Abstract
Systems biology provides new approaches for metabolic engineering through the development of models and methods for simulation and optimization of microbial metabolism. Here we explore the relationship between two modeling frameworks in common use namely, dynamic models with kinetic rate laws and constraint-based flux models. We compare and analyze dynamic and constraint-based formulations of the same model of the central carbon metabolism of E. coli. Our results show that, if unconstrained, the space of steady states described by both formulations is the same. However, the imposition of parameter-range constraints can be mapped into kinetically feasible regions of the solution space for the dynamic formulation that is not readily transferable to the constraint-based formulation. Therefore, with partial kinetic parameter knowledge, dynamic models can be used to generate constraints that reduce the solution space below that identified by constraint-based models, eliminating infeasible solutions and increasing the accuracy of simulation and optimization methods.
Keywords: Systems biology, Metabolic networks, Dynamic models, Constraint-based models
1. Introduction
The prevalence of systems approaches to biological problems has renewed interest in mathematical models as fundamental research tools for performing in silico experiments of biological systems (Kitano, 2002). In the context of metabolic engineering, models of metabolism play an important role in the simulation of cellular behavior under different genetic and environmental conditions (Stephanopoulos, 1998). Typical experiments include knockout simulations to study how metabolic flux distributions readjust throughout a given network. With the selection of an optimal set of knockouts or changes in enzyme expression levels, it is desirable to optimize the production of compounds of industrial interest (Burgard et al., 2003; Patil et al., 2005).
Systems of ordinary differential equations (ODEs) have been applied in different areas to model dynamical systems. In the context of metabolic networks, they describe the rate of change of metabolite concentrations. These dynamic models contain rate law equations for the reactions as well as their kinetic parameters and initial metabolite concentrations. Building this type of model requires insight into enzyme mechanism to select appropriate rate laws, as well as experimental data for parameter estimation. Therefore, their application has been more limited, but areas of application include central metabolic pathways of well-studied organisms such as E. coli (Chassagnole et al., 2002) and S. cerevisiae (Rizzi et al., 1997). There are, however, some recent efforts to overcome these limitations in the reconstruction of large-scale dynamic models, such as through the hybrid dynamic/static approach (Yugi et al., 2005), the ensemble modeling approach (Tran et al., 2008), and the application of approximative kinetic formats using stoichiometric models as a scaffold (Smallbone et al., 2010; Jamshidi and Palsson, 2010). Nevertheless, these techniques have so far been applied to very few organisms.
On the other hand, advances in genome sequencing have facilitated the reconstruction of genome-scale metabolic networks for several organisms, with over 50 reconstructions available to date (Oberhardt et al., 2011). Due to the lack of kinetic data at the genome scale, this type of model only accounts for reaction stoichiometry and reversibility. Analysis is performed under the assumption of steady state using a constraint-based formulation that is underdetermined, resulting in a continuous space of solutions for the reaction flux distributions. This uncertainty of the flux distributions requires additional conditions to determine unique solutions and predictions. Often this takes the form of an optimization based on a particular assumption, such as optimal biomass growth for wild-type (Edwards and Palsson, 2000) and minimization of cellular adjustments for knock-out strains (Segré et al., 2002; Shlomi et al., 2005). The inclusion of regulatory constraints, introduced by Covert and Palsson (2003), is a current approach to reduce the size of the solution space and eliminate infeasible solutions. One limitation of the constraint-based approach is the unability to express transient behavior. In order to simulate fermentation profiles, a few methods have been developed to integrate the variation of external concentrations while assuming an internal pseudo steady state (Oddone et al., 2009; Leighty and Antoniewicz, 2011).
The two most common model types in use, therefore, represent two extremes. The dynamic ODE formulation contains detailed mechanistic information that gives solutions of the transient dynamic approach to equilibrium from any given set of initial conditions (generally concentrations of enzymes and metabolites), as well as the steady state specified by metabolite concentrations that depend on total enzyme concentrations (for the usual case where they are treated as fixed) but often do not depend on the initial metabolite concentrations. Steady-state fluxes are readily computed from the steady-state concentrations and the rate laws. The constraint-based formulation seems minimalist by comparison: it has no mechanistic knowledge of any of the chemical reactions beyond their stoichiometry, its solutions have fluxes at steady state but no information regarding concentrations or dynamics, and rather than giving a unique solution, it produces a high-dimensional continuum of steady-state solutions (referred to as a flux cone). The dynamic formulation needs significant information (parameters in term of rate constants and total enzyme concentrations, as well as reaction mechanisms to give rate laws), but generally rewards that effort with unique and detailed solutions. The constraint-based formulation requires less (no parameters except maximum fluxes) but delivers less.
Because of these significant differences between dynamic and constraint-based formulations, they treat the effects of network perturbations, which might be undertaken as part of a metabolic engineering study, very differently. A dynamic formulation will make very specific predictions about the response to a gene knockout, for example, but generally such models lack information about gene regulatory changes that accompany metabolic changes, and so without foreknowledge to adjust relative enzyme concentrations, such predictions can be significantly in error. Constraint-based formulations can access all possible steady-state solutions but can only rely on relatively simple heuristics to select among them, and are uncertain how to include specific information on gene regulatory changes.
Here we explore further the relationship between these formulations by essentially considering the continuous ensemble of dynamic formulations obtained by varying parameters (principally rate constants and enzyme concentrations) and compare the steady-state solutions to those from the corresponding constraint-based formulation. We find an equivalence between the sets of steady states when only maximum flux constraints are present, but that more specific constraints and enzyme concentrations can be directly incorporated to define a reduced dynamic ensemble that is significantly more informative regarding possible steady-state solutions than the constraint-based formulation.
2. Methods
2.1. Models
We have used a dynamic model of the central carbon metabolism of E. coli (Chassagnole et al., 2002) available at the Biomodels database (Le Novere et al., 2006). The model was converted from its original SBML format into a MATLAB (The Mathworks; Natick, MA, USA) file that was used for all computations in this work. The model consists of a total of 18 metabolites and 31 reactions, including several enzymatic reactions, one exchange reaction, and a few lumped versions of biosynthetic pathways. Several types of rate laws are used, including constant-rate, mass-action, Hill cooperativity, allosteric regulation, and Michaelis-Menten with its variants for reversibility and inhibition, with a total of 125 parameters. We have not considered metabolite dilution or algebraic rules for co-metabolite variation, as they cannot be represented in the constraint-based model. Also, we changed the rate law of MurSynth from constant rate to Michaelis-Menten, as it leads to inconsistencies when its substrate (f6p) depletes. The model maintained its original steady state despite these changes.
