The C-terminal domain (CTD) is a highly conserved domain unique to RNA polymerase II. It serves as a critical platform for the complex medley of regulatory interactions that occur during transcription and RNA processing. This scholarly review discusses the large body of literature on the RNA polymerase II CTD, providing an up-to-date and very useful guide to the CTD's role in gene expression regulation.
Keywords: kinase, phosphatase, chromatin modification, splicing, 3′ end, termination
Abstract
The C-terminal domain (CTD) of the RNA polymerase II largest subunit consists of multiple heptad repeats (consensus Tyr1–Ser2–Pro3–Thr4–Ser5–Pro6–Ser7), varying in number from 26 in yeast to 52 in vertebrates. The CTD functions to help couple transcription and processing of the nascent RNA and also plays roles in transcription elongation and termination. The CTD is subject to extensive post-translational modification, most notably phosphorylation, during the transcription cycle, which modulates its activities in the above processes. Therefore, understanding the nature of CTD modifications, including how they function and how they are regulated, is essential to understanding the mechanisms that control gene expression. While the significance of phosphorylation of Ser2 and Ser5 residues has been studied and appreciated for some time, several additional modifications have more recently been added to the CTD repertoire, and insight into their function has begun to emerge. Here, we review findings regarding modification and function of the CTD, highlighting the important role this unique domain plays in coordinating gene activity.
Eukaryotes have three nuclear DNA-dependent RNA polymerases (RNAPs): RNAP I, RNAP II, and RNAP III. RNAP II, responsible for the synthesis of all mRNA as well as many noncoding RNAs (ncRNAs), consists of 12 polypeptides, of which Rpb1, which possesses the enzyme's catalytic activity, is the largest. Rpb1 also contains a C-terminal domain (CTD) composed of tandem heptad repeats that constitutes a unique feature of RNAP II and distinguishes it from all other polymerases. The CTD is conserved from fungi to humans, although the number of repeats and their deviation from the consensus vary, reflecting to a large degree the complexity of the organism. The CTD plays important roles at all steps of the transcription process, including enhancing or modulating the efficiency of all of the RNA processing reactions required for completion of synthesis of the mature RNA. The phosphorylation state of the CTD is critical in determining its activity.
Transcription by RNAP II is an immensely complicated process at each step from initiation to termination. Initiation requires the assembly of the preinitiation complex (PIC), composed of the general transcription factors (GTFs) TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH; the Mediator complex; and RNAP II with an unphosphorylated CTD (designated RNAP IIA) (for review, see Cramer 2004; Hirose and Ohkuma 2007; Sikorski and Buratowski 2009). But, concomitant with initiation and throughout the transcription cycle, the CTD becomes highly phosphorylated (RNAP IIO), with the Ser2 and Ser5 positions of the heptad being especially important sites of modification. A generalized model of CTD phosphorylation during transcription depicts that at the beginning of genes, the CTD is phosphorylated on Ser5 by the TFIIH-associated kinase CDK7 (cyclin-dependent kinase 7), and as RNAP II elongates, Ser2 is increasingly phosphorylated by CDK9 or pTEFb, while Ser5 phosphorylation is gradually removed by phosphatases. Therefore, a phosphorylation pattern emerges with Ser5 phosphorylation (Ser5-P) peaks around the transcription start site (TSS), and Ser2-P accumulates toward the ends of transcribed genes (for review, see Chapman et al. 2008; Egloff and Murphy 2008). But, as we discuss here, the situation is now known to be considerably more complex.
The CTD orchestrates multiple events during the transcription process. Genes transcribed by RNAP II are bound dynamically by nucleosomes. The CTD, modulated by phosphorylation, provides a means to recruit histone modifiers and chromatin remodeling complexes to influence transcription initiation, elongation, and termination (for review, see Spain and Govind 2011). RNA transcribed by RNAP II is processed to mature RNA through the steps of 5′ capping, intron removal, and 3′ end formation. These processes are frequently coupled with transcription, and all involve the CTD (for review, see Hirose and Manley 2000; Maniatis and Reed 2002; Proudfoot et al. 2002). Through different patterns of phosphorylation and other modifications, the CTD functions to coordinate these events during transcription (for review, see Phatnani and Greenleaf 2006; Buratowski 2009; Perales and Bentley 2009). It is striking that the simplicity of the CTD structure (i.e., YSPTSPS consensus repeats) is coupled with complex and dynamic patterns of modification to organize the numerous and even more complex nuclear events necessary to form mature transcripts. Below we review studies documenting the structure and function of the RNAP II CTD.
CTD architecture and modification
The structure of the RNAP II CTD displays several intriguing and idiosyncratic features. For example, vertebrate CTDs contain 52 tandem consensus Y1S2P3T4S5P6S7 heptapeptide repeats, whereas the budding yeast CTD has only 26, and other eukaryotes have intermediate numbers. In vertebrate CTDs, 21 out of 52 tandem repeats match the consensus perfectly, while the remaining 31 heptads have one or more substitutions, at position 2, 4, 5, and/or 7 (Corden et al. 1985; Chapman et al. 2008; Liu et al. 2010). Strictly conserved consensus heptads occur mostly in the N-terminal half of the CTD, whereas less conserved, divergent, heptads are found largely in the C-terminal half (Fig. 1). The most conserved residues are Tyr1 and Pro6, present in all 52 repeats, while Ser7 is the least conserved, found in only 26 repeats. A 10-residue sequence is present at the very C terminus, which functions to help stabilize the CTD (Chapman et al. 2004). The CTD extends from the core of the enzyme to form a tail-like structure that provides binding sites for various factors involving in RNA processing and transcription-coupled modifications. The structural length of the tail-like CTD is determined by the phosphorylation status, with the unphosphorylated yeast CTD likely forming a more compacted spiral (∼100 Å), and the phosphorylated CTD forming a more extended tail (∼650 Å), about four times the diameter of the surface of RNAP II (for review, see Meinhart et al. 2005).
Figure 1.
Comparison of select CTD sequences. The CTD sequences of fission and budding yeast, zebrafish, and humans are shown and aligned to display the context of the heptad repeats. All-consensus YSPTSPS repeats heptads are in red, and the numbers next to the parentheses indicate the repeat number. The CTD of fission yeast contains 29 heptads, 24 of which are all-consensus, whereas the CTD of budding yeast consists of 26 heptads, 19 of which are perfect consensus. The fish and human CTDs each consist of 52 repeats, with a 10- to 11-residue motif (in bold) at the very C terminus. Highlighting the conservation of the CTD among vertebrates, fish and human CTDs are 97% identical. Residues in the fish CTD that differ from humans are in yellow, and human residues that deviate from the consensus are in blue.
The CTD is subject to extensive modification. While the most widely studied of these is phosphorylation, glycosylation, ubiquitinylation, and methylation have also been reported (Kelly et al. 1993; Li et al. 2007; Sims et al. 2011). All five of the hydroxylated amino acids can be phosphorylated. Coupled with the fact that a number of the heptads are degenerate, the existence of different combinations of phosphorylated residues suggests that the CTD may form a remarkable number of distinct conformations or functional states. This potential complexity can be further increased by the activity of cis/trans prolyl isomerases. For example, Pin1 (Ess1 in yeast) specifically recognizes phosphoserine–proline bonds (Yaffe et al. 1997), and its activity can alter the structure (Verdecia et al. 2000) and function (Xu et al. 2003; Xu and Manley 2007; Xiang et al. 2010) of the CTD.
Genetic analyses in several organisms have provided insight into the number of heptads required for cell viability as well as into the importance of specific residues in the repeat and the spacing between heptads. In general, cells expressing Rpb1 with CTDs consisting of only ∼50% of the natural numbers of heptads are viable. For example, the CTD of budding yeast contains 26 repeats, but only eight heptads are required for cell viability and 13 are required for wild-type-like growth (West and Gorden 1995). Similarly, chicken DT40 cells expressing an Rpb1 with only 26 all-consensus repeats are fully viable (Hsin et al. 2011). Mouse cells require for viability a CTD with >26 repeats (Bartolomei et al. 1988), and mice homozygous for a deletion of 13 repeats of the CTD are viable, but with high neonatal mortality (Litingtung et al. 1999). Requirements for specific residues can vary from species to species. Thus, in Saccharomyces cerevisiae, substitution of all Tyr1 residues to phenylalanine (Phe) or of Ser2 or Ser5 to alanine (Ala) is lethal (West and Gorden 1995). However, surprisingly, in Schizosaccharomyces pombe, only Ser5 is absolutely essential, and cells with Tyr1 mutated to Phe or Ser2 replaced with Ala, although cold-sensitive, are viable (Schwer and Shuman 2011). It will be interesting to learn the requirement of these residues for viability in vertebrates. Both budding and fission yeast with Thr4 or Ser7 substituted with Ala are viable (Stiller et al. 2000; Schwer and Shuman 2011). However, DT40 cells expressing an Rpb1 with all Thr4 residues changed to valine (Val) are inviable (Hsin et al. 2011), as are human cells with Thr4-to-Ala substitutions (Hintermair et al. 2012). With respect to spacing between heptads, experiments in yeast defined a minimal functional unit that requires two consecutive heptads. Insertion of single Ala residues between such diheptad units was without effect, while insertion of five Alas resulted in slow growth, but cells remained viable, indicating a tolerance for considerable flexibility in the CTD (Stiller and Cook 2004; Liu et al. 2008).
CTD kinases
Several kinases have been implicated in phosphorylation of the CTD. For example, the CTD is phosphorylated by several CDKs, most notably CDK7 and CDK9. Ser5 is phosphorylated by CDK7, a component of TFIIH, and Ser2 is phosphorylated by CDK9, or P-TEFb in mammals (for review, see Meinhart et al. 2005; Hirose and Ohkuma 2007; Egloff and Murphy 2008; Buratowski 2009). CDK7 also appears to phosphorylate Ser7 (Akhtar et al. 2009; Glover-Cutter et al. 2009), and CDK9 is required for phosphorylation of Thr4 (Hsin et al. 2011). Surprisingly, a Polo-like kinase (Plk3) has also recently been implicated in Thr4 phosphorylation (Hintermair et al. 2012). Tyrosine residues are phosphorylated by Abl tyrosine kinases Abl1 and Abl2 (Baskaran et al. 1993, 1997).
CDK7 is a component of the GTF TFIIH. Both yeast (Kin28) and human CDK7 were initially discovered as TFIIH-associated kinase activities (Feaver et al. 1991; Lu et al. 1992), and the activity of CDK7/cyclin H was shown to be necessary for reconstituted in vitro transcription (Akoulitchev et al. 1995). Consistent with its presence in TFIIH, CDK7 functions during transcription initiation, phosphorylating Ser5 (Hengartner et al. 1998). In yeast, mutations that reduce Kin28 kinase activity abolish Ser5 phosphorylation at promoters (Komarnitsky et al. 2000; Rodriguez et al. 2000). More recently, evidence was presented that shows CDK7 also phosphorylates Ser7 in vitro, and inhibition of CDK7 activity decreases the levels of Ser7-P on a collection of genes in vivo. RNAP II in early transcription is thus phosphorylated on both Ser5 and Ser7 residues (Akhtar et al. 2009; Glover-Cutter et al. 2009; Kim et al. 2009).
