Abstract
The antioxidant response element (ARE) is a critical regulatory element for the expression of many phase II drug metabolizing enzymes (DME), phase III transporters, and anti-oxidant enzymes, mediated by the transcription factor Nrf2. The aim of this study was to examine the potential activation and synergism of Nrf2-ARE-mediated transcriptional activity between four common phytochemicals present in cruciferous vegetables, the indoles; indole-3-carbinol (I3C), 3,3’-diindolylmethane (DIM), and the isothiocyanates (ITCs); phenethyl isothiocyanate (PEITC) and sulforaphane (SFN). The cytotoxicity of the compounds was determined in human liver hepatoma cell line (HepG2-C8). The combination index was calculated to assess the synergistic effects on the induction of ARE-mediated gene expressions. qPCR was employed to measure the mRNA expressions of Nrf2 and Nrf2-mediated genes. I3C and DIM showed less cytotoxicity than SFN and PEITC. Compared to I3C, DIM was found to be a stronger inducer of ARE. Synergism was observed after combined treatments of I3C 6.25 µM + SFN 1 µM, I3C 6.25 µM + PEITC 1 µM and DIM 6.25 µM + PEITC 1 µM, while additive effect was observed for DIM 6.25 µM + SFN 1 µM. Induction of endogenous Nrf2, phase II genes (GSTm2, UGT1A1, and NQO1) and antioxidant genes (HO-1 and SOD1) was also observed. In summary, the indole I3C or DIM alone could induce or syngergistically induce in combination with the ITCs SFN or PEITC, Nrf2-ARE-mediated gene expression, which could potentially enhance cancer chemopreventive activity.
Keywords: Antioxidant response element (ARE); nuclear factor (erythroid-derived 2)-like 2 (NFE2L2 or Nrf2); indole-3-carbinol (I3C); 3,3’-diindolylmethane (DIM); isothiocyanates
Introduction
When cells are exposed to excessive oxidative stress, DNA would go through oxidative damage [1]. When coupled with chronic inflammation [2] with formation of DNA adducts, this would lead to enhanced genomic instability, neoplastic transformation, and ultimately driving cancer formation and tumorigenesis [3]. To counteract oxidative stress, induction of various cellular protective enzymes including phase II drug metabolizing enzymes (DME), phase III transporters and antioxidant enzymes occur [4–5]. Carcinogens are typically metabolized via oxidation and reduction by phase I DME [6]. The resulting products will subsequently undergo phase II conjugations catalyzed by phase II DME such as glutathione S-transferases (GST) and UDP-glucuronosyltransferases (UGT), resulting in the formation of conjugated products which are more water soluble and can be easily excreted in the bile or in the urine [6–7].
The induction of phase II DME can be largely attributed to the transcriptional control of the antioxidant response element (ARE) by the nuclear factor (erythroid-derived 2)-like 2 (NFE2L2 or Nrf2) [8]. Nrf2 is known as a key regulator of the ARE-mediated gene expression and therefore a potential target for cancer chemopreventive compounds [9–11]. Nrf2 is inhibited in the cytoplasm by the anchor protein Kelch-like ECH-associated protein-1 (Keap1) and in the presence of oxidative stress or chemical inducers, Nrf2 is released from Keap1 inhibition, translocates to the nucleus, dimerizes with small Mafs (sMaf) and binds to ARE consensus sequence [12]. Regulation of Nrf2 by cancer chemopreventive compounds would lead to the induction of phase II DME, phase III transporters, and anti-oxidative stress enzymes such as heme oxygenase 1 (HO-1). HO-1 catalyzes the degradation of heme to carbon monoxide, iron and biliverdin. HO-1 is critically essential in cellular defensive mechanisms and is implicated with various pathophysiological disease conditions including inflammation, atherosclerosis, neurodegenerative diseases and cancers [13].
The cruciferous vegetables contain abundant phytochemicals with potentially super cancer chemopreventive activities [14]. Cruciferous vegetables include broccoli, Brussels sprouts, cabbage, and cauliflower are rich in glucosinolates that can endogenously be converted into compounds including indoles [indole-3-carbinol (I3C) and 3,3’-diindolylmethane (DIM)] and isothiocyanates (ITCs) [phenethyl isothiocyanate (PEITC) and sulforaphane (SFN)] (Fig.1) upon ingestion [14]. Epidemiological studies indicate that dietary consumption of these compounds via high consumption of cruciferous vegetables would reduce cancer risk [15–17].
Fig. 1.
Chemical structures of phytochemicals used in the current study.
Phytochemicals, including indoles and ITCs possess potent cancer chemopreventive effects [18–21]. Previous studies show that the indoles would achieve the cancer chemopreventive effects potentially via multi-targets. For instance, they are capable of inducing antioxidant activity, regulate cellular proliferative genes, induce cell cycle arrest/apoptosis, regulate hormone metabolism, and stimulate the immune system [22–28]. ITCs can also elicit their chemopreventive effects via various mechanisms such as regulating DME phase I cytochrome P450s and phase II DME, Nrf2-Keap1 anti-oxidative stress, and anti-inflammatory NFkB pathways, as well as inducing cell cycle arrest/apoptosis [20–21].
The Nrf2-ARE signaling pathway has been shown to play important role in cancer chemoprevention [9–11]. In the present study, we investigated the transcriptional activation of Nrf2-ARE signaling pathway by the indoles (I3C and DIM) and the potential synergistic effect between the indoles and the ITCs (SFN and PEITC). To accomplish these goals, we utilized the human liver carcinoma cell line (HepG2-C8-ARE-luciferease cells; the original HepG2 cell stabilized with the ARE-luciferase reporter gene [29]), a metabolic competent cell line, which is a useful in vitro cell culture model to study regulation of DME [29]. Our results show that the indoles, I3C and DIM alone can transcriptionally activated Nrf2-ARE-mediated gene expression, and importantly, the indoles can also act synergistically in activating the Nrf2-ARE-mediated signaling when combined with the ITCs SFN or PEITC
Materials and Methods
Materials
I3C, DIM, and PEITC were purchased from Sigma Chemicals Co. (St Louis, USA). SFN was obtained from LKT Laboratories (St. Paul, USA).
Cell culture
Stably transfected single clone HepG2-ARE-C8 (HepG2-C8) cell line has been previously established in our laboratory using the pARE-TI-luciferase reporter gene [29–36]. The cells were maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum (FBS), 1.17 g/L sodium bicarbonate, and 100 unit/mL penicillin, 100 µg/ml streptomycin at 37 °C in a humidified incubator with 5% CO2.
MTS assay
The cytotoxicity of the phytochemicals was tested in HepG2-C8 cells using the CellTiter 96 aqueous non-radioactive cell proliferation assay MTS assay [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H- tetrazolium, inner salt; MTS] (Promega, Madison, WI). The cells were first cultured in 96-well plates for 24 h and then were treated with I3C, DIM, PEITC or SFN at various concentrations for 24 h. The cells were then treated with MTS for 1 h at 37°C. Absorbance of the formazan product was read at 490 nm with µQuant Biomolecular Spectrophotometer from Bio-Tek Instruments Inc. (Winooski, VT). Independent control studies were conducted using 1% and 10% FBS medium.
ARE-luciferase assay
HepG2-C8 cells were cultured in 12-well plates and each well contained 1 million cells in 1 ml of 10% FBS medium. The cells were treated with compounds for 24 h. The luciferase activity was determined using a luciferase kit from Promega (Madison, USA) according to the manufacturer’s instructions. Briefly, after treatments for 24 h, the cells were washed twice with ice-cold phosphate buffered-saline (PBS, pH 7.4) and harvested in 1× reporter lysis buffer and kept at −20°C overnight. After centrifugation at 4°C, 12,000 rpm for 5 min, a 10 µl aliquot of the supernatant was assayed for luciferase activity with a SIRIUS luminometer (Berthold Detection System GmbH, Pforzheim, Germany). The luciferase activity was normalized against protein concentration, determined by BCA protein assay (Pierce, Rockford, USA), and expressed as fold of induction over the luciferase activity of control vehicle-treated cells. At least two to three independent studies were conducted in triplicates.