A constraint-based version of the model was built by accounting only for the stoichiometry and reversibility constraints. The glucose uptake rate was allowed to vary between 0 and the maximum value in the dynamic model. The dynamic model also contains two other inputs (TrpSynth, MethSynth), with a constant rate, that were treated in the constraint-based version with constant fluxes.
2.2. Hit-and-Run sampler
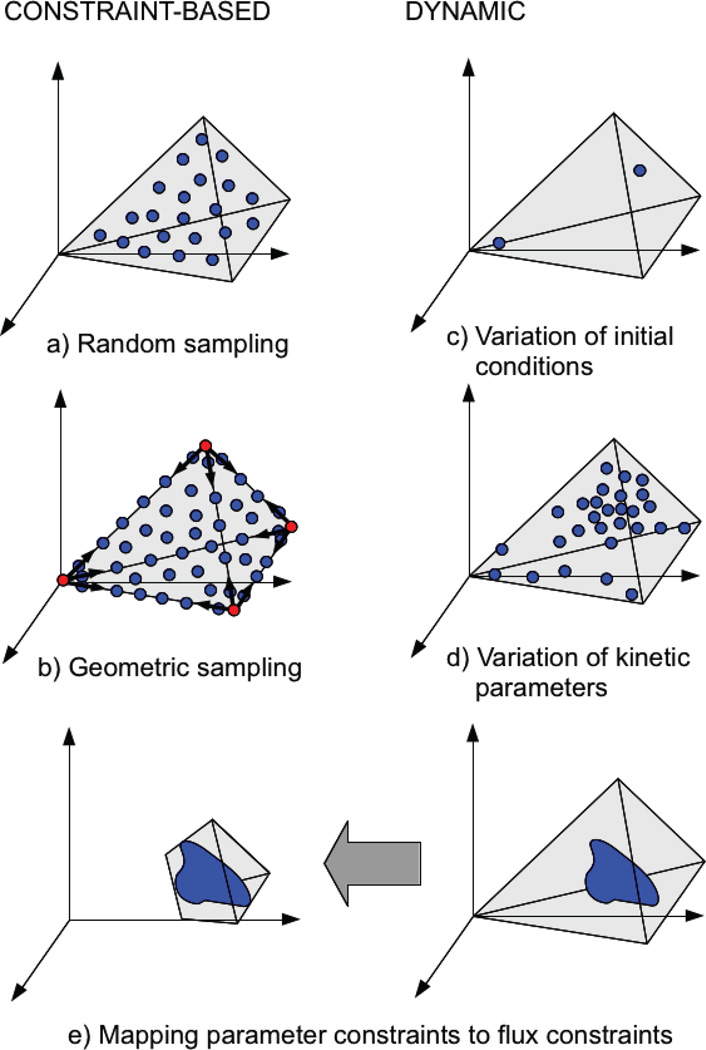
As a means of mapping out the feasible steady-state flux space for the constraint-based model, we implemented an algorithm for random sampling (Figure 1a) adapted to this problem following the concept of hit-and-run methods (Smith, 1984). The solution space of the constraint-based model is contained within the null space of the stoichiometric matrix. Starting with a point inside this coordinate space, the sampler started generating new points by iterative steps in one direction. Each point was then projected into the flux space and tested by checking the flux boundary constraints. Each time the test failed, meaning that it crossed the boundary of the flux cone, the point was discarded and a new direction was randomly chosen. Otherwise, the point’s projection in the flux space was stored. To facilitate uniform sampling of the whole space, the sampler only stored one point every 1000 iterations. Also, in order to adapt to cones of different sizes, it used a variable step size that increased (decreased) in case of successful (failed) iterations, which quickly converged to an average size.
Figure 1.
Overview of the methods applied in this work to the constraint-based and the dynamic model. The solution space of the constraint-based model has been sampled by (a) random sampling using a Hit-and-Run algorithm and (b) geometric sampling using the corners of the flux cone as starting points. The solution space of the dynamic model has been sampled by (c) varying the initial metabolite concentrations and (d) the kinetic parameters. (e) By constraining the kinetic parameters of the dynamic model we can delimit kinetically feasible flux regions and transfer them to the constraint-based model.
2.3. Geometric sampler
To provide improved mapping of the steady-state flux solution space for the constraint-based model, given the poor results obtained by the hit-and-run method at the edges of the flux cone, we designed and implemented a geometric sampler (Figure 1b) that started by searching the corners of the flux cone. It found the corners by solving linear programing problems within the model with randomized objective functions using the GLPK library (Makhorin, 2006). After finding the corners, it sampled along all possible edges between the corners, which defined the boundary of the cone. Then, it iteratively sampled from all edges in the direction of the center of the cone, defined as the mean of all corners. This method facilitated the visualization of the flux cone. However, in this case, the probability distribution of the points did not have any statistical meaning.
2.4. Parameter sampler
We developed a sampler to sample from the parameter space (concentrations and kinetic parameters) of the dynamic model to map out its allowed steady states. Metabolite concentrations and kinetic parameters are theoretically defined in an infinite semi-positive space. Therefore, in order to sample this type of space without constraints, we scaled each element individually (concentration or parameter) by a random factor with log-normal distribution (log10(X) ~ 𝒩(0, 1)). This distribution is defined over ℝ+, with nearly all values (99.73%) within 3 orders of magnitude above or below unity. This resulted in variation of the original values by several orders of magnitude. In order to perform constrained parameter variation within well-defined ranges, specified in terms of orders of magnitude (m), we scaled each parameter by a factor with uniform distribution in logarithmic scale (log10(X) − 𝒰(−m/2, m/2)). All kinetic parameters associated with binding and rate constants were varied, while other parameters such as Hill coefficients, co-metabolite levels, and dilution rate, were kept fixed.