The CTD can also be phosphorylated by a second PIC component, CDK8, a subunit of the Mediator. CDK8/Cyclin C was shown to phosphorylate the CTD on both Ser2 and Ser5 in vitro (Liao et al. 1995; Sun et al. 1998). Furthermore, the ability of TFIIH to stimulate transcription was shown to be repressed by CDK8, indicating that CDK8 functions as a negative transcription regulator, perhaps by its ability to phosphorylate the CTD (Hengartner et al. 1998; Akoulitchev et al. 2000). However, deleting SRB10 (yeast CDK8) does not affect the CTD phosphorylation level in cells (Rodriguez et al. 2000), and how much CDK8 in fact contributes to phosphorylating the CTD in vivo is unclear (for review, see Galbraith et al. 2010). CDK8/Srb10 indeed targets several other transcription factors. For example, phosphorylation of the yeast transcription factor Gcn4 by Srb10 leads to Gcn4 promoter clearance (Rosonina et al. 2012) and degradation (Chi et al. 2001), and a number of studies have provided evidence that CDK8 can play a coactivator role (for review, see Galbraith et al. 2010).
The CTD of elongating RNAP II is phosphorylated by P-TEFb. P-TEFb was initially found to overcome pausing of RNAP II near promoters and to stimulate transcription elongation in vitro (Marshall and Price 1995; Marshall et al. 1996). P-TEFb is composed of CDK9 and cyclin T, and CDK9 kinase activity is inhibited by 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) (Peng et al. 1998a,b), which was known to block transcription elongation in vitro (Marciniak and Sharp 1991). CDK9's ability to trigger the transition from transcription initiation to elongation was further substantiated by studies demonstrating that CDK9 stimulates transcript elongation from the HIV-1 promoter (Herrmann and Rice 1995; Fujinaga et al. 1998). The HIV transcriptional transactivator Tat binds to the transactivation response element in the nascent RNA to enhance elongation by recruiting P-TEFb to phosphorylate the CTD (Zhou et al. 2000).
P-TEFb is also involved in regulating the activities of two elongation factors, DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF). After transcription initiation, the two factors associate with RNAP II and pause the elongating RNAP II downstream from the TSS (Yamaguchi et al. 1999; Wu et al. 2003). To overcome this pausing, P-TEFb phosphorylates the two factors as well as the CTD, and elongation proceeds after dissociation of phosphorylated NELF from the RNAP II complex (Renner et al. 2001; Fujinaga et al. 2004; Cheng and Price 2007). Significantly, about one-third of genes in both fly and human cells appear to contain a paused RNAP II downstream from the TSS (Core et al. 2008; Nechaev et al. 2010). Pausing is thought to constitute a mechanism to obtain rapid and coordinated transcription during development and in response to external stimuli (Muse et al. 2007; Zeitlinger et al. 2007). The detailed molecular mechanisms responsible for pausing remain under investigation (for review, see Chiba et al. 2010; Levine 2011).
Budding yeast has two CDKs, Bur1 and Ctk1, that function similarly to CDK9 of higher eukaryotes. CTDK-1 (composed of Ctk1, the cyclin Ctk2, and a regulatory subunit, Ctk3), like pTEFb, is capable of stimulating transcription elongation by phosphorylating the CTD as shown by in vitro transcription assays (Sterner et al. 1995; Lee and Greenleaf 1997), and CTK1 deletion abolishes Ser2 phosphorylation in vivo (Cho et al. 2001). Bur1, together with the Bur2 cyclin, can phosphorylate the CTD in vitro (Yao et al. 2000; Murray et al. 2001). Both Ctk1 and Bur1 function in transcription elongation, but the main function of Bur1 in vivo may not be as a CTD kinase (Keogh et al. 2003). Instead, Bur1/Bur2 is involved in establishing histone H2B monoubiquitinylation and H3K4 trimethylation at promoters by recruiting a ubiquitin ligase and histone methyltransferase, and the bulk of Ser2 phosphorylation reflects the action of Ctk1 (Wood and Shilatifard 2006; although see Bartkowiak et al. 2010). Indeed, Bur kinase, recruited by the CTD phosphorylated on Ser5 by Kin28, phosphorylates Ser2 at promoter regions, and subsequent Ctk1 recruitment, stimulated by Bur1, phosphorylates Ser2 further downstream in the coding region (Qiu et al. 2009). This model is supported by a recent genome-wide analysis showing that Bur1 is recruited to promoter regions prior to Ctk1 (Zhang et al. 2012). The promoter-proximal preference of Bur1 explains why only a mild reduction of Ser2 phosphorylation was observed in cells with impaired Bur kinase, since the majority of Ser2 phosphorylation occurs more downstream (Keogh et al. 2003; Qiu et al. 2009).
The existence of two CDK9-like kinases in yeast begs the question: Is there a second Ser2 kinase in higher eukaryotes? Recently, Greenleaf and colleagues (Bartkowiak et al. 2010) reported that Drosophila CDK12, which is conserved in humans, can phosphorylate the CTD on Ser2 and proposed, based on phylogenetic relationships (Guo and Stiller 2004), that the ortholog of Bur1 is CDK9, whereas CDK12 is the counterpart of Ctk1. Furthermore, depletion of CDK12 or the associated cyclin CycK in human cells reduced both total Ser2 levels and expression of select genes, including several DNA damage response genes (Blazek et al. 2011). Global RNAP II transcription, however, was not affected detectably in the CDK12/CycK-depleted cells. Thus, the contribution of CDK12 to Ser2 phosphorylation and overall RNAP II transcription, while intriguing, requires further study.
Ser2 and Ser5 are both followed by Pro residues, which is characteristic of substrates of CDKs, such as CDK7 and CDK9. But this structure has also raised the question of whether the CTD might be phosphorylated by classical CDKs that function in cell cycle control. Indeed, the first CTD kinase identified was the mitotic regulator Cdc2 (Cisek and Corden 1989), although the significance of this finding was unclear for some time. However, Xu et al. (2003) subsequently showed that the CTD can be phosphorylated in M phase by Cdc2/cyclin B to generate a hyperphosphorylated RNAP II isoform dubbed RNAP IIOO. This hyperphosphorylation, which requires Pin1 (also known to be a mitotic regulator) (Lu et al. 1996), inhibits RNAP II, contributing to mitotic gene silencing. On the other hand, given the preference of CDKs for Ser/Thr–Pro substrates, it is perhaps surprising that CDK7 and CDK9 appear to phosphorylate Ser7 and Thr4, respectively, although in each case, precedents exist for targeting non-S/T-P substrates (Larochelle et al. 2006; Baumli et al. 2008).
CTD phosphatases
The pattern of CTD phosphorylation during the transcription cycle is highly dynamic and requires the activity of dedicated phosphatases as well as kinases. For example, as discussed above, the CTD is hypophosphorylated in the PIC and becomes highly phosphorylated on multiple residues during transcription. Thus, both to ready RNAP II for new rounds of transcription and to regulate the dynamic CTD phosphorylation status during transcription, specific CTD residues must be dephosphorylated throughout the transcription cycle. This involves the activities of two major phosphatases, Fcp1 and Ssu72, conserved from yeast to humans, and others such as the small CTD phosphatases (SCPs).
Fcp1 (TFIIF-associating CTD phosphatase 1) was initially described as an activity in HeLa cells (Chesnut et al. 1992) and in yeast (Archambault et al. 1997) that can dephosphorylate the CTD. Human Fcp1 was found to function on elongating RNAP II and to recycle RNAP II for PIC assembly (Cho et al. 1999), and inactivation of FCP1 results in impaired transcription and lethality in yeast (Kobor et al. 1999). Fcp1 is capable of dephosphorylating both Ser2-P and Ser5-P (Lin et al. 2002), although it appears to prefer Ser2-P (Cho et al. 2001; Hausmann and Shuman 2002; Ghosh et al. 2008). Consistent with this, while Fcp1 is present on active genes at both 5′ and 3′ ends (Cho et al. 2001; Calvo and Manley 2005), in global analyses, higher levels were found in promoter-distal regions (Zhang et al. 2012), and mutations in FCP1 led to increased levels of Ser2 phosphorylation (Bataille et al. 2012). Thus, Fcp1 is likely responsible for dephosphorylating the CTD at the end of the transcription cycle but may also function earlier in the transcription process.
Ssu72 was first found in a genetic screen as a suppressor of a defect in the GTF TFIIB (Sun and Hampsey 1996). Later, it was identified as a component of yeast cleavage and polyadenylation factor (CPF) and shown to function in 3′ end formation of both polyadenylated and nonpolyadenylated RNA (Dichtl et al. 2002a; He et al. 2003; Nedea et al. 2003). Essentially simultaneously, Ssu72 was shown to be a CTD phosphatase with a preference for Ser5-P (Ganem et al. 2003; Krishnamurthy et al. 2004). Consistent with both these observations, the genome-wide distribution of Ssu72 indicates that Ssu72 peaks at both the promoter and 3′ end of genes, with more present in 3′ end regions (Zhang et al. 2012), and depletion of Ssu72 leads to increased levels of Ser5-P toward the 3′ end of genes (Bataille et al. 2012). The interaction with TFIIB now appears to reflect the role of these factors in the phenomenon of gene looping (Singh and Hampsey 2007). The phosphatase activity of yeast Ssu72 is stimulated in vivo by Pta1, a component of CPF (Ghazy et al. 2009). This interaction is conserved in higher eukaryotes, as human Ssu72 activity is stimulated in vitro by a direct interaction with the human homolog of Pta1, Symplekin (Xiang et al. 2010). Although the phosphatase activity of Ssu72 is not required for its function in 3′ processing in yeast cell extracts (Ghazy et al. 2009), it is in HeLa extracts, but only when the 3′ processing reaction is coupled to transcription (Xiang et al. 2010). This suggests that Ser5 dephosphorylation is important for the role of the CTD in coupling transcription and polyadenylation (see below). An unexpected finding was that Ssu72 favors the CTD substrate with the Ser5–Pro6 peptide bond in the cis configuration, which contrasts with all other known CTD phosphatases (Xiang et al. 2010; Werner-Allen et al. 2011). Consistent with the fact that the cis configuration is energetically unfavored, dephosphorylation of the CTD by Ssu72 is enhanced by Pin1 (Xiang et al. 2010). In line with this, ESS1 and SSU72 interact genetically, and mutations in Ess1 result in accumulation of Ser5-P (Krishnamurthy et al. 2009; Bataille et al. 2012). Recently, Ssu72 was suggested to dephosphorylate Ser7-P, as depletion of Ssu72 and mutation in SSU72 also result in elevated levels of Ser7-P at the 3′ end of genes, and Ssu72 dephosphorylates both Ser5-P and Ser7-P on CTD substrates in vitro (Bataille et al. 2012; Zhang et al. 2012).
In searching for human Fcp1 homologs, three related proteins containing a region with homology with the Fcp1 phosphatase domain were identified (SCP1–3) and found to dephosphorylate the CTD (Yeo et al. 2003; Kamenski et al. 2004). SCPs preferentially dephosphorylate Ser5-P (Yeo et al. 2003; Zhang et al. 2006) and were shown to be recruited by the transcription factor REST/NRSF to suppress transcription of neural genes in nonneuronal cells by removing Ser5 phosphorylation on promoter-proximal RNAP II (Yeo et al. 2005). The SCPs, which are not present in lower eukaryotes, thus appear to play a role in tissue-specific transcriptional regulation.