RNA extraction and quantitative real-time PCR
The cells were treated similarly as the MTS and ARE-luciferase assays described above using 10% FBS medium. The incubation of the compounds with the cells was terminated at 6 h later. The mRNA expression was evaluated utilizing quantitative real-time polymerase chain reaction (qPCR). RNeasy kit from Qiagen was used for RNA extraction (Valencia, CA). Total RNA was reverse-transcribed to cDNA by TaqMan Reverse Transcription Reagents (Applied Biosystems Inc, Foster City, CA). SYBR Green (Applied Biosystems Inc, Foster City, CA) fluorescence was used to measure the product of qPCR. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the housekeeping gene, and the Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems Inc, Foster City, CA) was used as previously described [37] to detect quantitatively the induction of mRNA of Nrf2, phase II DME GSTm2, NAD(P)H dehydrogenase, quinone 1 (NQO1), UGT family, polypeptide A1 (UGT1A1) and antioxidant enzymes HO-1, superoxide dismutase 1 (SOD1). The primer pairs were designed using Primer Quest Oligo Design and Analysis Tool by Integrated DNA Technologies Inc. (Coralville, IA, USA) and the sequences are listed in Table 1. At least 4 wells of each treatment were performed and duplicate samples were carried out for each treatment.
Table 1.
Human oligonucleotide primers used for qPCR
| Gene | Association no. | Forward (5’) primer | Reverse (3’) primer |
|---|---|---|---|
| Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | NM_002046.3 | 5'-TCG ACA GTC AGC CGC ATC TTC TTT-3' | 5'-ACC AAA TCC GTT GAC TCC GAC CTT-3' |
| Glutathione S-transferase mu 2 (GSTm2) | NM_000848 | 5’-ACT AAA GCC AGC CTG ACC TTC CTT-3’ | 5’-AAT GCT GCT CCT TCA TGC AAC ACG-3’ |
| Hemeoxygenase-1 (HO-1) | NM_206866 | 5'- ACG CGT TGT AAT TAA GCC TCG CAC-3' | 5'-TTC CGC TGG TCA TTA AGG CTG AGT-3' |
| NAD(P)H dehydrogenase, quinone 1 (NQO1) | NM_001025434 | 5’-AAG GAT GGA AGA AAC GCC TGG AGA-3’ | 5’-GGC CCA CAG AAA GGC CAA ATT TCT-3’ |
| Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) | NM_001145413 | 5’-TGC TTT ATA GCG TGC AAA CCT CGC-3’ | 5’-ATC CAT GTC CCT TGA CAG CAC AGA-3’ |
| Superoxide dismutase 1 (SOD1) | NM_000454 | 5'-GCA GGG CAT CAT CAA TTT CGA GCA-3' | 5'- TGC AGG CCT TCA GTC AGT CCT TTA-3' |
| UDP-Glucuronosyltransferase 1 family, polypeptide A1 (UGT1A1) | NM_000463 | 5'-ATG ACC CGT GCC TTT ATC ACC CAT-3' | 5'-AGT CTC CAT GCG CTT TGC ATT GTC -3' |
Western blotting
The cells were treated similarly as the MTS, ARE-luciferase and qPCR assays described above using 10% FBS medium. HepG2-C8 cells were treated with the compound for 24 h. Cells were washed with ice-cold PBS (pH 7.4) and harvested in Cell Culture Lysis Reagent (Promega E153A, Madison, WI). The homogenate was centrifuged at 4°C, 12,000 rpm for 5 min. The supernatants were collected and 15 µg of total protein, as determined by BCA protein assay (Pierce, Rockford, USA) were mixed with 5 µl Laemmli’s SDS-Sample Buffer (Boston Bioproducts, Ashland, MA, USA) and denatured at 95°C, for 5 minutes. The samples and the protein standard (Bio-Rad, Hercules, CA, USA) were then loaded onto a polyacrylamide gel (Criterion Tris–HCl gel, Bio-Rad Lab, Hercules, CA, USA) and gel electrophoresis run at 130 mA for 60 min. Proteins were transferred onto a polyvinylidene difluoride (PVDF) membranes (Immobilon-P, Millipore, Bedford, MA, USA) over 1.5 h using a semi-dry transfer system (BioRad, Hercules, CA, USA). The membranes were blocked with 5% BSA solution for 1 h at room temperature and incubated with the primary antibody (1:1000, in 3% BSA in TBST) overnight at 4°C. Antibody against Actin (catalog no. sc-1616), NQO1 (catalog no. sc-16464) and SOD1 (catalog no. sc-11407) were purchased from Santa Cruz (Santa Cruz Biotechnology, Inc., CA, USA). Antibody against Nrf2 (catalog no. 2178-1) were purchased from Epitomics (Burlingame, CA, USA). After hybridization with primary antibody, membranes were washed with TBST (Tris-buffered-saline and Tween 20) four times. The immunoreactions were continued with the respective secondary antibodies (1:5000, in 3% BSA in TBST) purchased from Santa Cruz Biotechnology, Inc., CA, USA, for 1 h at room temperature. After washing four times with TBST, the immunocomplexes were determined using the enhanced chemiluminescent system to detect the horseradish peroxidase on the immunoblots (Thermo Scientific, Rockford, IL, USA) and the bands were visualized and captured by BioRad ChemiDoc XRS system (Hercules, CA, USA).
Combination index calculation
To determine the synergistic effect between the combination of two different compounds, the combination index (CI) can be calculated with the following formula: CI= d1/Dx,1 + d2/Dx,2 where d1 and d2 are doses of drugs 1 and 2 in combination, which produces an effect x. Dx,1 and Dx,2 are the doses of drug 1 and 2 that produce the same effect × when given alone. When the CI is equal to, less than or greater than 1, the combination dose will be additive, synergistic or antagonistic, respectively, as we have described previously [37–38]. This approach is based on the Loewe additivity model and although the exact mechanism of interaction may be unknown, this model is one of the most commonly used reference models for evaluating potential drug-drug interactions [39]. Using this CI calculation for the ARE-luciferase activity induced by I3C or DIM combined with PEITC or SFN, it is possible to identify whether the combination of these phytochemicals at certain concentrations would be synergistic, antagonistic or additive.
Statistical analysis
The results are presented as means ± standard error of the mean (SEM). MTS assay data were analyzed using one-way ANOVA with a post hoc multiple comparison analysis by Bonferroni. Luciferase assay and qPCR data were statistically analyzed using Student’s t-test. P value of less than 0.05 was considered to be statistically significance.
Results
Cell viability by MTS assay
To test the cell viability of I3C, DIM, SFN and PEITC, MTS assay was employed. DIM and I3C showed less toxicity than SFN and PEITC in 1% FBS medium (Fig. 2), SFN and PEITC showed similar cell viability inhibitory concentrations (IC50) of around 20 µM, whereas I3C and DIM had higher IC50 of 135 µM and 51 µM, respectively. Using 10% FBS, several previous publications show that DIM was more cytotoxic than I3C, hence we tested the same dosage in HepG2-C8 cells with 10% FBS. The cytotoxicity of HepG2-C8 was affected more with DIM than I3C, i.e. DIM showed an IC50 of around 85 µM while I3C showed an IC50 of 300 µM in 10% FBS medium (data not shown).
Fig. 2.
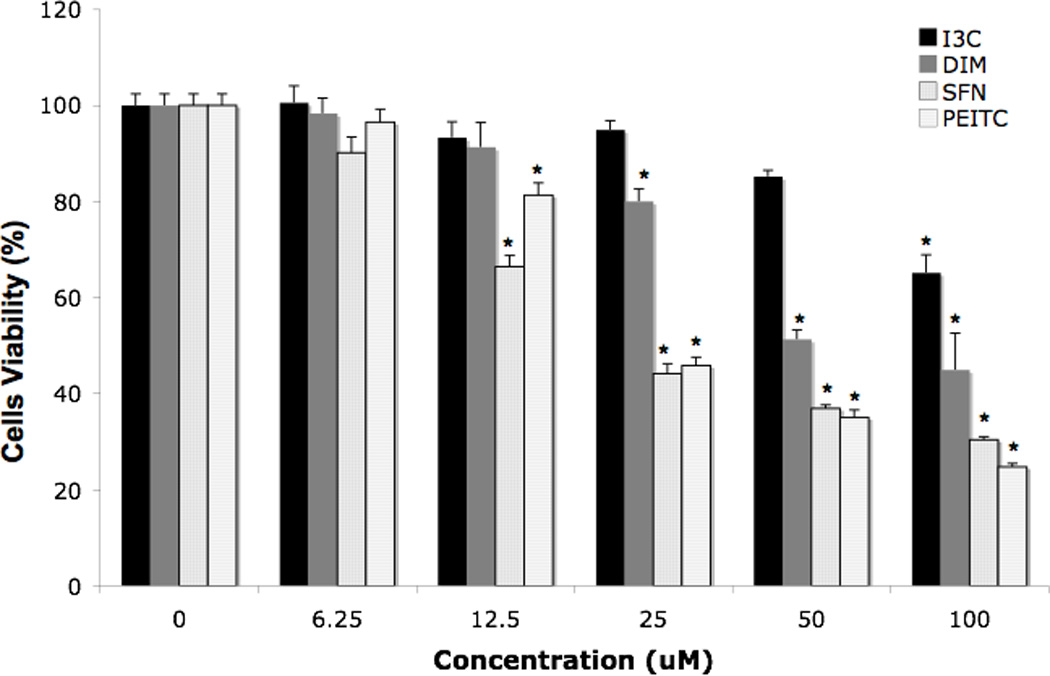
Effect of the compounds tested on the cell viability determined by MTS assay, using medium with 1% FBS. Results are expressed as the mean ± SEM. *p < 0.05, compared with corresponding value for 0.1% DMSO-treated cells.