2.5. Calculating steady states
For each simulation of the dynamic model, the steady state was calculated by numerically integrating the differential equations from time zero toward infinity with a stop condition when the steady state was reached. To avoid non-halting computations when the system diverged or was oscillatory, a second stop condition, based on a computational time limit, was also added.
2.6. Relative volume estimation
In order to estimate the volume of the cone after imposition of the kinetic parameter constraints, we started by sampling the dynamic model under those constraints. In this way the kinetic parameter ranges could be mapped to flux ranges (Figure 1e). Then, we used a random sample of the constraint-based model (obtained with the hit-and-run sampler) and calculated the fraction of points of that sample that were contained within the generated flux ranges. This fraction determined the relative volume of the subspace compared to the original space (Wiback et al., 2004).
2.7. Mapping steady-state fluxes to parameter space
In order to find the parameters that match any given steady-state flux distribution (v) we solved the system of non-linear equations v = r(x, p), where r(x, p) is a vector function that represents the reaction rate laws as a function of the metabolite concentrations (x) and the kinetic parameters (p). Given a vector of arbitrary steady-state concentrations the system can be solved for the kinetic parameters only. Furthermore, because each individual rate law has its own parameters, we can define a partition of the set of parameters , that effectively decouples the system of equations into n independent equations of the form vi = ri(x, si), si ∈ Si. These equations are underdetermined because there are usually several parameters per equation (with an average of 4). Therefore, each equation has multiple solutions. In order to obtain a single solution, the strategy implemented in this work consisted of mapping the variables into logarithmic space (which enforces positively defined solutions) and minimizing the length of the solution vector. Additional constraints could be placed on the parameters at this stage.
3. Results
In order to explore the gap between both types of formulations, we analyzed and compared the dynamic and constraint-based formulations of the same model of the central carbon metabolism of E. coli (Chassagnole et al., 2002) (see Methods section for model formulation).
Our goal is to compare the steady states achievable by the two model types. Intuitively the dynamic formulation has more constraints than the constraint-based one because the later only enforces the steady-state condition and maximum flux constraints. Therefore, any set of steady-state fluxes achieved by the dynamic formulation that do not violate the maximum flux constraints will automatically be a solution of the constraint-based formulation. Thus, here we focus on mapping solutions in the opposite direction: Is every solution of the constraint-based formulation also a steady-state solution of the dynamic one? Or, instead, does the extra information in the dynamic formulation effectively reduce the steady-state solution space so that it is a proper subset of the constraint-based formulation.
3.1. Solution space of the constraint-based model
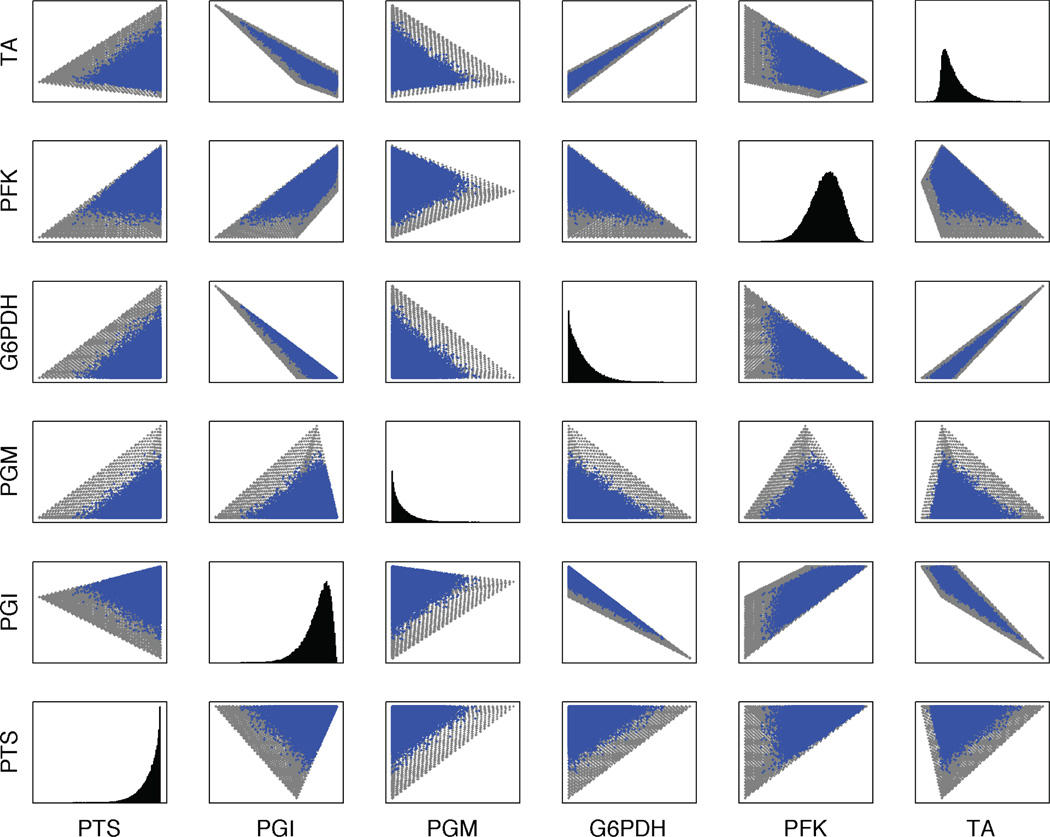
We implemented a Monte-Carlo based random sampler, which is a variation of the hit-and-run method (Smith, 1984) (see Methods) and applied it to the constraint-based model. The sampling distribution for each reaction (Figure 2, diagonal) forms skewed gaussian shaped curves, very similar to the results obtained by Wiback et al. (2004) for the human red blood cell model. However, more insight into the shape of the solution space can be revealed by plotting the sample two-dimensionally for every pair of reactions (Figure 2). We observe that, due to the random nature of this method, the edges of the flux cone are not sharply defined due to the low probability of samples in the tails of the distributions. To obtain a clearer delineation of the borders of the space, we implemented a geometric sampling approach that systematically identified first the vertices of the flux cone through solution of linear programs, then the edges through vertex connection, and finally explored the interior of the flux cone (see Methods). The full solution space of the constraint-based model became clear (Figure 2), and it could be compared to that from the dynamic model.