Finally, yeast Rtr1 (regulator of transcription 1) has also been reported to be a CTD phosphatase, functioning to dephosphorylate Ser5-P in vitro and in vivo (Mosley et al. 2009). Rtr1 associates with RNAP II and localizes to the 5′ ends of genes, and RTR1 deletion causes increased levels of Ser5-P and reduced RNAP II occupancy on a collection of genes (Mosley et al. 2009). Unlike Ssu72, Rtr1 is required only for Ser5-P, not Ser7-P, dephosphorylation (Kim et al. 2009). RPAP2, the human homolog of Rtr1, has also been reported to possess Ser5-P phosphatase activity (Egloff et al. 2012). However, the structure of Rtr1 was recently solved and did not reveal an apparent active site, and enzymatic assays with purified Rtr1 or RPAP2 and CTD substrates failed to detect activity (Xiang et al. 2012). Thus, the role of Rtr1 in CTD dephosphorylation is currently unclear.
Transcription elongation, chromatin, and the CTD
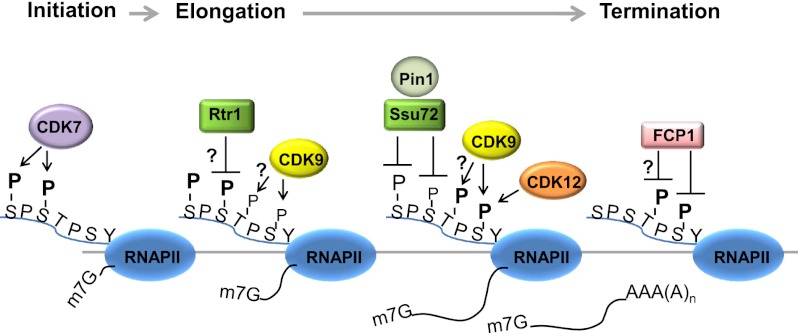
As alluded to above, the CTD undergoes dynamic changes in phosphorylation during transcription elongation. Very simply, the CTD is phosphorylated on Ser2 by CDK9, while Ser5-P is removed by Ser5 phosphatases, and, toward the end of genes, the CTD is dephosphorylated by Ser5 and Ser2 phosphatases. RNAP II with a hypophosphorylated CTD, RNAP IIA, can then be recycled for another round of transcription. This view has been expanded by inclusion of two additional phosphorylation sites, Ser7 and Thr4, requiring CDK7 and CDK9, respectively. Chromatin immunoprecipitation (ChIP) experiments in yeast revealed a resemblance of Ser7-P distribution to Ser5-P on select genes (Glover-Cutter et al. 2009; Kim et al. 2009), suggesting that, in general, Ser5 and Ser7 are both phosphorylated at the beginning of transcription. Thr4-P functions in facilitating histone mRNA 3′ end processing, and Thr4 phosphorylation requires CDK9, independent of its role in Ser2 phosphorylation (Hsin et al. 2011), suggesting that the Thr4-P pattern may be similar to Ser2-P, increasing toward the 3′ end of genes. Indeed, a recent study in human cells found that the genome-wide profiles of Thr4-P overlapped with, but shifted slightly 3′ to, Ser2-P (Hintermair et al. 2012). Therefore, a simple model of CTD phosphorylation is that Ser5 and Ser7 are phosphorylated at the beginning of transcription, and Ser2 and Thr4 phosphorylation occurs during elongation (Fig. 2).
Figure 2.
Dynamic modification of the CTD during the transcription cycle. At transcription initiation, CDK7 phosphorylates Ser5 and Ser7 residues. Later, during elongation, CDK9 phosphorylates Ser2 and perhaps Thr4, while the phosphate groups on Ser5 and Ser7 are gradually removed by phosphatases. For example, Rtr1, likely indirectly with another phosphatase, and Ssu72, with the aid of the prolyl isomerase Pin1, dephosphorylate Ser5-P early and late during elongation, respectively. Ssu72 also dephosphorylates Ser7-P. CDK12 likely also contributes to Ser2 phosphorylation during elongation of at least some genes. As RNAP II nears termination, Fcp1 dephosphorylates Ser2-P, regenerating unphosphorylated RNAP II that can be recycled for another round of transcription.
The above general model for the dynamics of CTD phosphorylation was, for the most part, based on analysis of a limited number of genes. This has more recently been reanalyzed by several genome-wide ChIP studies in yeast (Kim et al. 2010; Mayer et al. 2010; Tietjen et al. 2010; Bataille et al. 2012; Zhang et al. 2012). Although somewhat different conclusions were reached, probably reflecting use of different analytical methods and/or antibodies, so far the general model holds for the majority of genes (Bataille et al. 2012). In brief, these studies showed, with exceptions, that Ser5-P is enriched at the 5′ ends of genes and peaks around TSSs. As RNAP II elongates toward the 3′ end, Ser5-P is gradually removed, and Ser2-P increases, beginning to saturate at ∼600 nucleotides (nt) downstream from TSS, regardless of gene length, and sharply decreases at 100 nt downstream from the poly(A) addition site (Mayer et al. 2010). A crossover of Ser5-P to Ser2-P was observed at ∼450 nt downstream from the TSS. Therefore, longer genes have phosphorylation patterns more consistent with the model, and shorter genes have higher levels of Ser5-P and lower levels of Ser2-P. Overall, Ser7-P and Ser5-P patterns overlap to some extent, although profiles of Ser7-P vary, with some genes having discrete peaks at the 5′ and/or 3′ end (Kim et al. 2010). In one study, uniform levels of Ser7-P were observed along genes, although this may have reflected a technical limitation of the ChIP assay (Bataille et al. 2012). In any event, it is intriguing that long and short genes have different patterns of CTD phosphorylation at their 3′ ends. Given the role of the CTD in processes such as mRNA 3′ end formation and transcription termination, the existence of this pattern points to possible differences in the mechanisms underlying these processes on different genes dependent on length (see below).
Just as CTD phosphorylation patterns change along the length of transcribed genes, so do histone modifications in the chromatin of active genes. This raises the possibility that changes in the CTD phosphorylation array may contribute to differential patterns of histone modification as transcription proceeds, and, indeed, considerable evidence suggests that this is the case (for review, see Hirose and Ohkuma 2007; Egloff and Murphy 2008; Spain and Govind 2011). For example, in yeast, the histone methyltransferases Set1 and Set2 are recruited to actively transcribed genes in a CTD-dependent manner. Set1 is recruited to the 5′ ends of genes by Ser5-P, together with the Paf1 complex, and methylates histone H3 on Lys 4 (H3K4) (Krogan et al. 2003; Ng et al. 2003). Set1 establishes two distinct chromatin zones on genes, with trimethylated H3K4 at promoter regions and dimethylated H3K4 downstream (Kim and Buratowski 2009). H3K4 dimethylation and CTD Ser5-P are then involved in recruiting the Set3 complex, a histone deacetylase that deactylates histones H3 and H4, leading to reduced histone acetylation levels at the 5′ ends of genes, which promotes the association of RNAP II (Kim and Buratowski 2009; Govind et al. 2010).
Set2 functions toward the 3′ ends of genes. It binds to phosphorylated CTD with a preference for Ser2/5-P (Kizer et al. 2005) and methylates H3 on Lys 36 (H3K36) (Li et al. 2003; Xiao et al. 2003). After H3K36 methylation, the histone deacetylase Rpd3S is recruited to deacetylate H3 and H4, which results in reduced transcription elongation efficiency and prevents cryptic transcription in the coding region. The recruitment of Rpd3S is mediated by two subunits: Eaf3 and Rco (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005). However, initial Rpd3S recruitment is independent of H3K36 methylation and occurs through direct binding to Ser2, Ser5 phosphorylated CTD (Govind et al. 2010). Rpd3S then transfers to H3 by interaction of the Eaf3 and Rco subunits with methylated H3K36, which is required for activation of acetylation activity (Drouin et al. 2010; Govind et al. 2010). Although recruitment of Rpd3S to coding regions and subsequent histone deacetylation to prevent cryptic transcription in the wake of the elongating RNAP II are well established, Rpd3S is not found on all transcribed genes, suggesting the existence of other mechanisms to prevent aberrant transcription (Drouin et al. 2010).
H3K4 and H3K36 methylation patterns similar to those in yeast have been observed in higher eukaryotes (Bannister et al. 2005; Barski et al. 2007; Edmunds et al. 2008). Mammalian cells contain six Set1-like factors, of which Set1A and Set1B are responsible for the bulk of H3K4 methylation (for review, see Shilatifard 2012). A component of Set1A and Set1B, Wrd82, binds to the CTD phosphorylated on Ser5 (Lee and Skalnik 2008), which facilitates recruitment of these factors. Additionally, association of the mixed lineage leukemia (MLL) Set1-like factors MLL1 and MLL2 with Ser5-P has been reported (Hughes et al. 2004; Milne et al. 2005). MLL1/MLL2 are required for proper Hox gene expression, and translocation mutations of MLL are associated with the pathogenesis of leukemia (for review, see Shilatifard 2012). A human homolog of yeast Set2, Huntington-interacting protein (HYPB, also known as Setd2), interacts with Ser2/5 phosphorylated CTD through its C-terminal region (Li et al. 2005) and methylates H3K36 (Sun et al. 2005). The association of HYPB with phosphorylated CTD is enhanced by an elongation factor, Iws1, that is cotranscriptionally recruited by the histone chaperon and transcription elongation factor Spt6 (Yoh et al. 2007, 2008). Spt6 directly binds to the phosphorylated CTD via its SH2 domain with a preference for Ser2-P (Yoh et al. 2007; Sun et al. 2010). Interestingly, Spt6 has also been shown to be involved in facilitating splicing and nuclear mRNA export (Yoh et al. 2007, 2008).
RNA processing and the CTD
The CTD is now known to function in essentially all of the RNA processing reactions involved in maturation of very likely all transcripts produced by RNAP II. This includes not only mRNAs, but also small nuclear RNAs (snRNAs), microRNAs (miRNAs), and other ncRNAs. The CTD thus provides the basis for the coupling between transcription and RNA processing that is important for ensuring the efficiency and accuracy of the complex processing reactions required for production of functional RNAs. Here we concentrate on the role of the CTD in mRNA processing events, both in the interests of space and because these are illustrative of the function of the CTD more generally. Reviews dealing with the function of the CTD in other aspects of RNA processing and metabolism have been published recently (Munoz et al. 2010; Pawlicki and Steitz 2010).