ARE-luciferase activity
In the ARE transcriptional activation assay, the cells were treated with higher doses of DIM and I3C, 25 and 75 µM, since from the MTS assay the viability was not affected at these concentrations in 10% FBS medium (data not shown). To evaluate the transcriptional activation of ARE, ARE-luciferase reporter assay was performed [40]. SFN and PEITC were used as positive controls and 0.1% DMSO was used as negative controls. The ARE-luciferase activity was expressed as fold of induction over the negative vehicle control. All compounds alone and in combinations induced ARE-luciferase activity in HepG2-C8 cells with different potency (Fig. 3). DIM at 75 µM strongly induced the ARE-luciferase as compared to any other treatments (p<0.05). Interestingly, DIM 25 µM with SFN 1 µM (DIM25/SFN1) was synergistic but not for I3C 25 µM with SFN 1 µM (Fig. 3). Although there are three synergistic interactions at low doses of combination having ARE activities close to value 1, all of their CI were < 1, and p < 0.05. Specifically, synergistic effects were observed for the combinations of I3C 6.25 µM with SFN 1 µM (I3C6.25/SFN1, p value for CI = 0.045), I3C 6.25 µM with PEITC 1 µM (I3C6.25/PEITC1, p value for CI = 0.044) and DIM 6.25 µM with PEITC 1 µM (DIM6.25/PEITC1, p value for CI = 0.003). Additive effect was observed with DIM 6.25 µM with SFN 1 µM (DIM6.25/SFN1) whereas I3C 25 µM with SFN 1 µM (I3C25/SFN1) was antagonistic. DIM25/SFN1 treatment displayed the most synergism, and I3C6.25/SFN1, DIM6.25/PEITC1 and I3C6.25/PEITC1 were not so obvious, however, their CI values were less than one (i.e. synergistic). DIM6.25/SFN1 had a CI value of 1 (i.e. additive). I3C25/SFN1 had a CI value of more than 1, (i.e. antagonistic). The classification of synergistic, additive or antagonistic was based mathematically on the CI calculations that were derived from the dose response of single compound, and the response of the combinations at different doses. We had also tested using different cell density at similar drug concentrations in medium with 1% FBS, similar observations were obtained (data not shown). When doses of I3C and DIM lower than 25 µM were tested in 10% FBS medium, no significant induction was observed (data not shown). As there was an obvious dose response for single treatment with DIM (i.e. DIM25 and DIM75), and not for I3C25 and I3C75, however, the CI calculations for DIM25/SFN1 and I3C25/SFN1 showed CI of 0.7 and 3, respectively. Next, we verified the identified additive / synergistic combinations particularly at those lower concentrations but may be more physiologically relevance concentrations of indoles and ITCs using qPCR and Western blotting analyses for the Nrf2-ARE-mediated genes, as described below.
Fig. 3.
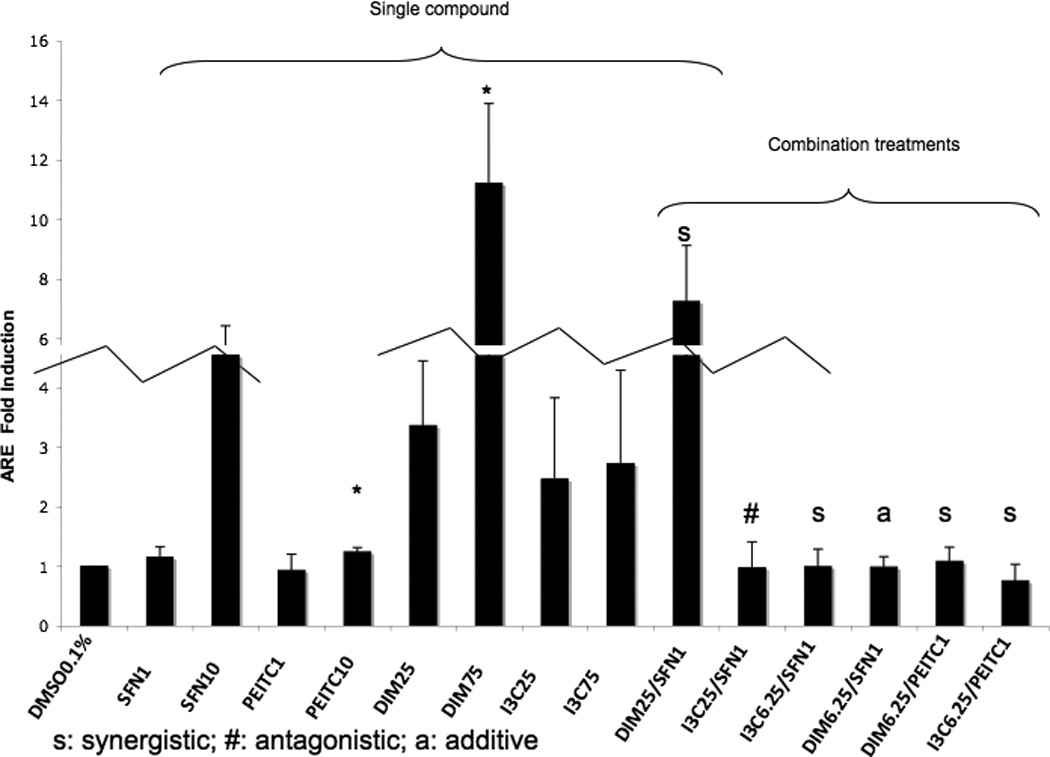
Luciferase activity in HepG2-C8 cells. All combinations are described in Materials and Methods. Note: “s” denotes synergistic; “#” denotes antagonistic; “a” denotes additive, though DIM6.25/PEITC1 is considered additive as the combination index (CI) is around 1, most of the mRNA levels were synergistically induced by this combination (eee Fig. 4). When doses of I3C and DIM lower than 25 µM were tested, no significant induction was observed. Therefore the single dose of 6.25 µM I3C and DIM is not presented. The CI for the combination studies was calculated as previously published to determine synergistic, additive or antagonistic effects [37, 39]. The changes in the fold of induction for synergistic and additive combination may not appear as robust nonetheless the qPCR results verify the findings (see Fig. 4). The broken lines are breaking the relatively higher fold changes into two corresponding connecting bars; the relative folds across all groups are maintained.
qPCR
To confirm that the cells treated with the agents induced endogenous phase II DME and antioxidant genes, we conducted qPCR to quantify the mRNA expression. Values higher than one were considered positive in comparison to cells treated with control 0.1% DMSO. The results for the induction of Nrf2, phase II DME and antioxidant genes are shown in Fig. 4. I3C alone at 25 µM did not show significant induction of Nrf2 and HO-1 mRNA (Fig. 4A and Fig. 4B). On the other hand, DIM 25 µM showed about 3 fold-induction for both of these genes. The higher dose, DIM 75 µM, induced only Nrf2 and HO-1 gene expression (Fig. 4A and Fig. 4B) which was somehow not as correlative to the dose-dependency activation of the ARE-luciferase above (Fig. 3). Interestingly, NQO1 gene expression was not significantly induced by SFN at any concentration, but it was greatly induced by PEITC even at very low concentration as low as 1 µM (Fig. 4C). Similar to PEITC 10 µM, increasing the concentrations of I3C and DIM from 25 µM to 75 µM, did not enhance NQO1 gene expression any further (Fig. 4C). Time course study using SFN and PEITC at 6 h treatment indicated that the lower concentration of PEITC was a faster ARE inducer compared to SFN at 6 h. In addition, 1 µM PEITC induced higher ARE activity than 10 µM PEITC (data not shown). At 12 h, both SFN and PEITC 10 uM had higher ARE induction than at lower 1 uM concentration (data not shown). We postulate that these observations could possibly due to the additional different mechanisms by which SFN and PEITC regulate gene expression, in addition to the common Nrf2-ARE mediated signaling pathway [21] and this will be further discussed later on.