Figure 2.
Pairwise projection of the sampling of the constraint-based solution space using the hit-and-run sampler (blue) and the geometric sampler (gray). The diagonal shows the probability distribution for each reaction relative to the hit-and-run sampling. Only the first six reactions are shown. The complete data are in Supplementary material. Note that the gray points are plotted underneath the blue ones, and that the geometric sampler delineates all of the space covered by the hit-and-run sampler, plus the additional spaces seen here.
3.2. Solution space of the dynamic model
Whereas the constraint-based model has no adjustable parameters (beyond maximum flux values not implemented here), the dynamic model has a large number of parameters that describe the specific chemistry being modeled, consisting of the rate laws, kinetic parameters (in which we include a fixed total concentration for each protein), and initial metabolite concentrations. This results in a single deterministic steady-state solution. To examine how this solution is influenced by the extra information, we varied the initial conditions and kinetic parameters, again by random sampling (see Methods).
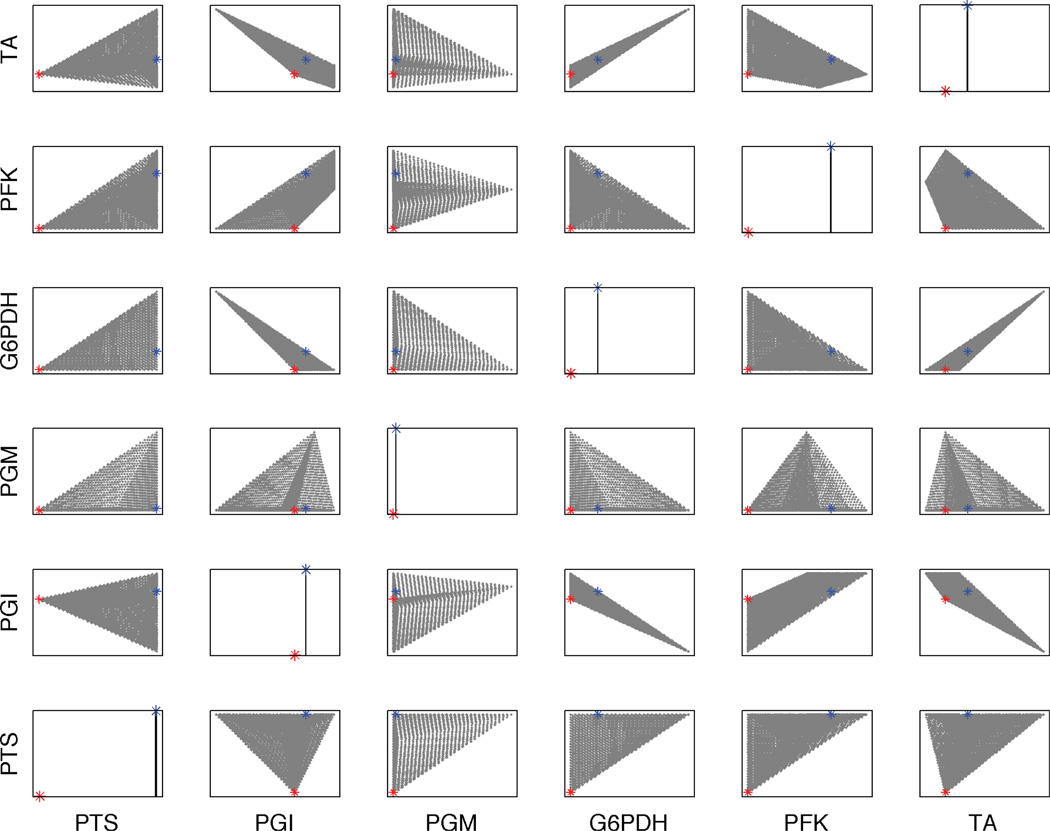
If the system has a unique steady state, then simulations will converge to the same steady state, independent of the initial concentrations. This network exhibits multistability; two distinct steady states were identified when the initial concentrations of metabolites were varied (Figure 3). This bi-stability is caused by a positive feedback loop that is formed when phosphoenolpyruvate (PEP), a product of glycolysis, is used as an energy source to import external glucose through the phosphotransferase system (PTS). During the transient phase of the system, the concentration of PEP may reach a critical level, whereby it becomes depleted before re-entering PTS. If this happens the cell is unable to capture its external substrate, and all internal metabolites eventually deplete as well, leading to a network with residual activity. This steady state (referred to here as secondary) occurs much less frequently than the steady state obtained with the original conditions (Figure 3, diagonal).
Figure 3.
Pairwise projection of the sampling of the solution space obtained for the dynamic model by sampling the initial metabolite concentrations, overlapping the complete solution space (gray) for better visualization. The blue dot shows the location of the original steady state. The red dot shows the location of the secondary steady state. Only the first six reactions are shown. The complete data are in Supplementary material. The diagonal gives the relative probabilities of the steady-state flux distribution.