Most of the earliest studies linking the CTD to RNA processing were performed in mammalian systems. Many of these used a “pseudogenetic” analysis that takes advantage of the RNAP II inhibitor α-amanitin. In this system, first introduced by Corden and colleagues (Gerber et al. 1995), α-amanitin, which binds Rpb1, is added to the cell culture to inhibit endogenous RNAP II after a plasmid encoding an exogenous, α-amanitin-resistant Rpb1 is introduced and expressed in the cells. Therefore, the only functional RNAP II will contain an α-amanitin-resistant Rpb1 subunit, and the effects of mutations, specifically of the CTD, can be determined. Although not without difficulties (see below), this system has provided a wealth of information about the function of the CTD and of CTD modifications. For example, Bentley and colleagues (McCracken et al. 1997a,b) used this method to provide the first direct evidence that the CTD indeed plays a role in mRNA processing. They showed, unexpectedly, that deletion of the CTD had no effects on transcription of a reporter plasmid but was required for efficient 5′ capping, splicing, and 3′ processing (McCracken et al. 1997a,b). While initial studies suggested that the C-terminal half of the CTD, rich in nonconsensus heptads, but not the more conserved N-terminal half, could support all three processing reactions (Fong and Bentley 2001), this was subsequently shown to reflect the presence of the 10-amino-acid nonconsensus motif following heptad 52: When the N-terminal half was fused with this motif, it fully supported processing (Fong et al. 2003). Indeed, another study indicated that only the number of heptads is important: Twenty-two repeats from either the conserved or divergent half of the CTD were found to be sufficient to stimulate splicing and 3′ end cleavage (Rosonina and Blencowe 2004). As mentioned above, the 10-amino-acid motif functions in Rpb1 stability (Chapman et al. 2004, 2005), likely explaining its requirement in processing. Intriguingly, the c-abl tyrosine kinase interacts with the CTD through this motif (Baskaran et al. 1999), although there is no evidence indicating the involvement of Abl kinase or Tyr1 phosphorylation in Rpb1 protein stability. More recently, the α-amanitin system has been used to determine the role of specific residues and their modification in CTD function. For example, it was used to provide evidence that Ser7 phosphorylation functions in the expression and 3′ formation of snRNAs (Egloff et al. 2007). Below, we discuss the role of the CTD and modifications in capping splicing and 3′ end formation and review the factors (listed in Table 1) involved in RNA processing that bind the CTD (Figs. 3, 4).
Table 1.
CTD-binding proteins
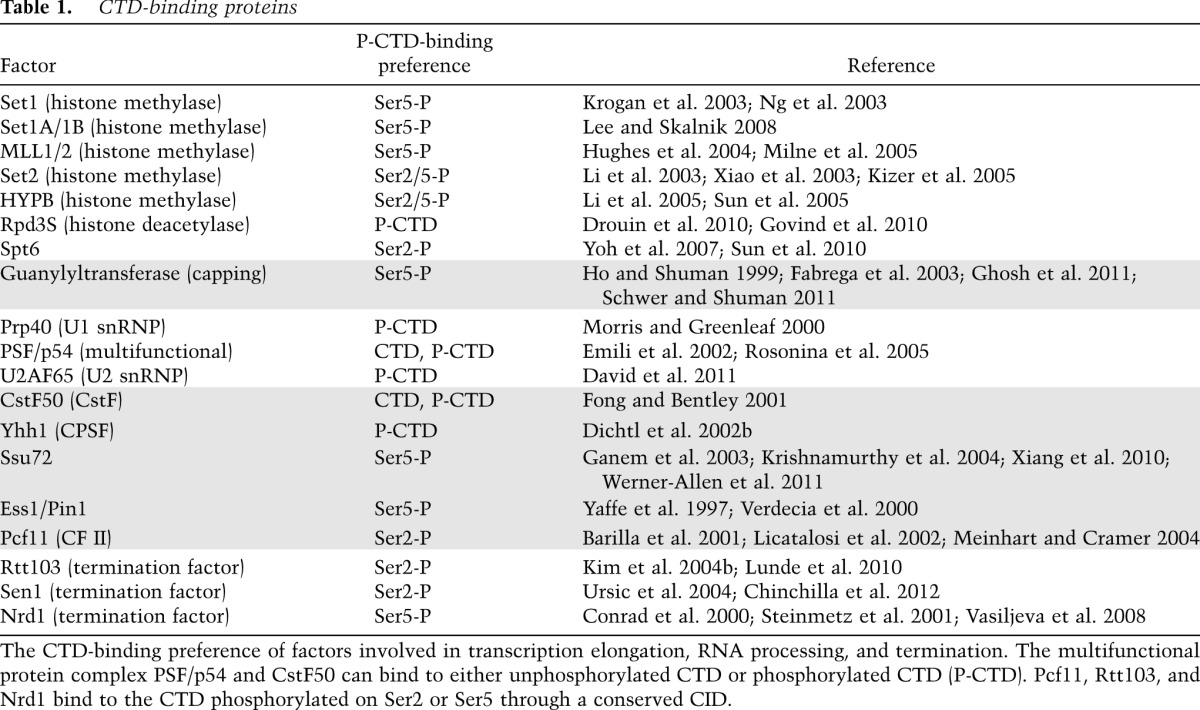
Figure 3.
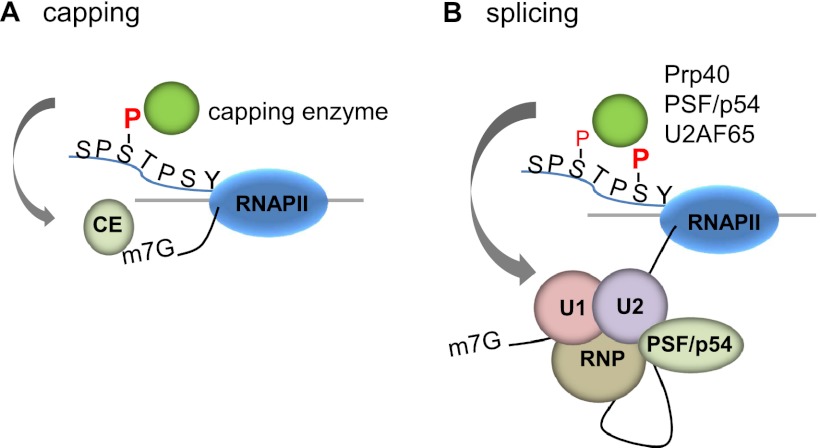
The CTD facilitates capping and splicing by recruitment of RNA processing factors. (A) Capping enzyme (CE) is recruited to the vicinity of nascent mRNA by the CTD phosphorylated on Ser5. (B) During transcription, the CTD is phosphorylated on Ser2, while the Ser5-P is dephosphorylated and is involved in recruiting the indicated splicing factors, which defines splice sites and facilitates assembly of the spliceosome. In this and subsequent figures, green spheres above the CTD represent relevant CTD-binding proteins, while assembled functional complexes are indicated below.
Figure 4.
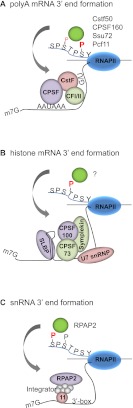
The CTD functions in 3′ processing of both polyadenylated and nonpolyadenylated RNAs. (A) At 3′ ends of polyadenylated mRNA, Ser2-P serves to recruit Pcf11, a component of CFII (human nomenclature is used for all factors). Other 3′ end factors, such as CstF50 and AAUAAA-binding factor CPSF-160 (Yhh1 in yeast), also bind the CTD, whereas Ssu72, with the aid of Pin1 (Ess1), must dephosphorylate Ser5-P. The AAUAAA element and G/U elements are bound by CPSF and CstF, respectively. Loading of some factors, including CPSF and CstF, may occur upstream, perhaps at the promoter (see the text). (B) The 3′ end of histone pre-mRNA contains a stem–loop motif bound by SLBP and a downstream element recognized by U7 snRNP. A complex containing CPSF73, CPSF100, and Symplekin is recruited for 3′ cleavage. Thr4-P facilitates this process, likely through a yet-to-be-identified factor. (C) The 3′ end of snRNA genes contains a 3′ box that interacts with the Integrator complex. RPAP2 binds to Ser7-P on the CTD and to recruit the Integrator, and Int 11, an Integrator subunit, cleaves the RNA.
Capping
5′ Capping, the first step of mRNA processing, occurs very early in transcription, essentially as soon as the newly synthesized RNA is extruded from RNAP II (Coppola et al. 1983; Jove and Manley 1984). Only RNAP II products are capped, and this specificity is due in large part to the CTD, which, when phosphorylated, recruits capping enzymes (Cho et al. 1997; McCracken et al. 1997a). In metazoans, a bifunctional capping enzyme with RNA triphosphatase and RNA guanylyltransferase activities binds to the phosphorylated CTD through the guanylyltransferase domain. This domain has two binding sites for phosphorylated CTD: one specific for the Ser2 phosphorylated CTD and the other, an allosteric activator site, for the Ser5 phosphorylated CTD (Ho and Shuman 1999), which stimulates formation of the enzyme–GMP intermediate (Ho and Shuman 1999; Ghosh et al. 2011). The interaction between guanylyltransferase and Ser5-P also occurs in yeast (Candida albicans) (Fabrega et al. 2003). Interestingly, the budding yeast capping enzyme contains another binding site, for the Rpb1 “foot domain,” which works synergistically with the Ser5-P-binding site to enhance interaction with RNAP II (Suh et al. 2010). Remarkably, it was recently shown that recruitment of capping enzymes is the sole essential function of Ser5, at least in fission yeast. As mentioned above, substitution of all Ser5 residues with Ala is lethal in S. pombe, but viability can be restored simply by tethering capping enzymes to the CTD (Schwer and Shuman 2011).
Splicing
Nearly all mammalian pre-mRNAs contain introns. In humans, the average size of an intron is ∼3 kb, but many are considerably larger (Lander et al. 2001). Furthermore, nearly all human genes produce transcripts in which one or more introns are subject to alternative splicing (for review, see Chen and Manley 2009). Given this complexity, coupled with the remarkable complexity of the spliceosome itself, it is perhaps not surprising that splicing and transcription are coupled, as this provides a mechanism to enhance the fidelity and efficiency of splicing as well as its regulation, and while other factors function in this coupling (e.g., Luco et al. 2011), it is also not surprising that the CTD plays a central role.
The CTD interacts with several proteins that function in splicing. Early on, RNAP IIO was found to associate with splicing complexes (Mortillaro et al. 1996). In vitro biochemical studies indicated that RNAP IIO, but not RNAP IIA, can enhance splicing of an exogenously supplied substrate by influencing an early step of spliceosome assembly (Hirose et al. 1999). Splicing of pre-mRNAs transcribed in vitro by RNAP II is accelerated compared with those transcribed by T7 RNAP (Ghosh and Garcia-Blanco 2000; Das et al. 2006), suggesting that the CTD can enhance the rate of splicing, likely by facilitating recruitment of splicing factors, and artificial tethering of the phosphorylated CTD to a pre-mRNA can also accelerate splicing in vitro (Millhouse and Manley 2005). Additionally, splicing factor SRSF2 (SC35) was shown to colocalize with nascent transcripts and associate with RNAP IIO in HeLa cells, but this colocalization and association were lost when the CTD was truncated (Misteli and Spector 1999). Additional splicing factors, including U1 snRNP-associated protein Prp40, involved in 5′ splice site recognition (Morris and Greenleaf 2000; Phatnani et al. 2004); PSF, which binds sequences downstream from the branch point (Emili et al. 2002; Rosonina et al. 2005); and U2AF, which helps define the 3′splice site (David et al. 2011), all appear to bind directly to the CTD (Fig. 3B). Together, these factors could help recruit the splicing machinery to both ends of the intron during transcription. Indeed, the U2AF interaction with the CTD recruits the Prp19 complex (PRP19C), an essential splicing factor required for activation of the spliceosome (David et al. 2011). An intriguing idea is that recruitment of splicing factors to nascent transcripts by the CTD is especially important for splicing of introns with weak splicing signals (for review, see David and Manley 2011).