Fig. 4.
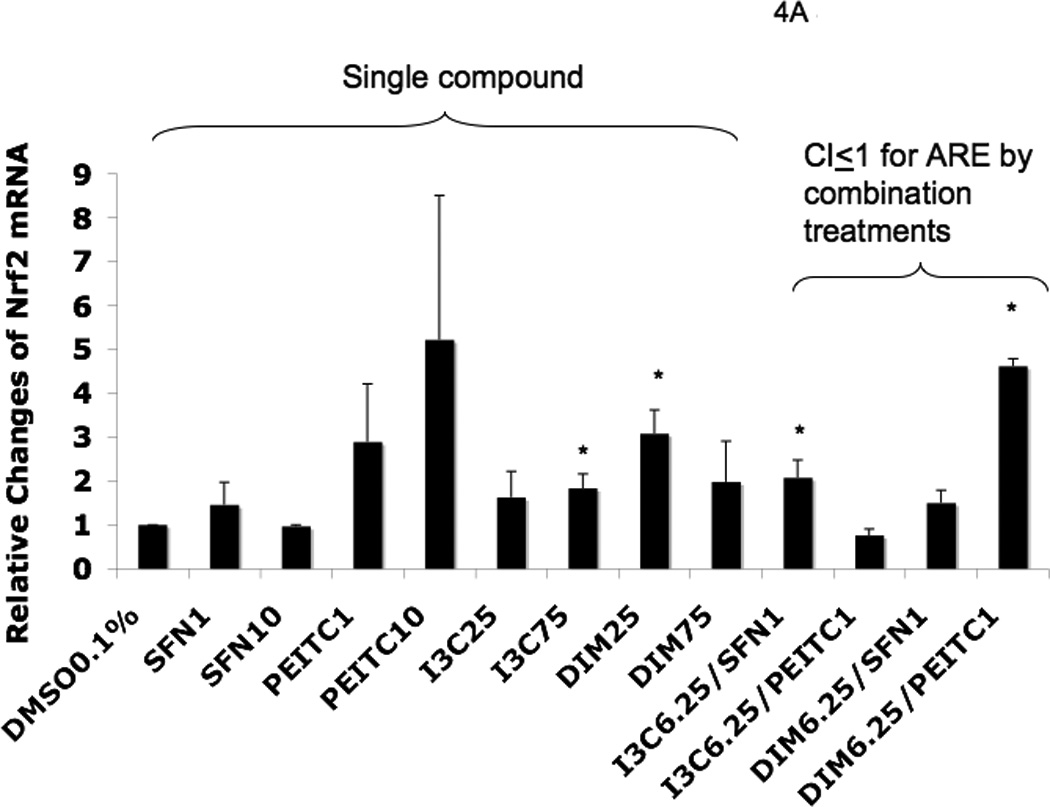
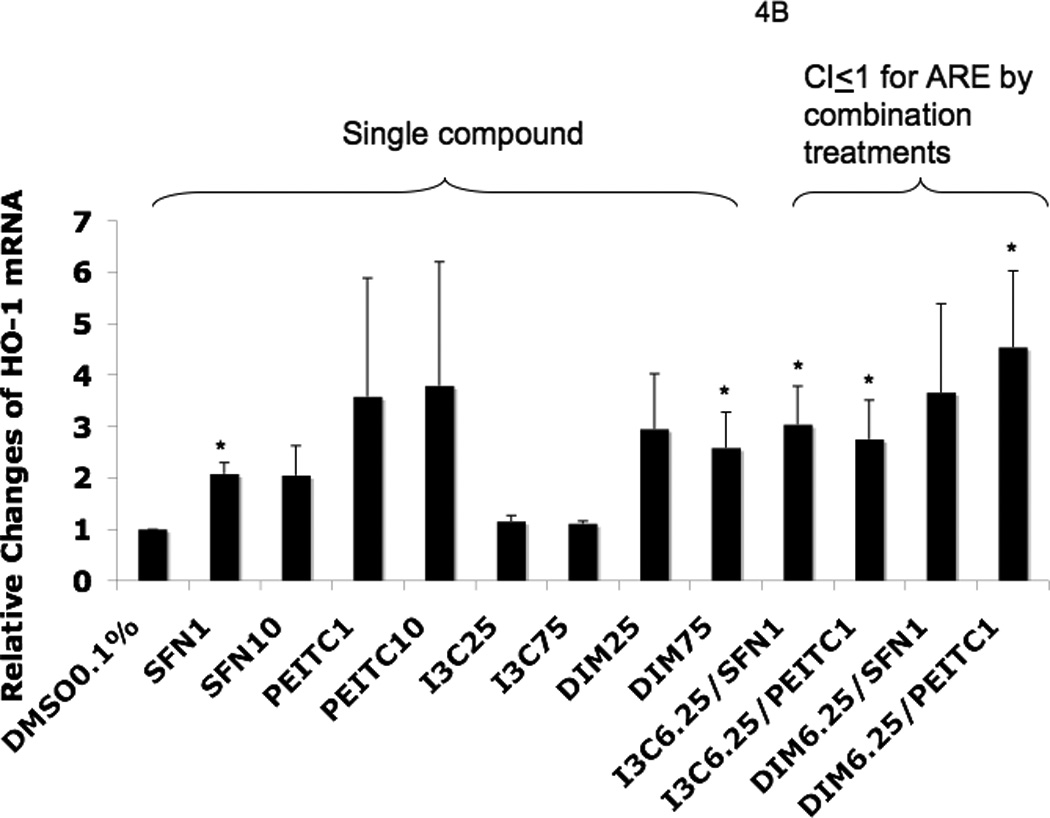
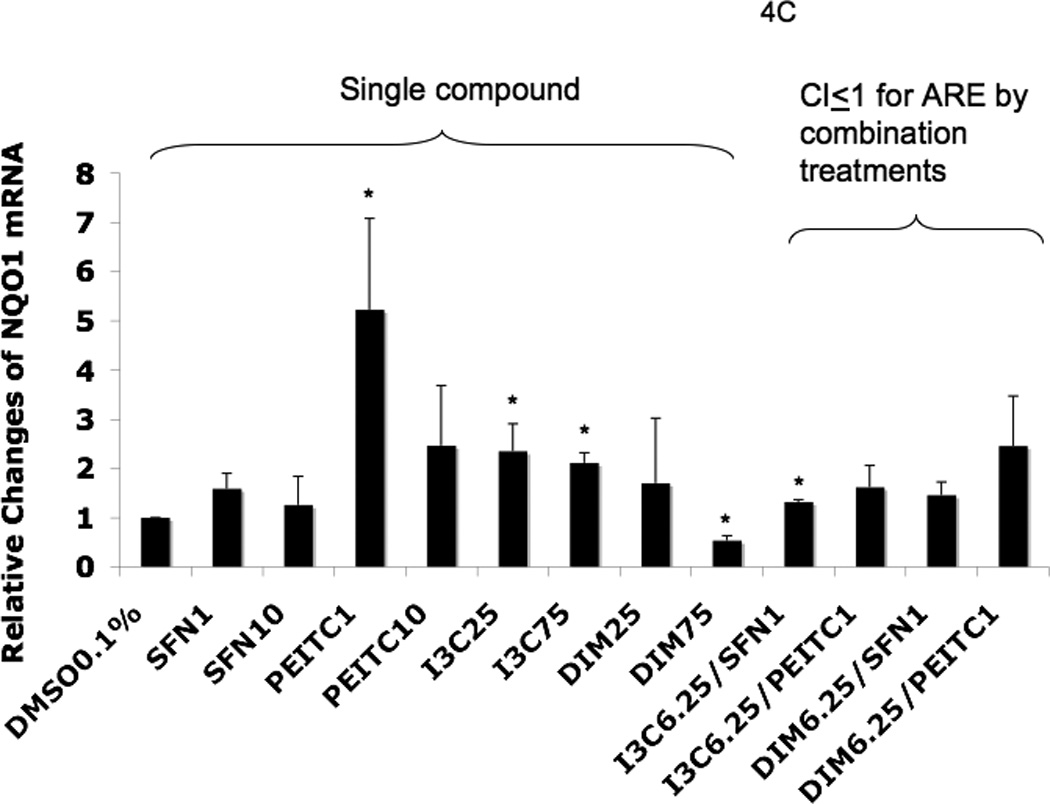
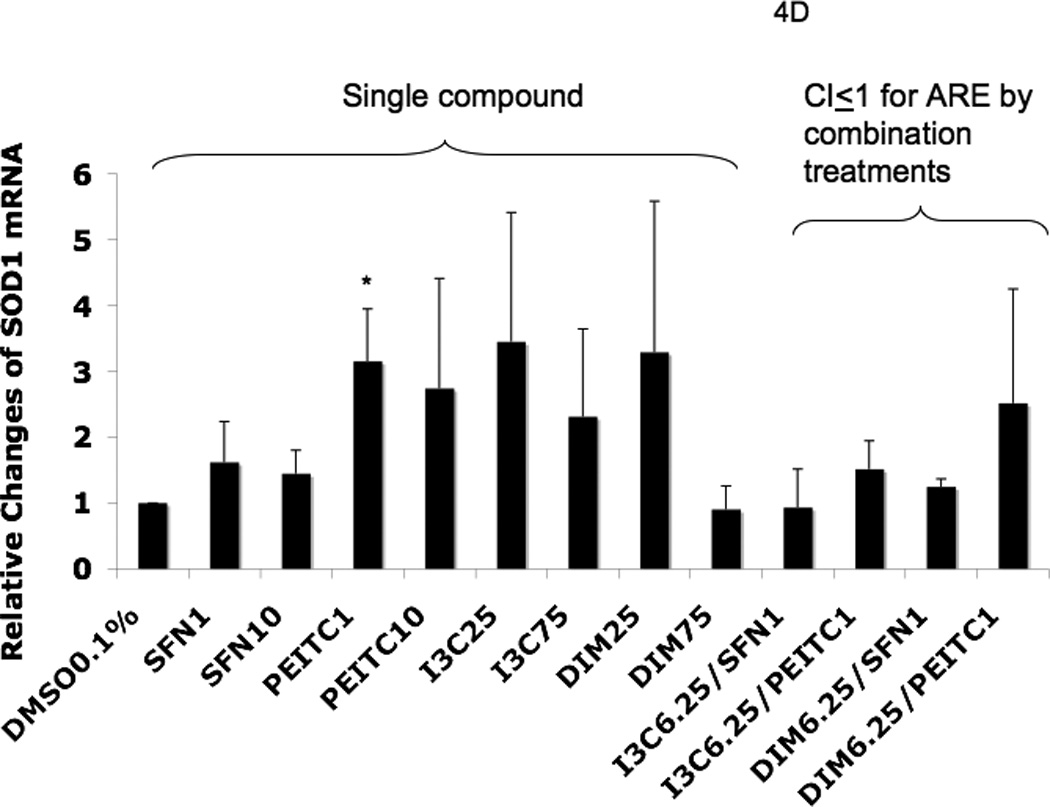
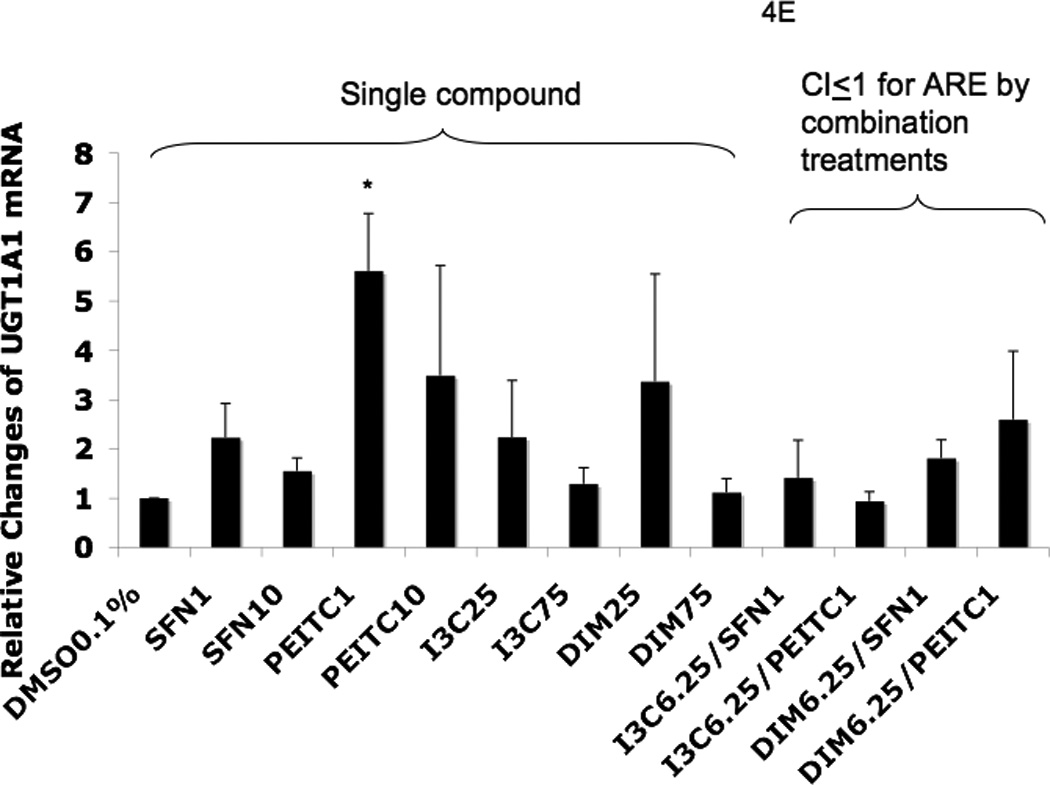
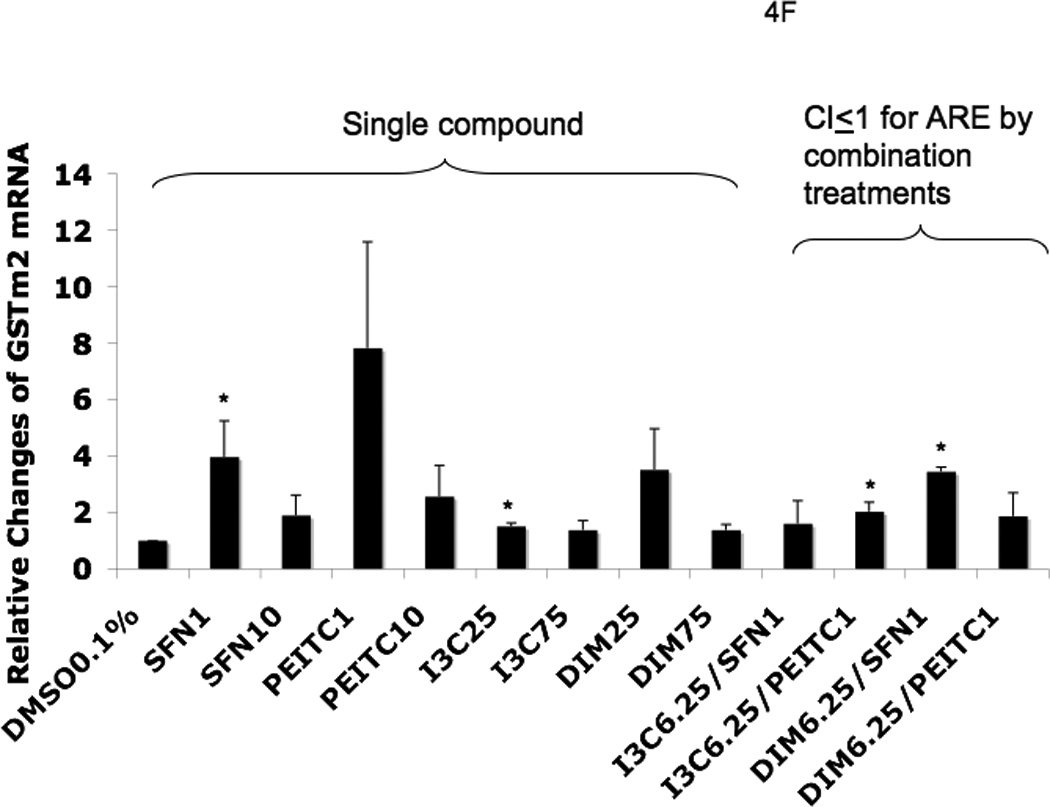
Real-time PCR (qPCR) results expressed in fold changes of mRNA over the control, using GAPDH as endogenous housekeeping gene. (A) relative expression level of Nrf2 mRNA. (B) relative expression level of HO-1 mRNA. (C) relative expression level of NQO1 mRNA. (D) relative expression level of SOD1 mRNA. (E) relative expression level of UGT1A1 mRNA. (F) relative expression level of GSTm2 mRNA. Results are expressed as mean ± SEM. The tested concentrations were in µM. *p < 0.05, compared with the 0.1% DMSO-treated control cells.