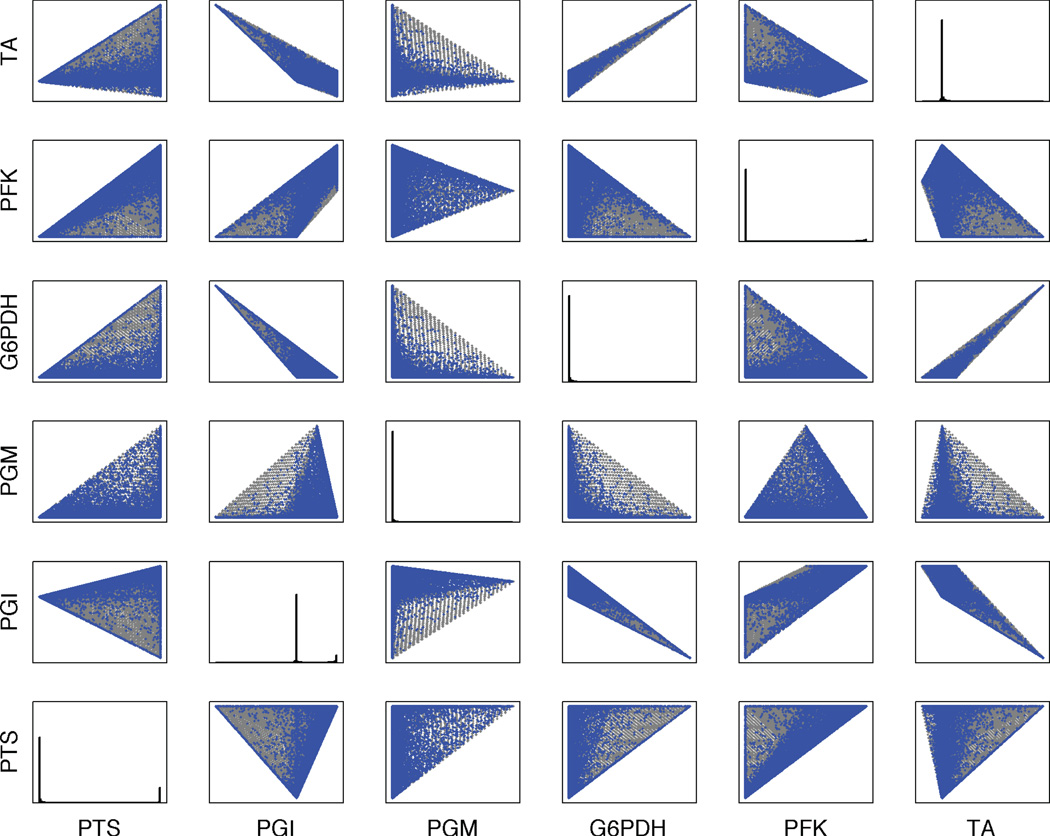
A random procedure was used to vary the kinetic model parameters, including binding and rate constants (because all enzyme concentrations are included in Vmax, which was varied, effectively enzyme concentrations were varied as well), but not Hill coefficients, co-metabolite concentrations, or the dilution rate (see Methods). A single set of initial concentrations was used (that for which the unperturbed model goes to the higher probability steady state). A projection of the resulting steady-state concentrations shows that the dynamic model, through parameter variation, appears to be able to produce the same steady states as the constraint-based model, but no additional steady states. This situation is tempered by two issues: (i) there are areas of light coverage in Figure 4 that one presumes are truly occupied, and (ii) even if the two-dimensional projection overlaps, this does not confirm that the full-dimensional flux cones for the two models overlap. To more stringently test the notion that the polytopes are identical, we generated a procedure to optimize parameters for the dynamic model to reproduce any desired steady-state solution (see Methods). We applied this to 10,000 randomly selected solutions from the constraint-based model and the resulting parameters recovered the desired steady state when run in the dynamic model every time. Thus, operationally the steady-state flux cones for ODE and constraint-based models are the same.
Figure 4.
Pairwise projection of the sampling of the steady-state solution space for the dynamic model obtained by sampling the kinetic parameters (blue). The corresponding space overlaps the solution space given by the stoichiometric model (gray). The diagonal shows the probability distribution for each reaction. Only the first six reactions are shown. The complete data are in Supplementary material.
3.3. Kinetically feasible solution space
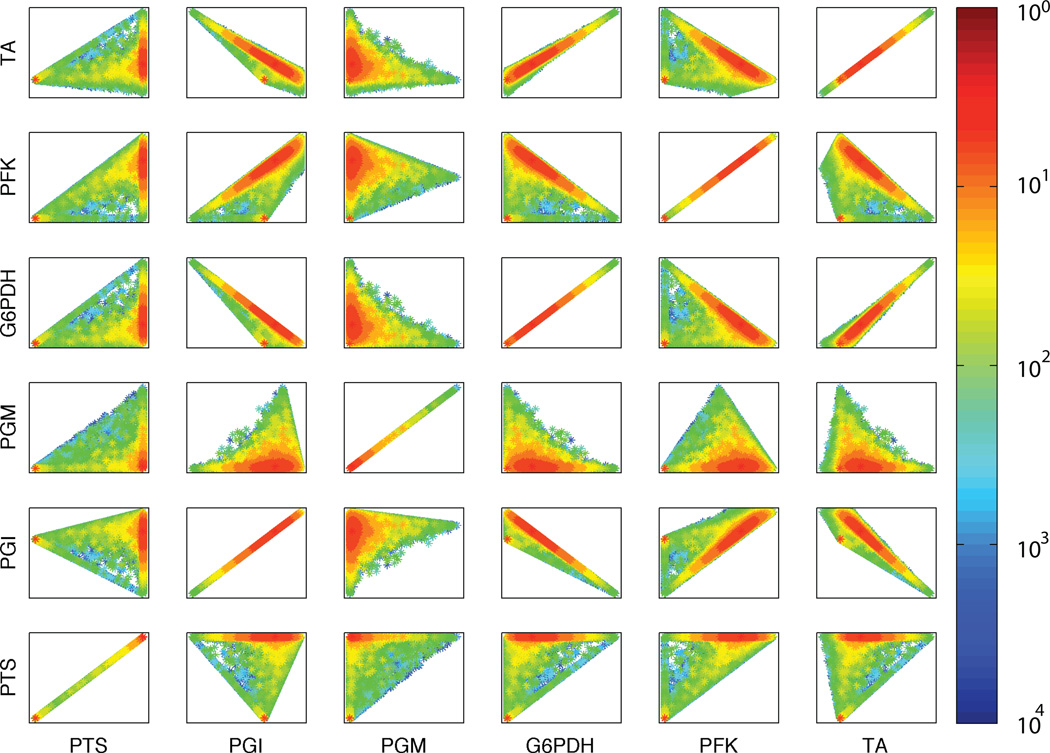
An ODE kinetic model of central carbon metabolism has exactly the same set of possible steady-state solutions as the corresponding flux balance model, as demonstrated in the previous section. The ODE model maps out the solution space through systematic variation of model parameters (binding constants, rate constants, and enzyme concentrations) with no constraints beyond non-negativity. Knowledge of actual parameter values or ranges, from experimental measurement or physical constraints, would lead to further constraints on the feasible parameter space. To explore how constraints on the feasible parameter space affect the range of steady-state solutions achievable in the ODE kinetic model, we sampled parameter combinations from constrained spaces and computed the steady states of the resulting models. The fluxes in those steady states are plotted in Figure 5 for parameter ranges from 100 up to 104-fold around the base parameter values. The results show that parameter variation of 100-fold or greater appears to produce the full set of steady-state flux solutions observed from the unconstrained non-negative parameters in the ODE model, which corresponds to the flux-balance steady states. Parameter constraints leading to less than 100-fold variation produced significant restriction of the steady-state fluxes.