Recruitment of splicing factors and other RNA-binding proteins to nascent transcripts can facilitate gene expression in additional ways. For example, in yeast, the cotranscriptional recruitment of PRP19C leads to corecruitment of the TREX complex, which functions in mRNA transport, thereby preparing a nascent transcript for transport to the cytoplasm as it is synthesized (Chanarat et al. 2011). These investigators also provided evidence that PRP19C enhances RNAP II elongation, which provides support for the view that splicing also communicates back to modulate transcription (for review, see Manley 2002). For example, the presence of splice sites on nascent transcripts can stimulate transcription, as reduced transcription can be seen when splice sites are impaired (Furger et al. 2002; Damgaard et al. 2008; Eberle et al. 2010); splicing factors, such as SKIP, Npl3, and SRSF2, can enhance transcriptional elongation (Bres et al. 2005; Dermody et al. 2008; Lin et al. 2008); and incorrect spliceosome recruitment and assembly can interfere with transcription termination (Martins et al. 2011). Finally, while histone modifications can help recruit splicing factors to active genes (Luco et al. 2010; for review, see Hnilicova and Stanek 2011), splicing can also influence chromatin structure; for example, by facilitating recruitment of HYPB and thereby increasing H3K36 methylation (de Almeida et al. 2011).
Cotranscriptional recruitment of RNA-binding proteins has an additional important effect in cells, which is to prevent genomic instability and DNA rearrangements during transcription (for review, see Li and Manley 2006; Aguilera and Garcia-Muse 2012). If nascent transcripts are not properly packaged or processed, they have the potential, especially in G-rich regions, to rehybridize with the template DNA strand, creating an R-loop structure in which the nontemplate strand is single-stranded. Such structures can be the targets of DNA double-strand breaks and rearrangements. The CTD plays an important role in this process, as prevention of R loops by SRSF1 in an in vitro reconstituted system was found to be dependent on the CTD (Li and Manley 2005).
3′ End processing
The CTD also plays an important role in 3′ end processing of RNAP II-produced transcripts. This has been most extensively studied in the case of polyadenylated mRNAs. Polyadenylation, a relatively simple two-step reaction consisting of an endonucleolytic cleavage followed by poly(A) tail synthesis, involves a remarkably complex set of protein factors (Shi et al. 2009), and the CTD functions in recruitment and/or stabilization of this complex on the pre-mRNA. The studies mentioned above using the α-amanitin system to provide evidence that 3′ end processing is impaired when Rpb1 is truncated also revealed that the CTD associates with two polyadenylation factors: CPSF and CstF (McCracken et al. 1997b). Hirose and Manley (1998) showed that the CTD is required for efficient 3′ end processing in vitro in the absence of transcription and that this function can be fulfilled by the CTD alone. Subsequent experiments revealed that 26 all-consensus repeats were required for reconstitution of CTD activity in vitro (Ryan et al. 2002), consistent with results in vivo. The CTD requirement is conserved in yeast, as CTD deletion was shown to cause defects in 3′ processing (Licatalosi et al. 2002).
Several 3′ processing factors interact with the CTD, consistent with it playing a role as a scaffolding factor (Fig. 4). For example, CstF-50, a component of CstF, and Yhh1, the yeast counterpart of the mammalian AAUAAA-binding CPSF-160, both interact with the CTD (Fong and Bentley 2001; Dichtl et al. 2002b), providing an explanation for the initial observations of McCracken et al. (1997b). Pcf11, which contains an N-terminal CTD interaction domain (CID) characteristic of several CTD-binding proteins (for review, see Corden and Patturajan 1997), binds the CTD in a manner enhanced by Ser2 phosphorylation (Barilla et al. 2001; Licatalosi et al. 2002; Meinhart and Cramer 2004). Indeed, Ser2-P plays an important role in facilitating polyadenylation. For example, impairment of Ser2 phosphorylation by deletion of CTK1 in yeast (Skaar and Greenleaf 2002; Ahn et al. 2004) or flavopiridol in metazoan cells (Ni et al. 2004) impairs recruitment of processing factors at the 3′ ends of genes and subsequent polyadenylation. However, at least in yeast, accumulation of these factors also requires an intact polyadenylation signal, indicating, not surprisingly, that interactions with the pre-mRNA are also important (Kim et al. 2004a). Extending these findings, genome-wide ChIP experiments showed that peaks of 3′ processing factors followed Ser2-P peaks, consistent with the view that both the poly(A) site in the RNA and CTD Ser2-P contribute to the recruitment/assembly of the polyadenylation complex (Kim et al. 2010; Mayer et al. 2010, 2012b). Together, these experiments point to an important role for the CTD and Ser2-P in polyadenylation of mRNA precursors.
The involvement of Ser2-P and the observed accumulation of processing factors at the 3′ ends of genes are consistent with the simple view that polyadenylation factors are recruited to genes near their site of action. However, considerable evidence also exists that polyadenylation factors are in fact recruited to genes at the promoter (for review, see Calvo and Manley 2003; see also Glover-Cutter et al. 2008; Nagaike et al. 2011 and references therein). Factors involved in this recruitment include the GTFs TFIID (Dantonel et al. 1997) and TFIIB (Sun and Hampsey 1996; Wang et al. 2010) and the PAF complex (Rozenblatt-Rosen et al. 2009) as well as transcriptional activators (for review, see Nagaike and Manley 2011). Indeed, as with splicing, these interactions may affect transcription, as poly(A) site mutations reduce transcription and reduce the levels of TFIIB and TFIID at promoters (Mapendano et al. 2010). While some of these interactions likely reflect gene looping (O'Sullivan et al. 2004; for review, see Hampsey et al. 2011), they highlight the importance of coupling transcription and polyadenylation via multiple mechanisms that extend beyond the CTD.
The 3′ ends of several types of RNAP II transcribed RNAs, including snRNA and histone mRNA, are not polyadenylated but also require the CTD. The CTD plays a role in snRNA 3′ end formation, and inhibition of Ser2 phosphorylation was shown to impair 3′ processing without affecting transcription (Jacobs et al. 2003; Medlin et al. 2003). Proper snRNA 3′ end formation is dependent on both the promoter and a 3′ box, located just downstream from the snRNA-encoding region (for review, see Egloff et al. 2008). The 3′ box is recognized by a multisubunit RNA 3′ end processing complex, the Integrator, and snRNAs are cleaved by the Int11 subunit (Baillat et al. 2005). The Integrator is recruited by interaction with the CTD phosphorylated on Ser2 and Ser7 (Egloff et al. 2010), consistent with earlier studies showing that mutation of all Ser7 residues to Ala impairs 3′ end processing of snRNAs but not of polyadenylated mRNAs (Egloff et al. 2007). Recently, it was shown that RPAP2, which functions in Ser5-P dephosphorylation (see above), binds to the CTD with Ser7-P and facilitates Integrator recruitment (Egloff et al. 2012). These results support the view that Ser5-P dephosphorylation is important for both snRNA and mRNA (Xiang et al. 2010) 3′ end formation. It is also noteworthy that methylation of a specific Arg residue in a nonconsensus heptad appears to dampen expression of certain snRNAs and small nucleolar RNAs (snoRNAs), as mutation of this residue to Ala resulted in enhanced accumulation of these RNAs in the α-amanitin assay (Sims et al. 2011). Although the mechanism remains to be determined, an intriguing possibility is that this modification in some way interferes with the positive effects of Ser2 and Ser7 phosphorylation.
Metazoan replication-dependent histone mRNAs are the only protein-coding transcripts that are not polyadenylated. The 3′ end of histone mRNA contains two cis-elements: a stem–loop structure that is recognized by stem–loop-binding protein (SLBP) and a downstream sequence bound by U7 snRNP. Together, SLBP and U7 snRNP recruit a cleavage complex containing polyadenylation factors CPSF73 (the endonuclease), CPSF100, and Symplekin to process histone mRNA 3′ ends (for review, see Marzluff et al. 2008). As with other RNAP II transcripts, the CTD also participates in histone 3′ end processing. This was first suggested by the finding that CDK9 is required for proper histone mRNA 3′ processing and SLBP recruitment (Pirngruber et al. 2009). Recently, using the DT40 system (see above), evidence was provided that Thr4-P is required for histone mRNA 3′ end processing (Hsin et al. 2011). Specifically, cells expressing Rpb1 with a mutant CTD in which all Thr4 residues were replaced by Val were found to be defective in histone mRNA 3′ end processing and recruitment of SLBP and CPSF100, while 3′ end formation of polyadenylated RNAs and overall transcription rates were essentially unaffected. Inhibition of CDK9 prevented Thr4 phosphorylation and inhibited histone 3′ processing, consistent with the results of Pirngruber et al. (2009). Hintermair et al. (2012) analyzed a related Thr4 mutant Rpb1 using the α-amanitin system. These investigators did not monitor 3′ processing but detected by RNAP II ChIP a global defect in elongation, characterized by promoter-proximal accumulation of initiated RNAP II. While Hsin et al. (2011) also observed some increase in promoter-proximal RNAP II on several genes, the effects were likely smaller than detected by Hintermair et al. (2012), as measurements of transcription rate and polyadenylated mRNA levels revealed at most modest effects.
The basis for the possible differences in the effects of Thr4 mutation is unclear, but one possibility may reflect features of the α-amanitin system. Although, as illustrated in the above discussion, a great deal has been learned about the functions of the CTD using this assay, there are several potential drawbacks. One of these is that transiently transfected reporter genes are often used to observe phenotypes of mutant CTDs because endogenous RNAs are stabilized by α-amanitin (Meininghaus et al. 2000; Chapman et al. 2004). Such reporter genes are not properly packaged into chromatin, which may affect results. Another drawback reflects the toxicity of α-amanitin. Thus, while stably transformed cells expressing α-amanitin-resistant Rpb1 avoid the problems of transient transfection, such cells initially grow slowly with reduced viability in the presence of α-amanitin (Meininghaus et al. 2000; Chapman et al. 2004, 2005), indicating that they must undergo certain changes to survive. Indeed, 24-h α-amanitin treatment was recently shown to bring about accelerated degradation of several proteins, including the transcription elongation factor DSIF, and this may complicate interpretation of experimental results (Tsao et al. 2012).
The CTD and transcription termination
Stopping RNAP II is much more difficult than any other RNAP. This undoubtedly reflects not only the need to transcribe successfully immensely long genes, which can extend >1 Mb in vertebrates, but also the diversity of sequence landscapes presented by the thousands of genes that RNAP II must negotiate, which can pose a variety of challenges to the elongating polymerase. Consistent with this, the mechanisms leading to RNAP II termination are complex and remain incompletely understood. While considerable progress has been made in deciphering these mechanisms in recent years (for review, see Richard and Manley 2009; Kuehner et al. 2011), here we discuss RNAP II termination from the perspective of the CTD and its role in the process (Fig. 5; Table 1).
Figure 5.