Among the combination treatments, DIM 6.25 µM with PEITC 1 µM had the greatest induction of SOD1 (Fig. 4D) and UGT1A1 (Fig. 4E), whereas DIM 6.25 µM with SFN 1 µM induced GSTm2 the most (Fig. 4F). These results confirmed the synergistic and additive effects of the combinations generated from the ARE-luciferase studies, respectively. In comparison to the other genes, with the same combinations, synergism was observed for HO-1, which was induced the most (Fig. 4B).
Western Blot
Fig. 5 shows the selected protein biomarkers of Nrf2 and one of the Nrf2-downstream targets SOD1 were examined using Western blotting. It was hypothesized that the combination of low doses of indoles and ITCs could enhance Nrf2/ARE-mediated Nrf2 and Nrf2-target antioxidant enzymes such as SOD1. I3C and DIM alone at various concentrations was able to induce the protein levels of Nrf2 and SOD1 in a dose-dependent manner (Fig. 5). The combinations of low doses of indoles and ITCs were also able to induce higher protein expression of SOD1 compared to the individual agent at higher concentrations and higher induction of Nrf2 and SOD1 proteins was also observed (Fig. 5, representative of three separate experiments with similar results), which corroborated with the synergistic effects (CI ≤ 1) for the combination treatments indentified in the ARE-luciferase assay (Fig. 3). In contrast, Nrf2 protein expression for DIM6.25/SFN1 treatment which was showed as additive using the CI calculation (Fig. 3), showed slightly less than 1 but yet the SOD1 expression was almost 2 folds compared to DMSO 0.1% control. These results suggest that differential signaling pathways were activated by the indoles and the ITC at different concentrations with different combinations and that some time, endogenous gene expression would vary from simple single promoter transcriptional reporter gene assay, of which the latter would provide a quick screen for potential in vivo activites.
Fig. 5.
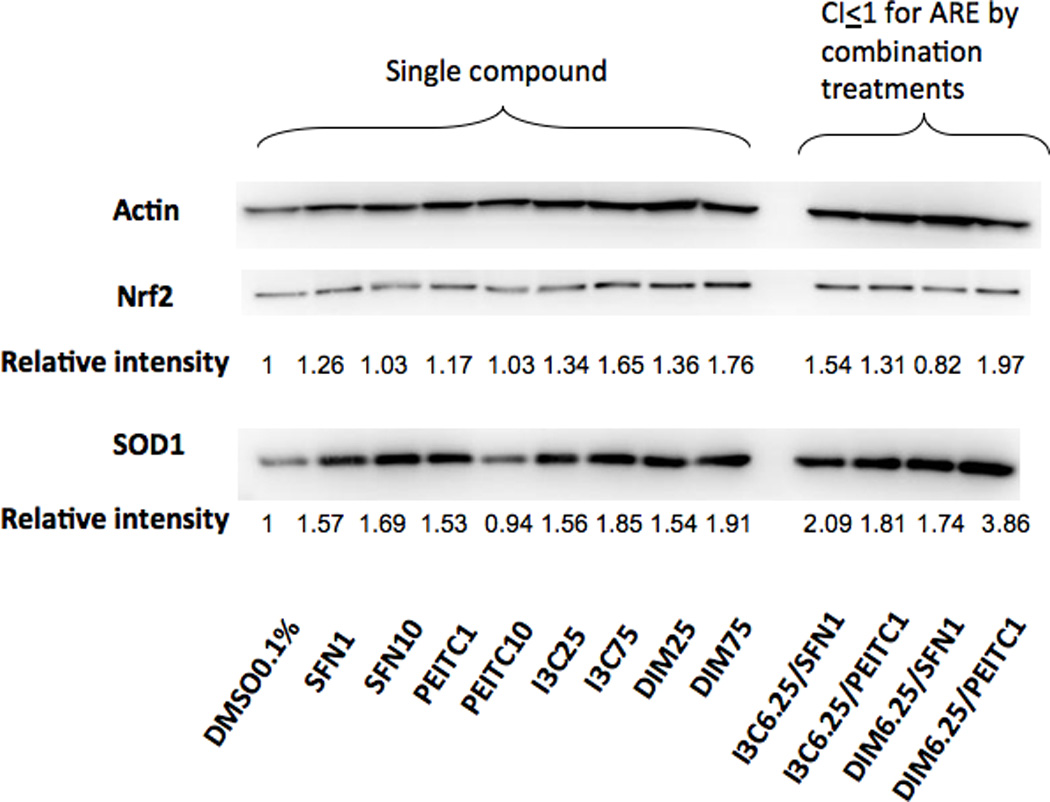
Effects of SFN, PEITC, I3C, DIM and their combinations on Nrf2 and SOD1 protein expression in HepG2-C8 cells by Western blotting using Actin as housekeeping protein. The combinations of low doses of indoles and ITCs were able to induce protein expression of Nrf2 and SOD1 and the synergism was evident for both Nrf2 and SOD1. The tested concentrations were in µM. Representative images of three independent experiments are shown.
Discussion and Conclusion
In general, diseases prevention including cancer chemoprevention could conceivably be achieved with increased consumption of fruits and vegetables containing rich sources of many phytochemicals [19]. The exact mechanisms by which phytochemicals could prevent diseases such as cancer are not clear, but would appear to potentially involve one of the signaling pathways, the Nrf2-ARE-mediated anti-oxidative stress pathway [8, 12]. Similarly, the involvement of Nrf2-ARE-mediated signaling by I3C and DIM remains unclear. Figure 6 shows the schematic diagram of the proposed mechanism by which Nrf2-ARE and its downstream targeting enzymes are induced by chemicals/phytochemicals, which has been previously proposed and reviewed [8, 12]. In the current study, we investigated the transcriptional activation of Nrf2-ARE mediated gene expression, as well as the potential synergistic effects of the indoles and the ITC compounds.
Fig. 6.
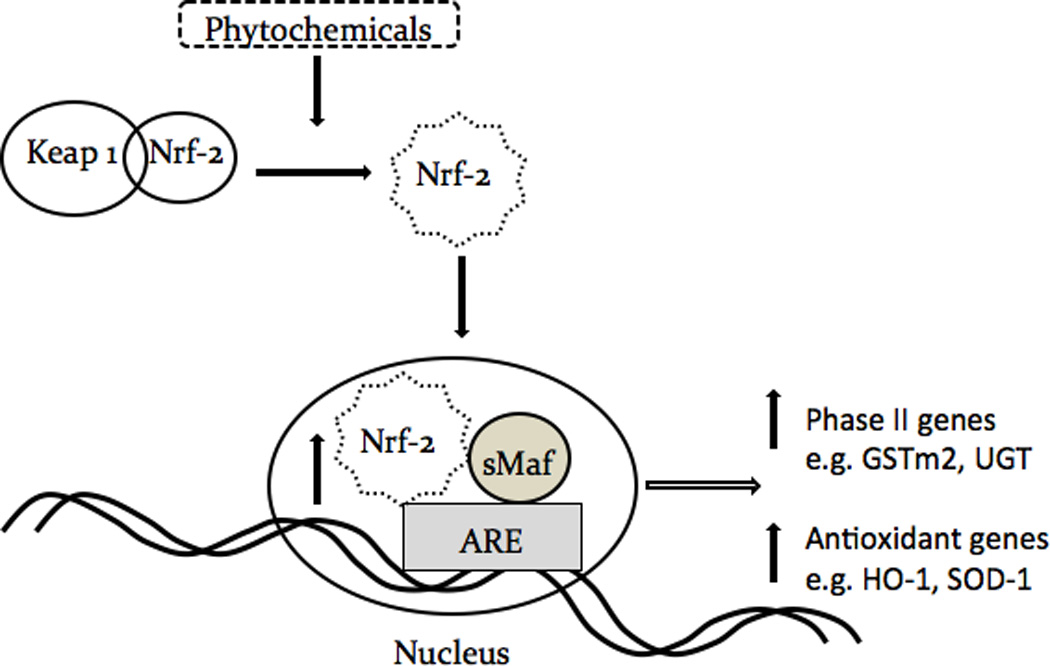
Schematic diagram of the proposed simplified pathway shows indole and isothiocyanate phytochemicals inducing Nrf2-ARE signaling through activation of the ARE and producing anti-oxidative and phase II detoxifying genes.