Figure 5.
Pairwise projection, in heat-map form, of the solution space reachable by the dynamic model as a function of the variation, in terms of orders of magnitude, of the kinetic parameter space. The diagonal shows the variation for each flux independently. Only the first six reactions are shown. The complete data are in Supplementary material.
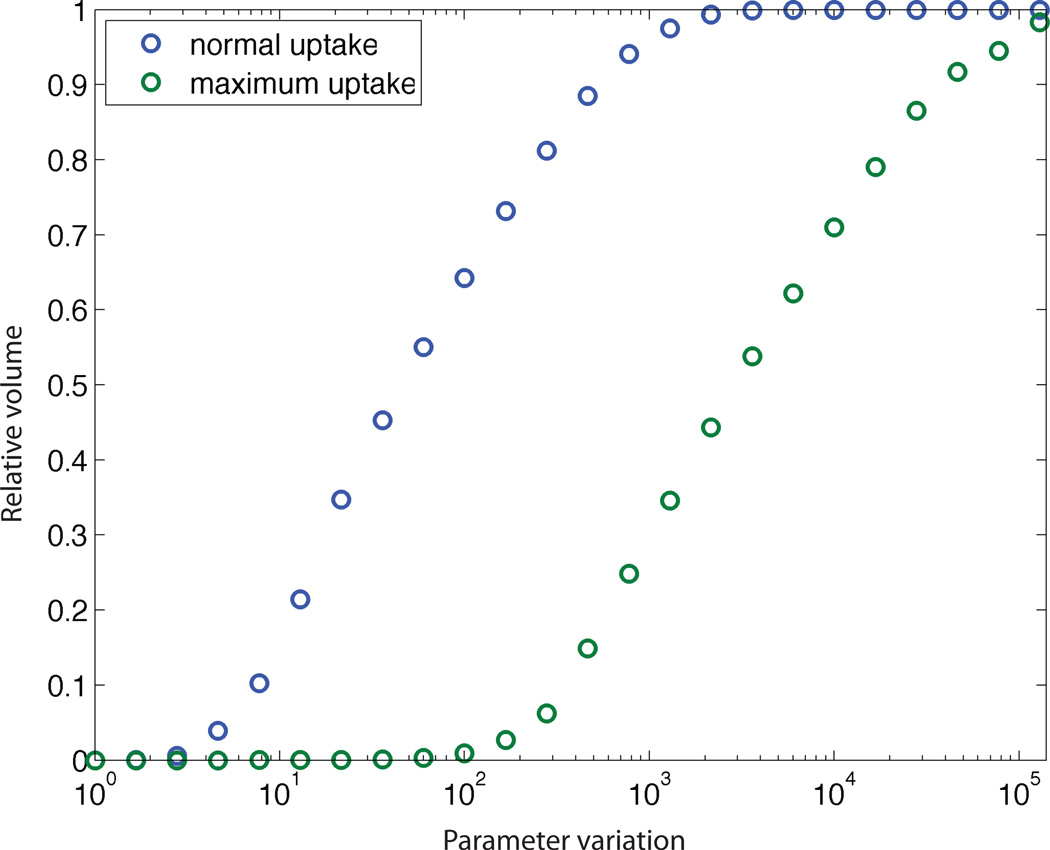
The solution-space volume reduction due to parameter constraints is plotted quantitatively in Figure 6. The ratio of the solution flux cone with constrained and unconstrained parameters is shown as a function of the constrained parameter ranges. The results (labeled “normal uptake”), show that reduction of parameter uncertainty to a 10-fold range leads to a reduction in the solution flux space to 10% of its unconstrained volume. Moreover, because the size of the original space depends on a control variable of the system, namely the glucose uptake rate, we increased glucose uptake from 1.28 mmol gDW−1 h−1, the value in the original model, to 10.50 mmol gDW−1 h−1, the maximum value for E. coli under aerobic conditions (Varma and Palsson, 1994). The results, shown in Figure 6 as “maximum uptake”, show a similar sigmoid shape but shifted toward greater parameter variation. Under these conditions the flux cone of solutions is reduced to 10% of its unconstrained volume with 300-fold parameter variation.
Figure 6.
Relative volume of the kinetically feasible solution space, compared to the original space, as a function of the parameter variation, in terms of orders of magnitude. The volume was calculated for the original glucose uptake rate in the model and also for the maximum uptake rate.
4. Discussion
We have analyzed and compared dynamic and constraint-based formulations of the same model for the central carbon metabolism of E. coli (Chassagnole et al., 2002). The constraint-based version does not account for metabolite concentrations, and it does not express transient behavior. Therefore, the formulations can only be compared in their common domain, which is the steady-state flux distribution.
The constraint-based model defines a solution space for the steady-state flux distribution (called the flux cone). This space is difficult to visualize due to its high dimensionality. We addressed this problem by developing sampling and projection approaches that facilitate the visualization of the shape of the solution space.
The steady state of the dynamic model contains the same constraints as the constraint-based model (stoichiometry, thermodynamic reversibility, and maximum uptake rates) and also any additional constraints imposed by the kinetic rate laws, kinetic parameters, and initial metabolite concentrations. Therefore, its solution space is a subset of the constraint-based solution space.
For a predefined set of initial conditions and parameter values, the dynamic model usually determines one steady-state solution. In fact, the initial metabolite concentrations of dynamic models determine their transient behavior, but, for the steady-state flux determination, they serve only to determine which steady state is chosen in the case of multistability. In this case, sampling the metabolite concentration space revealed a second steady-state characterized by a flux distribution with lower values of the fluxes and an accumulation of external glucose.
