmonary tuberculosis. BMJ. 1948;2:769–782. [PMC free article] [PubMed] [Google Scholar]
- British Medical Research Council. Treatment of pulmonary tuberculosis with streptomycin and para-aminosalicylic acid; a Medical Research Council investigation. BMJ. 1950;2:1073–1085. [PMC free article] [PubMed] [Google Scholar]
- British Medical Research Council. Treatment of pulmonary tuberculosis with isoniazid; an interim report to the Medical Research Council by their Tuberculosis Chemotherapy Trials Committee. BMJ. 1952;2:735–746. [PMC free article] [PubMed] [Google Scholar]
- Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci USA. 2005;102:3395–3400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A. The case for pathogen-specific therapy. Expert Opin Pharmacother. 2009;10:1699–1703. doi: 10.1517/14656560903066837. [DOI] [PubMed] [Google Scholar]
- Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, Majumder PK, Baselga J, Rosen N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TT, Gish RG, de Man R, Gadano A, Sollano J, Chao YC, Lok AS, Han KH, Goodman Z, Zhu J, et al. 463022 Study Group. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N Engl J Med. 2006;354:1001–1010. doi: 10.1056/NEJMoa051285. [DOI] [PubMed] [Google Scholar]
- Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- Connolly LE, Edelstein PH, Ramakrishnan L. Why is long-term therapy required to cure tuberculosis? PLoS Med. 2007;4:e120. doi: 10.1371/journal.pmed.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348:1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- Critical Path Institute. Critical path to TB drug regimens initiative. 2010 http://c-path.org/pdf/CPTRWorkScope2010.pdf.
- de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB, Jr, Saad F, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Favero A, Menichetti F, Martino P, Bucaneve G, Micozzi A, Gentile G, Furno P, Russo D, D’Antonio D, Ricci P, et al. A multicenter, double-blind, placebo-controlled trial comparing piperacillin-tazobactam with and without amikacin as empiric therapy for febrile neutropenia. Clin Infect Dis. 2001;33:1295–1301. doi: 10.1086/322646. [DOI] [PubMed] [Google Scholar]
- DiPersio JF, Micallef IN, Stiff PJ, Bolwell BJ, Maziarz RT, Jacobsen E, Nademanee A, McCarty J, Bridger G, Calandra G 3101 Investigators. Phase III prospective randomized double-blind placebo-controlled trial of plerixafor plus granulocyte colony-stimulating factor compared with placebo plus granulocyte colony-stimulating factor for autologous stem-cell mobilization and transplantation for patients with non-Hodgkin’s lymphoma. J Clin Oncol. 2009;27:4767–4773. doi: 10.1200/JCO.2008.20.7209. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Eron JJ, Benoit SL, Jemsek J, MacArthur RD, Santana J, Quinn JB, Kuritzkes DR, Fallon MA, Rubin M North American HIV Working Party. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. N Engl J Med. 1995;333:1662–1669. doi: 10.1056/NEJM199512213332502. [DOI] [PubMed] [Google Scholar]
- FDA; U.S.D.o.H.a.H. Services, F.a.D. Administration, and C.f.D.E.a.R. (CDER) Guidance for Industry: Codevelopment of Two or More Unmarketed Investigational Drugs for Use in Combination. 2010 http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM236669.pdf.
- Fischl MA, Richman DD, Grieco MH, Gottlieb MS, Volberding PA, Laskin OL, Leedom JM, Groopman JE, Mildvan D, Schooley RT, et al. The efficacy of azidothymidine (AZT) in the treatment of patients with AIDS and AIDS-related complex. A double-blind, placebo-controlled trial. N Engl J Med. 1987;317:185–191. doi: 10.1056/NEJM198707233170401. [DOI] [PubMed] [Google Scholar]
- Fox W, Ellard GA, Mitchison DA. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council tuberculosis units, 1946–1986, with relevant subsequent publications. Int J Tuberc Lung Dis. 1999;3(10, Suppl 2):S231–S279. [PubMed] [Google Scholar]
- Godin-Heymann N, Bryant I, Rivera MN, Ulkus L, Bell DW, Riese DJ, 2nd, Settleman J, Haber DA. Oncogenic activity of epidermal growth factor receptor kinase mutant alleles is enhanced by the T790M drug resistance mutation. Cancer Res. 2007;67:7319–7326. doi: 10.1158/0008-5472.CAN-06-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- Guise T. Examining the metastatic niche: targeting the microenvironment. Semin Oncol. 2010;37(Suppl 2):S2–S14. doi: 10.1053/j.seminoncol.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, Richman DD, Valentine FT, Jonas L, Meibohm A, et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- Gulick RM, Lalezari J, Goodrich J, Clumeck N, DeJesus E, Horban A, Nadler J, Clotet B, Karlsson A, Wohlfeiler M, et al. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med. 2008;359:1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer SM, Squires KE, Hughes MD, Grimes JM, Demeter LM, Currier JS, Eron JJ, Jr, Feinberg JE, Balfour HH, Jr, Deyton LR, et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- Hochhaus A, O’Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, Goldman JM, Müller MC, Radich JP, Rudoltz M, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–1061. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- Hughes TP, Hochhaus A, Branford S, Müller MC, Kaeda JS, Foroni L, Druker BJ, Guilhot F, Larson RA, O’Brien SG, et al. Long-term prognostic significance of early molecular response to imatinib in newly diagnosed chronic myeloid leukemia: an analysis from the International Randomized Study of Interferon and STI571 (IRIS) Blood. 2010;116:3758–3765. doi: 10.1182/blood-2010-03-273979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M, Moiraghi B, Shen Z, Mayer J, Pasquini R, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362:2260–2270. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- Kantarjian HM, Giles F, Gattermann N, Bhalla K, Alimena G, Palandri F, Ossenkoppele GJ, Nicolini FE, O’Brien SG, Litzow M, et al. Nilotinib (formerly AMN107), a highly selective BCR-ABL tyrosine kinase inhibitor, is effective in patients with Philadelphia chromosome-positive chronic myelogenous leukemia in chronic phase following imatinib resistance and intolerance. Blood. 2007;110:3540–3546. doi: 10.1182/blood-2007-03-080689. [DOI] [PubMed] [Google Scholar]
- Lander AD. Pattern, growth, and control. Cell. 2011;144:955–969. doi: 10.1016/j.cell.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann J. Para-aminosalicylic acid in the treatment of tuberculosis. Lancet. 1946;1:15–16. doi: 10.1016/s0140-6736(46)91185-3. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661–662. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- Mackinnon S. Donor leukocyte infusions. Baillieres Clin Haematol. 1997;10:357–367. doi: 10.1016/s0950-3536(97)80012-1. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci USA. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Marcellin P, Heathcote EJ, Buti M, Gane E, de Man RA, Krastev Z, Germanidis G, Lee SS, Flisiak R, Kaita K, et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med. 2008;359:2442–2455. doi: 10.1056/NEJMoa0802878. [DOI] [PubMed] [Google Scholar]
- Martinez-Picado J, Martínez MA. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res. 2008;134:104–123. doi: 10.1016/j.virusres.2007.12.021. [DOI] [PubMed] [Google Scholar]
- Maskell R, Okubadejo OA, Payne RH, Pead L. Human infections with thymine-requiring bacteria. J Med Microbiol. 1978;11:33–45. doi: 10.1099/00222615-11-1-33. [DOI] [PubMed] [Google Scholar]
- Moyed HS, Bertrand KP. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK, Prior JL, Piwnica-Worms D, Bridger G, Ley TJ, DiPersio JF. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood. 2009;113:6206–6214. doi: 10.1182/blood-2008-06-162123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Richman DD, Havlir D, Corbeil J, Looney D, Ignacio C, Spector SA, Sullivan J, Cheeseman S, Barringer K, Pauletti D, et al. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Thomas L, Bondarenko I, O’Day S, MDJW, Garbe C, Lebbe C, Baurain JF, Testori A, Grob JJ, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- Saglio G, Kim DW, Issaragrisil S, le Coutre P, Etienne G, Lobo C, Pasquini R, Clark RE, Hochhaus A, Hughes TP, et al. Nilotinib versus imatinib for newly diagnosed chronic myeloid leukemia. N Engl J Med. 2010;362:2251–2259. doi: 10.1056/NEJMoa0912614. [DOI] [PubMed] [Google Scholar]
- Schapiro JM, Winters MA, Stewart F, Efron B, Norris J, Kozal MJ, Merigan TC. The effect of high-dose saquinavir on viral load and CD4+ T-cell counts in HIV-infected patients. Ann Intern Med. 1996;124:1039–1050. doi: 10.7326/0003-4819-124-12-199606150-00003. [DOI] [PubMed] [Google Scholar]
- Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- Scher HI, Beer TM, Higano CS, Anand A, Taplin ME, Efstathiou E, Rathkopf D, Shelkey J, Yu EY, Alumkal J, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1–2 study. Lancet. 2010;375:1437–1446. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke RT, Kaufman RJ, Alt FW, Kellems RF. Gene amplification and drug resistance in cultured murine cells. Science. 1978;202:1051–1055. doi: 10.1126/science.715457. [DOI] [PubMed] [Google Scholar]
- Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner SA, Mulder J, Loveday C, Christopherson C, et al. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaggs BJ, Gorre ME, Ryvkin A, Burgess MR, Xie Y, Han Y, Komiso-poulou E, Brown LM, Loo JA, Landaw EM, et al. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc Natl Acad Sci USA. 2006;103:19466–19471. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen R, Sköld O. Plasmid-borne or chromosomally mediated resistance by Tn7 is the most common response to ubiquitous use of trimeth-oprim. Antimicrob Agents Chemother. 1985;27:933–937. doi: 10.1128/aac.27.6.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O’Brien S, Nicaise C, Bleickardt E, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson JS, Hooper DC, McHugh GL, Bozza MA, Swartz MN. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob Agents Chemother. 1990;34:1938–1943. doi: 10.1128/aac.34.10.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol Rev. 2008;32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- Zolopa AR, Shafer RW, Warford A, Montoya JG, Hsu P, Katzenstein D, Merigan TC, Efron B. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann Intern Med. 1999;131:813–821. doi: 10.7326/0003-4819-131-11-199912070-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]