As shown in Fig. 3, SFN is a stronger inducer than PEITC in the ARE-luciferease transcription assay. However, in contrast PEITC induced higher mRNA levels of endogenous Nrf2 and Nrf2-mediated genes than SFN (Fig. 4). Previous reports show that HepG2 cells treated by PEITC [41] and SFN [42] have shown different time course and concentration-dependent apoptosis. Since, there is no existing report on the direct comparison of SFN and PEITC in the activities of Nrf2-ARE induction, therefore, we performed time course studies on the induction of ARE-luciferase activities by SFN and PEITC. As early as 6 h PEITC 1 µM induced higher ARE activities than SFN 1 µM (data not shown). The slower inducing effect of SFN correlated with our previous report that SFN reached its peak induction at 18 h after treatment [35]. Moreover, there are also differences between SFN and PETIC with respect to treatment time for ARE activities (24 h, Fig. 3), mRNA (6 h, Fig. 4) and protein (24 h, Fig. 5). It appears that the kinetic profiles for SFN and PEITC in inducing ARE, Nrf2 and Nrf2-mediated genes are quite different. These findings suggest that, in addition to the Nrf2-ARE mediated signaling pathway, other pathways such as the activation of the mitogen-activated protein kinases (MAPKs), could also be involved (reviewed in [21]).
In our current study, synergism was observed for different combinations between the indoles and the ITCs at some concentrations (Fig 3). In order to confirm that the observations that the phytochemicals were promoting the induction of Nrf2, phase II DME and antioxidant genes, we performed qPCR and Western blotting. Since preventing diseases including cancer initiation could be achieved by protecting cells and tissues against oxidative stress-mediated damages, an effective mechanism of defense against such damages would be via the induction of cellular phase II DME/detoxifying and antioxidant enzymes such as UGT, GST, NQO1, SOD1 and HO-1 [43]. The inductions of these enzymes are mediated by the Nrf2-ARE signaling pathway. In this context, our current study quantifies the gene expression of Nrf2, HO-1, SOD1, NQO1, UGT and GSTm2 and the induction of these genes is shown in Fig. 4. DIM 6.25 µM plus PEITC 1 µM showed the most robust overall synergistic effect as compared to the other treatments (Fig. 4). Overall, it appears that DIM 6.25 µM with PEITC 1 µM would be the best combination under our experimental conditions, since we observed synergistic induction for all the genes studied (except GSTm2) and the fold of induction was also relatively higher as compared to the single agent treatment and the other combinations tested (Fig. 4). In addition, Nrf2 and SOD1 proteins also show potential synergism after 24 h of treatment with DIM 6.5 µM plus PEITC 1 µM (Fig. 5).
It is highly likely that the metabolism in HepG2 cells would occur during the course of the studies, as reported by others [44–46], although our main focus is on the interactions between the indoles and ITCs with the Nrf2-ARE mediated phase II DME and antioxidant gene expression. It has been reported that I3C could be converted to DIM in culture medium and metabolized in breast cancer cells [44–45], the current study with equal doses of I3C and DIM at 25 µM and 75 µM did not produce equal ARE induction activities, indicating that the effects of I3C by itself might be active, although not as potent as DIM (Fig. 3). This is interesting observation indeed, since many more clinical studies have been performed using I3C than with DIM [47]. In summary, we have shown that the indoles (I3C and DIM) and ITCs (PEITC and SFN) could induce Nrf2 and its downstream genes synergistically at certain combinations. The indoles and ITCs are found abundantly in our daily consumed crucifers. The potential of indoles to induce Nrf2-ARE-mediated phase II DME / antioxidant genes and the potential of synergism with ITCs has been suggested previously but has not been studied in detail. In the present study, we found that both I3C and DIM could induce Nrf2-ARE-mediated luciferase reporter gene with DIM being the more potent inducer. Furthermore, both the indoles I3C and DIM displayed synergism with the ITCs PEITC and SFN in inducing Nrf2-ARE-mediated reporter gene as well as inducing endogenous phase II DME and antioxidant genes. The results of our current study would suggest potential synergistic cancer chemopreventive effects of indoles and ITCs in vivo as well as in human.
Acknowledgements
The source of financial support: Supported in part by the RISE Program of Rutgers University/UMDNJ to M.C. and NIH-R01-CA094828 to A.N.T.K. We thank the Kong lab’s members for helpful discussions.
References
- 1.Weinberg F, Chandel NS. Reactive oxygen species-dependent signaling regulates cancer. Cell Mol Life Sci. 2009;66:3663–3673. doi: 10.1007/s00018-009-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 3.Liu RH. Potential synergy of phytochemicals in cancer prevention: mechanism of action. J Nutr. 2004;134:3479S–3485S. doi: 10.1093/jn/134.12.3479S. [DOI] [PubMed] [Google Scholar]
- 4.Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- 5.Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 6.Yu S, Kong AN. Targeting carcinogen metabolism by dietary cancer preventive compounds. Curr Cancer Drug Targets. 2007;7:416–424. doi: 10.2174/156800907781386669. [DOI] [PubMed] [Google Scholar]
- 7.Shen G, Kong AN. Nrf2 plays an important role in coordinated regulation of Phase II drug metabolism enzymes and Phase III drug transporters. Biopharm Drug Dispos. 2009;30:345–355. doi: 10.1002/bdd.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48:91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khor TO, Yu S, Kong AN. Dietary cancer chemopreventive agents - targeting inflammation and Nrf2 signaling pathway. Planta Med. 2008;74:1540–1547. doi: 10.1055/s-0028-1088303. [DOI] [PubMed] [Google Scholar]
- 10.Nair S, Li W, Kong AN. Natural dietary anti-cancer chemopreventive compounds: redox-mediated differential signaling mechanisms in cytoprotection of normal cells versus cytotoxicity in tumor cells. Acta Pharmacol Sin. 2007;28:459–472. doi: 10.1111/j.1745-7254.2007.00549.x. [DOI] [PubMed] [Google Scholar]
- 11.Kwak MK, Kensler TW. Targeting NRF2 signaling for cancer chemoprevention. Toxicol Appl Pharmacol. 2010;244:66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abraham NG, Kappas A. Pharmacological and clinical aspects of heme oxygenase. Pharmacol Rev. 2008;60:79–127. doi: 10.1124/pr.107.07104. [DOI] [PubMed] [Google Scholar]
- 14.Verhoeven DT, Verhagen H, Goldbohm RA, van den Brandt PA, van Poppel G. A review of mechanisms underlying anticarcinogenicity by brassica vegetables. Chem Biol Interact. 1997;103:79–129. doi: 10.1016/s0009-2797(96)03745-3. [DOI] [PubMed] [Google Scholar]
- 15.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol Res. 2007;55:224–236. doi: 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan R, Lok K, Woo J. Prostate cancer and vegetable consumption. Mol Nutr Food Res. 2009;53:201–216. doi: 10.1002/mnfr.200800113. [DOI] [PubMed] [Google Scholar]
- 17.Tang L, Zirpoli GR, Guru K, Moysich KB, Zhang Y, Ambrosone CB, et al. Consumption of raw cruciferous vegetables is inversely associated with bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:938–944. doi: 10.1158/1055-9965.EPI-07-2502. [DOI] [PubMed] [Google Scholar]