Instead, we also verified, as expected, that the location of the steady-state solution(s) inside the solution space is determined by the kinetic parameters, because by varying the kinetic parameters, the solution moves inside the solution space. The sampling of the kinetic parameter space revealed that, with unconstrained parameter values, the solutions of the dynamic model cover the whole steady-state solution space identified by the constraint-based model. This overlapping may seem unintuitive, as one would expect the rate laws to impose one additional layer of constraint into the steady-state solution space. However, besides having observed this with our sampling approaches, we also observe that, given any valid steady-state flux distribution, one can find kinetic parameter values that make the rate laws produce those steady-state flux values by solving each equation separately. This separation is only possible because the parameters are specific for each rate law, which defines a partition over the parameter set. The running example contains an average of 4 parameters per rate law, yielding many degrees of freedom for each equation. Thus, it is not surprising that, generally, parameter values can be found that satisfy the equations.
Interestingly, we found that by varying only one class of rate constants (Vmax = kcat[E]0), the dynamical model formulation was able to achieve all of the same steady states as the constraint-based model (data not shown). This is an important observation, because it suggests that by changing only the expression levels of proteins ([E]0’s), which can be achieved through regulation, a cell can adapt to reach essentially any possible steady state, without the need to introduce mutations that change rate constants. This observation reflects the adaptability of cell under different conditions and is in agreement with observations that microorganisms can undergo adaptive evolution to attain their optimal theoretical yields when placed under conditions where they originally performed sub-optimally (Ibarra et al., 2002).
The observations stated above show that, in theory, a dynamic model can be fitted to any steady-state flux distribution inside the constraint-based solution space. However, there are physical limitations to the values of the kinetic parameters. Also, by querying parameter databases such as BRENDA (Schomburg et al., 2002) and SABIO-RK (Rojas et al., 2007), it is possible to observe that for each kinetic parameter there is a range of possible values determined by experimental conditions (such as temperature and pH) in which the cells are able to grow. Therefore, we evaluated how the imposition of parameter ranges map into flux ranges within the steady-state solution space. Although the rate laws do not constrain the solution space by themselves, they influence the probability distribution of the steady-state solutions. This is evidenced by the imposition of the kinetic parameter constraints. As the constraints become tighter, the solutions of lower probability disappear and the reachable solution space becomes smaller. Our results show that the impact of these constraints depends on the size of the solution space of the genome-scale model, which is mainly determined by the uptake rate of the limiting substrates, and on the allowable ranges of the kinetic parameters in the dynamic model.
The subset of the solution spaced obtained by constrained variation of kinetic parameters reveals that it is possible to map parameter ranges into flux ranges. This can be performed by sampling the parameter space and determining the respective steady-states. The generated flux ranges can be directly added into the FBA formulation as flux bounds. A similar sampling procedure, although with a different goal, is performed in the ensemble modeling approach (Tan et al., 2010).
5. Conclusions
In this work we have explored the solution spaces of both dynamic and constraint-based models in order to bring together top-down and bottom-up approaches, and we have proposed methods of treating each as well as their interrelation.
Dynamic model reconstruction is a bottom-up approach for iteratively building large-scale metabolic pathways with kinetic detail. Due to a lack of experimental data, differences in experimental conditions, and measuremental uncertainty, the kinetic parameters are often unavailable or defined within certain ranges.
On the other hand, genome-scale reconstruction is a top-down approach that takes advantage of available high-throughput data to build models of metabolic networks that account for stoichiometry and thermodynamic constraints. These models are analyzed under a steady-state assumption through the constraint-based approach. Reducing the solution space of constraint-based models so as to eliminate infeasible solutions is an important topic. Several approaches are in use, including the imposition of regulatory constraints (Covert and Palsson, 2003), the experimental determination of some fluxes (Wiechert, 2001), and the imposition of thermodynamic reversibility constraints (Beard et al., 2002; Hoppe et al., 2007). The results obtained in this work are complementary to those efforts and can be used in combination with any of them. For the case of thermodynamic constraints, their application can be two fold. When applied to the dynamic model, the estimation of the Gibbs free energy can be used to determine the value of the equilibrium constant, which further constrains the kinetic parameters. When applied to the constraint-based model, it can be used to constrain the direction of reversible reactions and, consequently, the solution space.
Taking advantage of the information available in dynamic models of central pathways can increase the accuracy of genome-scale constraint-based models by imposition of kinetic feasibility constraints, even if the dynamic model is not fully determined. Furthermore, sampling the solution space of the dynamic model can be used as an experimental design tool to determine which kinetic parameters have greater influence in defining the volume of the solution space.
Increasing the accuracy of constraint-based models can influence simulation methods such as metabolic flux analysis (MFA) (Wiechert, 2001), flux balance analysis (FBA) (Edwards and Palsson, 2000), minimization of metabolic adjustment (MOMA) (Segré et al., 2002) and regulatory on/off minimization (ROOM) (Shlomi et al., 2005). Tools that implement these methods (Rocha et al., 2010) can be extended to include kinetic constraints.
The constraint-based approach has been recently applied to other kinds of biological networks, namely gene regulatory and signaling networks (Gianchandani et al., 2006; Lee et al., 2008). The availability of models for all kinds of networks will facilitate the creation of integrated cellular models that account for all types of intracellular phenomena under the same mathematical framework. Because those models can be either constraint-based or dynamic, understanding relationships between the two as discussed in this paper will have an even greater impact. In fact, although the use of common frameworks (either constraint-based or dynamic) for representing different kinds of biological phenomena is a step towards the use of integrated models, the development of tools that promote the integration of the two most important representation frameworks is also necessary for true integration. The current contribution is a step in that direction.
Constriant-based and full kinetic models share the same set of steady states.
Partial rate-constant knowledge can further constrain kinetic models.
Such reductions can be transferred from kinetic to constraint-based formulations.