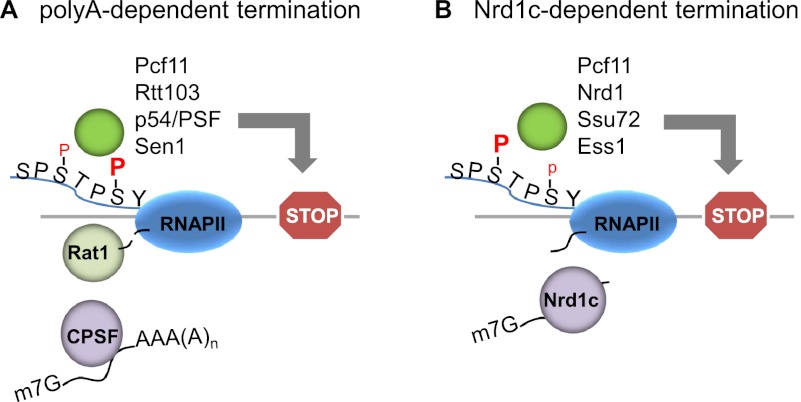
The CTD facilitates different termination mechanisms for protein-coding and noncoding genes. (A) Poly(A)-dependent termination pathway. RNA is cleaved by 3′ end processing factors at the polyadenylation site. The CTD with Ser2-P is involved in recruiting factors, including Pcf11, Rtt103, p54/PSF, and Sen1, to facilitate termination of long polyadenylated transcripts. Pcf11 and Rtt103 are required for the recruitment of exoribonuclease Rat1 in yeast, while Xrn2 is recruited by p54/PSF in humans. Sen1 (Senataxin in humans) may function on some of these genes by resolving RNA–DNA hybrids. (B) Nrd1c-dependent termination pathway. The Nrd1 complex (Nrd1–Nab3–Sen1) interacts via Nrd1 with the CTD phosphorylated on Ser5, which is present at the 3′ ends of short genes, such as snoRNAs and CUTs. Ssu72 and Ess1 are also required to dephosphorylate Ser5-P, although the exact mechanism of Nrd1c-dependent termination awaits further studies.
One of the earliest insights into the mechanism of RNAP II termination was the discovery in mammalian systems that a functional polyadenylation signal is required for subsequent termination (for review, see Proudfoot 1989). This suggested that 3′ processing of the mRNA precursor is required for termination, and subsequent experiments, principally in yeast and using transcriptional run on and/or RNAP II ChIP to measure termination, revealed that a number of 3′ cleavage factors are also necessary for termination (Birse et al. 1998; Dichtl et al. 2002b; Ganem et al. 2003; Nedea et al. 2003; Kim et al. 2010; Zhang et al. 2012). Given the requirement of the CTD for 3′ processing, it is not surprising that the CTD is also required for termination (McCracken et al. 1997b), with the N-terminal half being sufficient (Park et al. 2004), as determined using the α-amanitin system. One CTD-binding cleavage factor, Pcf11, may play a particularly important role in termination. A mutant yeast Pcf11 that retains 3′ cleavage activity but is defective in CTD binding was found to be defective in termination (Sadowski et al. 2003). In vitro studies suggested that both yeast and fly Pcf11 can disassociate RNAP II and the transcribed RNA from the DNA by bridging the CTD to RNA (Zhang et al. 2005; Zhang and Gilmour 2006). However, the dissociation activity is not specific to Pcf11 (Zhang et al. 2004), and Pcf11 cleavage activity, but not CTD-binding ability, was subsequently found to be important for termination of protein-coding genes (Kim et al. 2006), leaving the precise role of Pcf11 in termination unclear.
The discovery that 3′ cleavage is necessary for termination led to a model in which a 5′-to-3′ exoribonuclease degrades the downstream nascent RNA and in some way signals termination to the elongating RNAP II (Connelly and Manley 1988). Eventually, confirmation of this model came from the discovery that termination indeed requires such a nuclease: Rat1 in yeast (Kim et al. 2004b), and Xrn2 in humans (West et al. 2004). The CTD plays a role in Rat1/Xrn2 recruitment. Pcf11 is required in yeast for Rat1 association with active genes (Luo et al. 2006) and in HeLa cells for efficient degradation of the downstream RNA (West and Proudfoot 2008). In line with this, the genome-wide profile of yeast Pcf11 closely overlaps that of Rat1 (Kim et al. 2010). Rat1 recruitment to the CTD also requires the associated protein Rtt103, which specifically interacts with CTD Ser2-P (Kim et al. 2004b). Rtt103 and Pcf11 both achieve high affinity for the CTD phosphorylated on Ser2 by cooperatively binding to neighboring Ser2-P residues (Lunde et al. 2010). In humans, Xrn2 associates with p54/PSF (Kaneko et al. 2007), a multifunctional protein dimer that binds to the CTD (Emili et al. 2002; Rosonina et al. 2005) and was identified as a component of the 3′ processing complex (Shi et al. 2009). p54/PSF may facilitate termination by recruiting Xrn2, as accumulation of the downstream cleaved RNA and termination defects were observed in vitro and in vivo after p54 was depleted from HeLa cells (Kaneko et al. 2007).
The RNA–DNA helicase Sen1—well established to function in termination of snoRNAs in yeast (see below)—also appears to function in the termination of some mRNA genes. Sen1 binds to the CTD phosphorylated on Ser2 (Ursic et al. 2004; Chinchilla et al. 2012), which likely facilitates its recruitment to multiple coding as well as noncoding genes, where it tends to accumulate toward the 3′ end (Chinchilla et al. 2012). However, in cells expressing a catalytically inactive mutant, Sen1, only a small set of protein-coding genes, usually short in length, are defective in termination, as shown by a genome-wide Rpb1 ChIP analysis (Steinmetz et al. 2006). The human homolog of Sen1, Senataxin, has also been implicated in termination (Suraweera et al. 2009), perhaps by resolving RNA–DNA hybrids (R loops) formed at pause sites downstream from the polyadenylation signal, allowing degradation of the cleaved RNA by Xrn2 (Skourti-Stathaki et al. 2011).
Termination of transcription on genes encoding small ncRNAs shares some features in common with mRNA-encoding genes but also displays differences. The requirement of Pcf11 in termination extends to yeast snoRNA genes. However, here the CTD-binding activity of Pcf11 is necessary for termination, while the 3′ cleavage function is dispensable (Kim et al. 2006). Termination on snoRNA genes requires a protein complex, the Nrd1 complex (Nrd1c), which consists of Nrd1, Nab3, and Sen1 (Steinmetz and Brow 1996; Conrad et al. 2000; Steinmetz et al. 2001; Kim et al. 2006). Nrd1 and Nab3 are RNA-binding proteins, and their interaction with nascent RNAs helps recruit Nrd1c to target genes (Steinmetz and Brow 1998; Conrad et al. 2000; Steinmetz et al. 2001). However, Nrd1, like Sen1, binds the CTD, except displaying a preference for Ser5-P (Conrad et al. 2000; Steinmetz et al. 2001; Vasiljeva et al. 2008). This serves to help target Nrd1c to short genes, such as snoRNA genes, and genes encoding CUTs (cryptic unstable transcripts). CUTs are short noncoding RNAP II-produced nonpolyadenylated transcripts found in yeast that are rapidly degraded by the nuclear exosome (for review, see Colin et al. 2011). Consistent with the requirement of Nrd1c, CUT termination is critically dependent on CTD phosphorylation status, requiring high levels of Ser5-P and low Ser2-P (Gudipati et al. 2008). However, the Ser5-P must be removed, as Ssu72 and Ess1 are required for snoRNA/CUT termination (Kim et al. 2006; Singh et al. 2009; Zhang et al. 2012). Precisely how Nrd1c brings about termination is not well understood and may vary among different genes (for review, see Richard and Manley 2009; Kuehner et al. 2011).
Nrd1c also associates with mRNA-encoding genes (Nedea et al. 2003; Kim et al. 2006). Genome-wide ChIP of Nrd1 revealed frequent colocalization with Pcf11 and, unexpectedly, Ser7-P at the 3′ ends of genes (Kim et al. 2010). However, termination of a collection of protein-coding genes is not affected by mutations in Nrd1 (Kim et al. 2006). Perhaps the function of Nrd1c on such genes is to provide an alternative pathway to terminate aberrant transcription (Rondon et al. 2009) and/or a quality control point to degrade erroneous transcripts (Honorine et al. 2011).
An interesting question is whether a Nrd1c-like pathway for termination exists in higher eukaryotes. Although, as mentioned above, a homolog of Sen1, Senataxin, exists in human cells, functional homologs of Nrd1 and Nab3 have not been identified, and there is no evidence for the existence of a Nrd1c-like complex. SCAF8, which shares sequence similarity with Nrd1, was in fact the first CID protein identified (Yuryev et al. 1996; Patturajan et al. 1998). While SCAF8 CTD binding has been extensively characterized (Becker et al. 2008), its function is unknown but does not appear to involve termination. The absence of Nrd1c in metazoans is consistent with the facts that snoRNAs tend to be produced by a different mechanism (i.e., cleavage from introns) (Richard and Kiss 2006), and while RNAP II transcribed ncRNAs are ubiquitous (for review, see Jacquier 2009), it is not clear whether CUT RNAs are indeed produced in higher eukaryotes. In the future, it will be interesting to determine whether additional mechanisms of termination exist in metazoans and, if so, how the CTD might participate.
Perspectives
A great deal has been learned over the course of the past two decades about the structure and function of the CTD, the nature and role of CTD modifications in gene expression, and how the CTD mediates the interplay between transcription and RNA processing. However, the goal of completely understanding the functions and importance of this deceptively simple domain remains a challenge. We mention here just a few examples. What is the function of Tyr1 phosphorylation, and how do other modifications, such as methylation, acetylation, and ubiquitination, affect CTD function and gene expression? Intriguingly, evidence that Tyr1-P occurs in budding yeast and may play a general role in facilitating elongation by blocking premature 3′ end formation was recently presented (Mayer et al. 2012a). These findings suggest an important, perhaps general, role for Tyr1-P in elongation. This is perhaps surprising given that Tyr phosphorylation is rare in yeast (e.g., Gnad et al. 2009), and further characterization of Tyr1-P in yeast and mammalian systems will therefore be informative. Although a general model outlines changes in CTD phosphorylation along the length of transcribed genes, how common is regulated CTD modification in control of specific genes, and how is this achieved? A well-documented example is recruitment of P-TEFb by the oncogenic transcription factor c-Myc (Eberhardy and Farnham 2002; Kanazawa et al. 2003; Rahl et al. 2010), but is this a common mechanism. Are other CTD modifiers targets for gene-specific recruitment? How many more CTD modifiers remain to be discovered, and might they work in gene- or cell-specific ways? For example, the Cdc14 cell cycle phosphatase (Clemente-Blanco et al. 2011) and the bromodomain transcriptional regulator Brd4, apparently functioning as an atypical protein kinase (Devaiah et al. 2012), have both been reported to target the CTD. Future studies will continue to unravel the complex workings of the CTD and, we suspect, consolidate its role as the conductor of the gene expression symphony.
Acknowledgments
We thank Manley laboratory members Emanuel Rosonina, Patricia Richard, Dafne Campigli Di Giammartino, and Kehui Xiang for discussion and critical reading of this manuscript. Work from our laboratory was supported by grants from the NIH.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.200303.112.