- 18.Nishino H, Murakoshi M, Mou XY, Wada S, Masuda M, Ohsaka Y, et al. Cancer prevention by phytochemicals. Oncology. 2005;69(Suppl 1):38–40. doi: 10.1159/000086631. [DOI] [PubMed] [Google Scholar]
- 19.Aggarwal BB, Shishodia S. Molecular targets of dietary agents for prevention and therapy of cancer. Biochem Pharmacol. 2006;71:1397–1421. doi: 10.1016/j.bcp.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Juge N, Mithen RF, Traka M. Molecular basis for chemoprevention by sulforaphane: a comprehensive review. Cell Mol Life Sci. 2007;64:1105–1127. doi: 10.1007/s00018-007-6484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. Aaps J. 2010;12:87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar FH, Li Y. Indole-3-carbinol and prostate cancer. J Nutr. 2004;134:3493S–3498S. doi: 10.1093/jn/134.12.3493S. [DOI] [PubMed] [Google Scholar]
- 23.Aggarwal BB, Ichikawa H. Molecular targets and anticancer potential of indole-3-carbinol and its derivatives. Cell Cycle. 2005;4:1201–1215. doi: 10.4161/cc.4.9.1993. [DOI] [PubMed] [Google Scholar]
- 24.Nachshon-Kedmi M, Yannai S, Haj A, Fares FA. Indole-3-carbinol and 3,3'-diindolylmethane induce apoptosis in human prostate cancer cells. Food Chem Toxicol. 2003;41:745–752. doi: 10.1016/s0278-6915(03)00004-8. [DOI] [PubMed] [Google Scholar]
- 25.Hong C, Kim HA, Firestone GL, Bjeldanes LF. 3,3'-Diindolylmethane (DIM) induces a G(1) cell cycle arrest in human breast cancer cells that is accompanied by Sp1-mediated activation of p21(WAF1/CIP1) expression. Carcinogenesis. 2002;23:1297–1305. doi: 10.1093/carcin/23.8.1297. [DOI] [PubMed] [Google Scholar]
- 26.Ali S, Banerjee S, Ahmad A, El-Rayes BF, Philip PA, Sarkar FH. Apoptosis-inducing effect of erlotinib is potentiated by 3,3'-diindolylmethane in vitro and in vivo using an orthotopic model of pancreatic cancer. Mol Cancer Ther. 2008;7:1708–1719. doi: 10.1158/1535-7163.MCT-08-0354. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Weng JR, Tsai CH, Kulp SK, Chen CS. Indole-3-carbinol as a chemopreventive and anti-cancer agent. Cancer Lett. 2008;262:153–163. doi: 10.1016/j.canlet.2008.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khwaja FS, Wynne S, Posey I, Djakiew D. 3,3'-diindolylmethane induction of p75NTR-dependent cell death via the p38 mitogen-activated protein kinase pathway in prostate cancer cells. Cancer Prev Res (Phila Pa) 2009;2:566–571. doi: 10.1158/1940-6207.CAPR-08-0202. [DOI] [PubMed] [Google Scholar]
- 29.Yu R, Mandlekar S, Lei W, Fahl WE, Tan TH, Kong AN. p38 mitogen-activated protein kinase negatively regulates the induction of phase II drug-metabolizing enzymes that detoxify carcinogens. J Biol Chem. 2000;275:2322–2327. doi: 10.1074/jbc.275.4.2322. [DOI] [PubMed] [Google Scholar]
- 30.Chen C, Yu R, Owuor ED, Kong AN. Activation of antioxidant-response element (ARE), mitogen-activated protein kinases (MAPKs) and caspases by major green tea polyphenol components during cell survival and death. Arch Pharm Res. 2000;23:605–612. doi: 10.1007/BF02975249. [DOI] [PubMed] [Google Scholar]
- 31.Wu TY, Khor TO, Saw CL, Loh SC, Chen AI, Lim SS, et al. Anti-inflammatory/Anti-oxidative stress activities and differential regulation of Nrf2-mediated genes by non-polar fractions of tea Chrysanthemum zawadskii and licorice Glycyrrhiza uralensis. Aaps J. 2011;13:1–13. doi: 10.1208/s12248-010-9239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prawan A, Keum YS, Khor TO, Yu S, Nair S, Li W, et al. Structural influence of isothiocyanates on the antioxidant response element (ARE)-mediated heme oxygenase-1 (HO-1) expression. Pharm Res. 2008;25:836–844. doi: 10.1007/s11095-007-9370-9. [DOI] [PubMed] [Google Scholar]
- 33.Jeong WS, Keum YS, Chen C, Jain MR, Shen G, Kim JH, et al. Differential expression and stability of endogenous nuclear factor E2-related factor 2 (Nrf2) by natural chemopreventive compounds in HepG2 human hepatoma cells. J Biochem Mol Biol. 2005;38:167–176. doi: 10.5483/bmbrep.2005.38.2.167. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Pung D, Leong V, Hebbar V, Shen G, Nair S, et al. Induction of detoxifying enzymes by garlic organosulfur compounds through transcription factor Nrf2: effect of chemical structure and stress signals. Free Radic Biol Med. 2004;37:1578–1590. doi: 10.1016/j.freeradbiomed.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Kim BR, Hu R, Keum YS, Hebbar V, Shen G, Nair SS, et al. Effects of glutathione on antioxidant response element-mediated gene expression and apoptosis elicited by sulforaphane. Cancer Res. 2003;63:7520–7525. [PubMed] [Google Scholar]
- 36.Yuan X, Xu C, Pan Z, Keum YS, Kim JH, Shen G, et al. Butylated hydroxyanisole regulates ARE-mediated gene expression via Nrf2 coupled with ERK and JNK signaling pathway in HepG2 cells. Mol Carcinog. 2006;45:841–850. doi: 10.1002/mc.20234. [DOI] [PubMed] [Google Scholar]
- 37.Saw CL, Huang Y, Kong AN. Synergistic anti-inflammatory effects of low doses of curcumin in combination with polyunsaturated fatty acids: docosahexaenoic acid or eicosapentaenoic acid. Biochem Pharmacol. 2010;79:421–430. doi: 10.1016/j.bcp.2009.08.030. [DOI] [PubMed] [Google Scholar]
- 38.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 39.Lee JJ, Kong M, Ayers GD, Lotan R. Interaction index and different methods for determining drug interaction in combination therapy. J Biopharm Stat. 2007;17:461–480. doi: 10.1080/10543400701199593. [DOI] [PubMed] [Google Scholar]
- 40.Yu R, Lei W, Mandlekar S, Weber MJ, Der CJ, Wu J, et al. Role of a mitogen-activated protein kinase pathway in the induction of phase II detoxifying enzymes by chemicals. J Biol Chem. 1999;274:27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 41.Rose P, Whiteman M, Huang SH, Halliwell B, Ong CN. beta-Phenylethyl isothiocyanate-mediated apoptosis in hepatoma HepG2 cells. Cell Mol Life Sci. 2003;60:1489–1503. doi: 10.1007/s00018-003-3150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh CT, Yen GC. Effect of sulforaphane on metallothionein expression and induction of apoptosis in human hepatoma HepG2 cells. Carcinogenesis. 2005;26:2138–2148. doi: 10.1093/carcin/bgi185. [DOI] [PubMed] [Google Scholar]
- 43.Hu R, Saw CL, Yu R, Kong AN. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxid Redox Signal. 2010;13:1679–1698. doi: 10.1089/ars.2010.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradlow HL, Zeligs MA. Diindolylmethane (DIM) spontaneously forms from indole-3-carbinol (I3C) during cell culture experiments. In Vivo. 2010;24:387–391. [PubMed] [Google Scholar]
- 45.Staub RE, Feng C, Onisko B, Bailey GS, Firestone GL, Bjeldanes LF. Fate of indole-3-carbinol in cultured human breast tumor cells. Chem Res Toxicol. 2002;15:101–109. doi: 10.1021/tx010056m. [DOI] [PubMed] [Google Scholar]
- 46.Egner PA, Kensler TW, Chen JG, Gange SJ, Groopman JD, Friesen MD. Quantification of sulforaphane mercapturic acid pathway conjugates in human urine by high-performance liquid chromatography and isotope-dilution tandem mass spectrometry. Chem Res Toxicol. 2008;21:1991–1996. doi: 10.1021/tx800210k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minich DM, Bland JS. A review of the clinical efficacy and safety of cruciferous vegetable phytochemicals. Nutr Rev. 2007;65:259–267. doi: 10.1301/nr.2007.jun.259-267. [DOI] [PubMed] [Google Scholar]