Supplementary Material
Acknowledgments
This research was supported by PhD grants SFRH/BD/35215/2007 and SFRH/BD/25506/2005 from the Fundação para a Ciência e a Tecnologia (FCT) and the MIT–Portugal Program through the project “Bridging Systems and Synthetic Biology for the Development of Improved Microbial Cell Factories” (MIT-Pt/BS-BB/0082/2008).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beard D, Liang S, Qian H. Energy balance for analysis of complex metabolic networks. Biophysical Journal. 2002;83(1):79–86. doi: 10.1016/S0006-3495(02)75150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgard A, Pharkya P, Maranas C. OptKnock: A bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnology and Bioengineering. 2003;84(6):647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- Chassagnole C, Noisommit-Rizzi N, Schmid J, Mauch K, Reuss M. Dynamic modeling of the central carbon metabolism of Escherichia coli. Biotechnology and Bioengineering. 2002;79(1):53–73. doi: 10.1002/bit.10288. [DOI] [PubMed] [Google Scholar]
- Covert M, Palsson B. Constraints-based models: Regulation of gene expression reduces the steady-state solution space. Journal of Theoretical Biology. 2003;221(3):309–325. doi: 10.1006/jtbi.2003.3071. [DOI] [PubMed] [Google Scholar]
- Edwards J, Palsson B. Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions. BMC Bioinformatics. 2000;1(1):1. doi: 10.1186/1471-2105-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianchandani E, Papin J, Price N, Joyce A, Palsson B. Matrix formalism to describe functional states of transcriptional regulatory systems. PLoS Computational Biology. 2006;2(8):e101. doi: 10.1371/journal.pcbi.0020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe A, Hoffmann S, Holzhütter H. Including metabolite concentrations into flux balance analysis: thermodynamic realizability as a constraint on flux distributions in metabolic networks. BMC Systems Biology. 2007;1:23. doi: 10.1186/1752-0509-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra R, Edwards J, Palsson B. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420(6912):186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- Jamshidi N, Palsson B. Mass action stoichiometric simulation models: Incorporating kinetics and regulation into stoichiometric models. Biophysical Journal. 2010;98:175–185. doi: 10.1016/j.bpj.2009.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Systems biology: A brief overview. Science. 2002;295(5560):1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Le Novere N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B, Snoep J, Hucka M. BioModels Database: A free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Research. 2006;34(suppl 1):D689. doi: 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Gianchandani E, Eddy J, Papin J. Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Computational Biology. 2008;4(5) doi: 10.1371/journal.pcbi.1000086. e1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighty R, Antoniewicz M. Dynamic metabolic flux analysis (DMFA): A framework for determining fluxes at metabolic non-steady state. Metabolic Engineering. 2011;13(6):745–755. doi: 10.1016/j.ymben.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Makhorin A. GLPK - The GNU Linear Programming Toolkit. 2006 URL http://www.gnu.org/software/glpk/ [Google Scholar]
- Oberhardt M, Puchałka J, dos Santos V, Papin J. Reconciliation of genome-scale metabolic reconstructions for comparative systems analysis. PLoS Computational Biology. 2011;7(3) doi: 10.1371/journal.pcbi.1001116. e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddone G, Mills D, Block D. A dynamic, genome-scale flux model of Lactococcus lactis to increase specific recombinant protein expression. Metabolic Engineering. 2009;11(6):367–381. doi: 10.1016/j.ymben.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Patil K, Rocha I, Forster J, Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinformatics. 2005;6(1):308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi M, Baltes M, Theobald U, Reuss M. In vivo analysis of metabolic dynamics in Saccharomyces cerevisiae: II. Mathematical model. 1997;55(4):592–608. doi: 10.1002/(SICI)1097-0290(19970820)55:4<592::AID-BIT2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Rocha I, Maia P, Evangelista P, Vilaça P, Soares S, Pinto J, Nielsen J, Patil K, Ferreira E, Rocha M. OptFlux: An open-source software platform for in silico metabolic engineering. BMC Systems Biology. 2010;4(1):45. doi: 10.1186/1752-0509-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas I, Golebiewski M, Kania R, Krebs O, Mir S, Weidemann A, Wittig U. SABIO-RK: A database for biochemical reactions and their kinetics. BMC Systems Biology. 2007;1(Suppl 1):S6. [Google Scholar]
- Schomburg I, Chang A, Schomburg D. BRENDA, enzyme data and metabolic information. Nucleic Acids Research. 2002;30(1):47–49. doi: 10.1093/nar/30.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segré D, Vitkup D, Church G. Analysis of optimality in natural and perturbed metabolic networks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(23):15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Berkman O, Ruppin E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(21):7695–7700. doi: 10.1073/pnas.0406346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallbone K, Simeonidis E, Swainston N, Mendes P. Towards a genome-scale kinetic model of cellular metabolism. BMC Systems Biology. 2010;4(1):6. doi: 10.1186/1752-0509-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. Efficient Monte Carlo procedures for generating points uniformly distributed over bounded regions. Operations Research. 1984;32(6):1296–1308. [Google Scholar]
- Stephanopoulos G. Metabolic engineering. Biotechnology and Bioengineering. 1998;58(2–3):119–120. [PubMed] [Google Scholar]
- Tan Y, Lafontaine Rivera J, Contador C, Asenjo J, Liao J. Reducing the allowable kinetic space by constructing ensemble of dynamic models with the same steady-state flux. Metabolic Engineering. 2010;13(1):60–75. doi: 10.1016/j.ymben.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Tran L, Rizk M, Liao J. Ensemble modeling of metabolic networks. Biophysical Journal. 2008;95(12):5606–5617. doi: 10.1529/biophysj.108.135442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Palsson B. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Applied and Environmental Microbiology. 1994;60(10):3724–3731. doi: 10.1128/aem.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiback S, Famili I, Greenberg H, Palsson B. Monte Carlo sampling can be used to determine the size and shape of the steady-state flux space. Journal of Theoretical Biology. 2004;228(4):437–447. doi: 10.1016/j.jtbi.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Wiechert W. 13C metabolic flux analysis. Metabolic Engineering. 2001;3(3):195–206. doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- Yugi K, Nakayama Y, Kinoshita A, Tomita M. Hybrid dynamic/ static method for large-scale simulation of metabolism. Theoretical Biology and Medical Modelling. 2005;2(1):42. doi: 10.1186/1742-4682-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.