References
- Aguilera A, Garcia-Muse T 2012. R loops: From transcription byproducts to threats to genome stability. Mol Cell 46: 115–124 [DOI] [PubMed] [Google Scholar]
- Ahn SH, Kim M, Buratowski S 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 34: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoulitchev S, Makela TP, Weinberg RA, Reinberg D 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377: 557–560 [DOI] [PubMed] [Google Scholar]
- Akoulitchev S, Chuikov S, Reinberg D 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407: 102–106 [DOI] [PubMed] [Google Scholar]
- Archambault J, Chambers RS, Kobor MS, Ho Y, Cartier M, Bolotin D, Andrews B, Kane CM, Greenblatt J 1997. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc Natl Acad Sci 94: 14300–14305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123: 265–276 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T 2005. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736 [DOI] [PubMed] [Google Scholar]
- Barilla D, Lee BA, Proudfoot NJ 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc Natl Acad Sci 98: 445–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, Adelman K, Lis JT, Greenleaf AL 2010. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24: 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Halden NF, Cullen CR, Corden JL 1988. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol 8: 330–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran R, Dahmus ME, Wang JYJ 1993. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc Natl Acad Sci 90: 11167–11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran R, Chiang GG, Mysliwiec T, Kruch GD, Wang JYJ 1997. Tyrosine phosphorylation of RNA polymerase II carboxyl-terminal domain by the Abl-related gene product. J Biol Chem 272: 18905–18909 [DOI] [PubMed] [Google Scholar]
- Baskaran R, Escobar SR, Wang JYJ 1999. Nuclear c-Abl is a COOH-terminal repeated domain (CTD)-tyrosine (CTD)-tyrosine kinase-specific for the mammalian RNA polymerase II: Possible role in transcription elongation. Cell Growth Differ 10: 387–396 [PubMed] [Google Scholar]
- Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F 2012. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 45: 158–170 [DOI] [PubMed] [Google Scholar]
- Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN 2008. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J 27: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker R, Loll B, Meinhart A 2008. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J Biol Chem 283: 22659–22669 [DOI] [PubMed] [Google Scholar]
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301 [DOI] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, Cimermancic P, Ule J, Peterlin BM 2011. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 25: 2158–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bres V, Gomes N, Pickle L, Jones KA 2005. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev 19: 1211–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Manley JL 2003. Strange bedfellows: Polyadenylation factors at the promoter. Genes Dev 17: 1321–1327 [DOI] [PubMed] [Google Scholar]
- Calvo O, Manley JL 2005. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J 24: 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Chanarat S, Seizl M, Strasser K 2011. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev 25: 1147–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Palancade B, Lang A, Bendaude O, Eick D 2004. The last CTD repeat of the mammalian RNA polymerase II large subunit is important for its stability. Nucleic Acids Res 32: 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Conrad M, Eick D 2005. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol Cell Biol 25: 7665–7674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Hintermair C, Eick D 2008. Molecular evolution of the RNA polymerase II CTD. Trends Genet 24: 289–296 [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Price DH 2007. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem 282: 21901–21912 [DOI] [PubMed] [Google Scholar]
- Chesnut JD, Stephens JH, Dahmus ME 1992. The interaction of RNA polymerase II with the adenovirus-2 major late promoter is precluded by phosphorylation of the C-terminal domain of subunit IIa. J Biol Chem 267: 10500–10506 [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang XL, Young RA, Annan RS, Carr SA, Deshaies RJ 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev 15: 1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Yamamoto J, Yamaguchi Y, Handa H 2010. Promoter-proximal pausing and its release: Molecular mechanisms and physiological functions. Exp Cell Res 316: 2723–2730 [DOI] [PubMed] [Google Scholar]
- Chinchilla K, Rodriguez-Molina JB, Ursic D, Finkel JS, Ansari AZ, Culbertson MR 2012. Interactions of Sen1, Nrd1, and Nab3 with multiple phosphorylated forms of the Rpb1 C-terminal domain in Saccharomyces cerevisiae. Eukaryot Cell 11: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev 11: 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D 1999. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev 13: 1540–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 15: 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek LJ, Corden JL 1989. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature 339: 679–684 [DOI] [PubMed] [Google Scholar]
- Clemente-Blanco A, Sen N, Mayan-Santos M, Sacristan MP, Graham B, Jarmuz A, Giess A, Webb E, Game L, Eick D, et al. 2011. Cdc14 phosphatase promotes segregation of telomeres through repression of RNA polymerase II transcription. Nat Cell Biol 13: 1450–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin J, Libri D, Porrua O 2011. Cryptic transcription and early termination in the control of gene expression. Genet Res Int 2011: 653494 doi: 10.4061/2011/653494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S, Manley JL 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev 2: 440–452 [DOI] [PubMed] [Google Scholar]
- Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola JA, Field AS, Luse DS 1983. Promoter-proximal pausing by RNA polymerase II in vitro: Transcripts shorter than 20 nucleotides are not capped. Proc Natl Acad Sci 80: 1251–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL, Patturajan M 1997. A CTD function linking transcription to splicing. Trends Biochem Sci 22: 413–416 [DOI] [PubMed] [Google Scholar]
- Corden JL, Cadena DL, Ahearn JM Jr, Dahmus ME 1985. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci 82: 7934–7938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P 2004. RNA polymerase II structure: From core to functional complexes. Curr Opin Genet Dev 14: 218–226 [DOI] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell 29: 271–278 [DOI] [PubMed] [Google Scholar]
- Dantonel J-C, Murthy KGK, Manley JL, Tora L 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389: 399–402 [DOI] [PubMed] [Google Scholar]
- Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev 20: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Manley JL 2011. The RNA polymerase C-terminal domain: A new role in spliceosome assembly. Transcription 2: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Boyne AR, Millhouse SR, Manley JL 2011. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65–Prp19 complex. Genes Dev 25: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. 2011. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 18: 977–983 [DOI] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME 2008. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE 3: e3273 doi: 10.1371/journal.pone.0003273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ III, Singer DS 2012. BRD4 is an atypical kinase that phosphorylates Serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci 109: 6927–6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W 2002a. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol Cell 10: 1139–1150 [DOI] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Sadowski M, Hubner W, Weiser S, Keller W 2002b. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J 21: 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F 2010. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet 6: e1001173 doi: 10.1371/journal.pgen.1001173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ 2002. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem 277: 40156–40162 [DOI] [PubMed] [Google Scholar]
- Eberle AB, Hessle V, Helbig R, Dantoft W, Gimber N, Visa N 2010. Splice-site mutations cause Rrp6-mediated nuclear retention of the unspliced RNAs and transcriptional down-regulation of the splicing-defective genes. PLoS ONE 5: e11540 doi: 10.1371/journal.pone.0011540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL 2008. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 27: 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Murphy S 2008. Cracking the RNA polymerase II CTD code. Trends Genet 24: 280–288 [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S 2007. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S 2008. Expression of human snRNA genes from beginning to end. Biochem Soc Trans 36: 590–594 [DOI] [PubMed] [Google Scholar]
- Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S 2010. The Integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem 285: 20564–20569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S 2012. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell 45: 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8: 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrega C, Shen V, Shuman S, Lima CD 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell 11: 1549–1561 [DOI] [PubMed] [Google Scholar]
- Feaver WJ, Gileadi O, Li Y, Kornberg RD 1991. CTD kinase associated with yeast RNA polymerase-II initiation factor-b. Cell 67: 1223–1230 [DOI] [PubMed] [Google Scholar]
- Fong N, Bentley DL 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes Dev 15: 1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Bird G, Vigneron M, Bentley DL 2003. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J 22: 4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Cujec TP, Peng J, Garriga J, Price DH, Grana X, Peterlin BM 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J Virol 72: 7154–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ 2002. Promoter proximal splice sites enhance transcription. Genes Dev 16: 2792–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Donner AJ, Espinosa JM 2010. CDK8: A positive regulator of transcription. Transcription 1: 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem C, Devaux F, Torchet C, Jacq C, Quevillon-Cheruel S, Labesse G, Facca C, Faye G 2003. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J 22: 1588–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber HP, Hagmann M, Seipel K, Georgiev O, West MAL, Litingtung Y, Schaffner W, Corden JL 1995. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature 374: 660–662 [DOI] [PubMed] [Google Scholar]
- Ghazy MA, He X, Singh BN, Hampsey M, Moore C 2009. The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensable for pre-mRNA 3′-end processing. Mol Cell Biol 29: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Garcia-Blanco MA 2000. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA 6: 1325–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Shuman S, Lima CD 2008. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol Cell 32: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Shuman S, Lima CD 2011. Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell 43: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL 2008. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15: 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL 2009. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol 29: 5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, de Godoy LM, Cox J, Neuhauser N, Ren S, Olsen JV, Mann M 2009. High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics 9: 4642–4652 [DOI] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell 39: 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D 2008. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol 15: 786–794 [DOI] [PubMed] [Google Scholar]
- Guo Z, Stiller JW 2004. Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genomics 5: 69 doi: 10.1186/1471-2164-5-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Singh BN, Ansari A, Laine JP, Krishnamurthy S 2011. Control of eukaryotic gene expression: Gene loops and transcriptional memory. Adv Enzyme Regul 51: 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Shuman S 2002. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. J Biol Chem 277: 21213–21220 [DOI] [PubMed] [Google Scholar]
- He X, Khan AU, Cheng H, Pappas DL Jr, Hampsey M, Moore CL 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev 17: 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell 2: 43–53 [DOI] [PubMed] [Google Scholar]
- Herrmann CH, Rice AP 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: Candidate for a Tat cofactor. J Virol 69: 1612–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, et al. 2012. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J 31: 2784–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 2000. RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Hirose Y, Ohkuma Y 2007. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem 141: 601–608 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev 13: 1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilicova J, Stanek D 2011. Where splicing joins chromatin. Nucleus 2: 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Honorine R, Mosrin-Huaman C, Hervouet-Coste N, Libri D, Rahmouni AR 2011. Nuclear mRNA quality control in yeast is mediated by Nrd1 co-transcriptional recruitment, as revealed by the targeting of Rho-induced aberrant transcripts. Nucleic Acids Res 39: 2809–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, Sheth A, Manley JL 2011. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science 334: 683–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. 2004. Menin associates with a Trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 13: 587–597 [DOI] [PubMed] [Google Scholar]
- Jacobs EY, Ogiwara I, Weiner AM 2003. Role of the C-terminal domain of RNA polymerase II in U2 snRNA transcription and 3′ processing. Mol Cell Biol 24: 846–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A 2009. The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10: 833–844 [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Jove R, Manley JL 1984. In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J Biol Chem 259: 8513–8521 [PubMed] [Google Scholar]
- Kamenski T, Heilmeier S, Meinhart A, Cramer P 2004. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol Cell 15: 399–407 [DOI] [PubMed] [Google Scholar]
- Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM 2003. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene 22: 5707–5711 [DOI] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL 2007. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev 21: 1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Dahmus ME, Hart GW 1993. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem 268: 10416–10424 [PubMed] [Google Scholar]
- Keogh MC, Podolny V, Buratowski S 2003. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol 23: 7005–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137: 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S 2004a. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogen NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt J, Buratowski S 2004b. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S 2006. Distinct pathways for snoRNA and mRNA termination. Mol Cell 24: 723–734 [DOI] [PubMed] [Google Scholar]
- Kim M, Suh H, Cho EJ, Buratowski S 2009. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J Biol Chem 284: 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL 2010. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD 2005. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 25: 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor MS, Archambault J, Lester W, Holstege FCP, Gileadi O, Jansma DB, Jennings EG, Kouyoumdjian F, Davidson AR, Young RA, et al. 1999. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell 4: 55–62 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M 2004. Ssu72 is an RNA polymerase II CTD phosphatase. Mol Cell 14: 387–394 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S, Ghazy MA, Moore C, Hampsey M 2009. Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries. Mol Cell Biol 29: 2925–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Larochelle S, Batliner J, Gamble MJ, Barboza NM, Kraybill BC, Blethrow JD, Shokat KM, Fisher RP 2006. Dichotomous but stringent substrate selection by the dual-function Cdk7 complex revealed by chemical genetics. Nat Struct Mol Biol 13: 55–62 [DOI] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL 1997. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem 272: 10990–10993 [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG 2008. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 28: 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M 2011. Paused RNA polymerase II as a developmental checkpoint. Cell 145: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378 [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL 2006. Cotranscriptional processes and their influence on genome stability. Genes Dev 20: 1838–1847 [DOI] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR III, Workman JL 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278: 8897–8903 [DOI] [PubMed] [Google Scholar]
- Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P 2005. Solution structure of the Set2–Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci 102: 17636–17641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang Z, Wang B, Zhang J, Zhao Y, Jin Y 2007. Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol Cell Biol 27: 5296–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao SM, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, Viljoen M, van Vuuren HJJ, Young RA 1995. A kinase–cyclin pair in the RNA polymerase II holoenzyme. Nature 374: 193–196 [DOI] [PubMed] [Google Scholar]
- Licatalosi DS, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Lin PS, Dubois MF, Dahmus ME 2002. TFIIF-associating carboxyl-terminal domain phosphatase dephosphorylates phosphoserines 2 and 5 of RNA polymerase II. J Biol Chem 277: 45949–45956 [DOI] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15: 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y, Lawler AM, Sebald SM, Lee E, Gearhart JD, Westphal H, Corden JL 1999. Growth retardation and neonatal lethality in mice with a homozygous deletion in the C-terminal domain of RNA polymerase II. Mol Gen Genet 261: 100–105 [DOI] [PubMed] [Google Scholar]
- Liu P, Greenleaf AL, Stiller JW 2008. The essential sequence elements required for RNAP II carboxyl-terminal domain function in yeast and their evolutionary conservation. Mol Biol Evol 25: 719–727 [DOI] [PubMed] [Google Scholar]
- Liu P, Kenney JM, Stiller JW, Greenleaf AL 2010. Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Mol Biol Evol 27: 2628–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zawel L, Fisher L, Egly JM, Reinberg D 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358: 641–645 [DOI] [PubMed] [Google Scholar]
- Lu KP, Hanes SD, Hunter T 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380: 544–547 [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Reichow SL, Kim M, Suh H, Leeper TC, Yang F, Mutschler H, Buratowski S, Meinhart A, Varani G 2010. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat Struct Mol Biol 17: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Johnson AW, Bentley DL 2006. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: Implications for a unified allosteric-torpedo model. Genes Dev 20: 954–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Manley JL 2002. Nuclear coupling: RNA processing reaches back to transcription. Nat Struct Biol 9: 790–791 [DOI] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH 2010. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 40: 410–422 [DOI] [PubMed] [Google Scholar]
- Marciniak RA, Sharp PA 1991. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J 10: 4189–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 270: 12335–12338 [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271: 27176–27183 [DOI] [PubMed] [Google Scholar]
- Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M 2011. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat Struct Mol Biol 18: 1115–1123 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P 2012a. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336: 1723–1725 [DOI] [PubMed] [Google Scholar]
- Mayer A, Schreieck A, Lidschreiber M, Leike K, Martin DE, Cramer P 2012b. The spt5 C-terminal region recruits yeast 3′ RNA cleavage factor I. Mol Cell Biol 32: 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Amgen EST Program, Shuman S, et al. 1997a. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev 11: 3306–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997b. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S 2003. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J 22: 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Cramer P 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430: 223–226 [DOI] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P 2005. A structural perspective of CTD function. Genes Dev 19: 1401–1415 [DOI] [PubMed] [Google Scholar]
- Meininghaus M, Chapman RD, Horndasch M, Eick D 2000. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J Biol Chem 275: 24375–24382 [DOI] [PubMed] [Google Scholar]
- Millhouse S, Manley JL 2005. The C-terminal domain of RNA polymerase II functions as a phosphorylation-dependent splicing activator in a heterologous protein. Mol Cell Biol 25: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL 2005. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci 102: 14765–14770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell 3: 697–705 [DOI] [PubMed] [Google Scholar]
- Morris DP, Greenleaf AL 2000. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 275: 39935–39943 [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci 93: 8253–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP 2009. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell 34: 168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ, de la Mata M, Kornblihtt AR 2010. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem Sci 35: 497–504 [DOI] [PubMed] [Google Scholar]
- Murray S, Udupa R, Yao S, Hartzog G, Prelich G 2001. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol Cell Biol 21: 4089–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Manley JL 2011. Transcriptional activators enhance polyadenylation of mRNA precursors. RNA Biol 8: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL 2011. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell 41: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J 2003. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem 278: 33000–33010 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell 13: 55–65 [DOI] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ 2004. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Park NJ, Tsao DC, Martinson HG 2004. The two steps of poly(A)-dependent termination, pausing and release, can be uncoupled by truncation of the RNA polymerase II carboxyl-terminal repeat domain. Mol Cell Biol 24: 4092–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patturajan M, Wei X, Berezney R, Corden JL 1998. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol Cell Biol 18: 2406–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA 2010. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol 20: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Marshall NF, Price DH 1998a. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J Biol Chem 273: 13855–13860 [DOI] [PubMed] [Google Scholar]
- Peng J, Zhu Y, Milton JT, Price DH 1998b. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev 12: 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D 2009. ‘Cotranscriptionality’: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936 [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Jones JC, Greenleaf AL 2004. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: Novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 43: 15702–15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA 2009. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep 10: 894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ 1989. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci 14: 105–110 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ 2002. Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Hinnebusch AG 2009. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell 33: 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH 2001. A highly purified RNA polymerase II elongation control system. J Biol Chem 276: 42601–42609 [DOI] [PubMed] [Google Scholar]
- Richard P, Kiss T 2006. Integrating snoRNP assembly with mRNA biogenesis. EMBO Rep 7: 590–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL 2009. Transcription termination by nuclear RNA polymerases. Genes Dev 23: 1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol 20: 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ 2009. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell 36: 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Blencowe BJ 2004. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA 10: 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ 2005. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol 25: 6734–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Duncan SM, Manley JL 2012. Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev 26: 350–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M 2009. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci 106: 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Murthy KGK, Kaneko S, Manley JL 2002. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol Cell Biol 22: 1684–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M, Dichtl B, Hubner W, Keller W 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J 22: 2167–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Shuman S 2011. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell 43: 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, Frank J, Manley JL 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 2012. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski TW, Buratowski S 2009. The basal initiation machinery: Beyond the general transcription factors. Curr Opin Cell Biol 21: 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ III, Eick D, Reinberg D 2011. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BN, Hampsey M 2007. A transcription-independent role for TFIIB in gene looping. Mol Cell 27: 806–816 [DOI] [PubMed] [Google Scholar]
- Singh N, Ma Z, Gemmill T, Wu X, Defiglio H, Rossettini A, Rabeler C, Beane O, Morse RH, Palumbo MJ, et al. 2009. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol Cell 36: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar DA, Greenleaf AL 2002. The RNA polymerase II CTD kinase CTDK-I affects pre-mRNA 3′ cleavage/polyadenylation through the processing component Pti1p. Mol Cell 10: 1429–1439 [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42: 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain MM, Govind CK 2011. A role for phosphorylated Pol II CTD in modulating transcription coupled histone dynamics. Transcription 2: 78–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol 16: 6993–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA 1998. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci 95: 6699–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell 24: 735–746 [DOI] [PubMed] [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL 1995. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin–cyclin-dependent kinase complex. Mol Cell Biol 15: 5716–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, Cook MS 2004. Functional unit of the RNA polymerase II C-terminal domain lies within heptapeptide pairs. Eukaryot Cell 3: 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, McConaughy BL, Hall BD 2000. Evolutionary complementation for polymerase II CTD function. Yeast 16: 57–64 [DOI] [PubMed] [Google Scholar]
- Suh MH, Meyer PA, Gu M, Ye P, Zhang M, Kaplan CD, Lima CD, Fu J 2010. A dual interface determines the recognition of RNA polymerase II by RNA capping enzyme. J Biol Chem 285: 34027–34038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Hampsey M 1996. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol 16: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D 1998. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell 2: 213–222 [DOI] [PubMed] [Google Scholar]
- Sun XJ, Wei J, Wu XY, Hu M, Wang L, Wang HH, Zhang QH, Chen SJ, Huang QH, Chen Z 2005. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 280: 35261–35271 [DOI] [PubMed] [Google Scholar]
- Sun M, Lariviere L, Dengl S, Mayer A, Cramer P 2010. A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD). J Biol Chem 285: 41597–41603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera A, Lim Y, Woods R, Birrell GW, Nasim T, Becherel OJ, Lavin MF 2009. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum Mol Genet 18: 3384–3396 [DOI] [PubMed] [Google Scholar]
- Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, et al. 2010. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 17: 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao DC, Park NJ, Nag A, Martinson HG 2012. Prolonged α-amanitin treatment of cells for studying mutated polymerases causes degradation of DSIF160 and other proteins. RNA 18: 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D, Chinchilla K, Finkel JS, Culbertson MR 2004. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res 32: 2441–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A 2008. The Nrd1–Nab3–Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP 2000. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nature 7: 639–643 [DOI] [PubMed] [Google Scholar]
- Wang Y, Fairley JA, Roberts SG 2010. Phosphorylation of TFIIB links transcription initiation and termination. Curr Biol 20: 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P 2011. Cis-proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem 286: 5717–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML, Gorden JL 1995. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ 2008. Human Pcf11 enhances degradation of RNA polymerase II-associated nascent RNA and transcriptional termination. Nucleic Acids Res 36: 905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ 2004. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525 [DOI] [PubMed] [Google Scholar]
- Wood A, Shilatifard A 2006. Bur1/Bur2 and the Ctk complex in yeast: The split personality of mammalian P-TEFb. Cell Cycle 5: 1066–1068 [DOI] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev 17: 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Behav MM, Manley JL, Tong L 2010. Crystal structure of the human symplekin–Ssu72–CTD phosphopeptide complex. Nature 467: 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Manley JL, Tong L 2012. The yeast regulator of transcription protein Rtr1 lacks an active site and phohphatase activity. Nat Commun 3: 946 doi: 10.1038/ncomms1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 17: 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YX, Manley JL 2007. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev 21: 2950–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YX, Hirose Y, Zhou XZ, Lu KP, Manley JL 2003. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev 17: 2765–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirchner MW, Fischer G, et al. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science 278: 1957–1960 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97: 41–51 [DOI] [PubMed] [Google Scholar]
- Yao S, Neiman A, Prelich G 2000. BUR1 and BUR2 encode a divergent cyclin-dependent kinase–cyclin complex important for transcription in vivo. Mol Cell Biol 20: 7080–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M, Lin PS, Dahmus ME, Gill GN 2003. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J Biol Chem 278: 26078–26085 [DOI] [PubMed] [Google Scholar]
- Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN 2005. Small CTD phosphatases function in silencing neuronal gene expression. Science 307: 596–600 [DOI] [PubMed] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA 2007. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev 21: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA 2008. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev 22: 3422–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci 93: 6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gilmour DS 2006. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell 21: 65–74 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wu CH, Gilmour DS 2004. Analysis of polymerase II elongation complexes by native gel electrophoresis. Evidence for a novel carboxyl-terminal domain-mediated termination mechanism. J Biol Chem 279: 23223–23228 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Fu J, Gilmour DS 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev 19: 1572–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kim Y, Genoud N, Gao J, Kelly JW, Pfaff SL, Gill GN, Dixon JE, Noel JP 2006. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell 24: 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ 2012. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J Biol Chem 287: 8541–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol 20: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]