The Hippo signaling pathway plays a role in cell proliferation, apoptosis, and tumorigenesis. In this study, Mo et al. identify upstream activators of the Hippo pathway. They present evidence that links protease-activated receptor (PAR) signaling through G12/13 to the dephosphorylation of the transcription factors YAP and TAZ through Rho-mediated suppression of Lats1/2. This work shows that thrombin acts as a signal for the Hippo pathway through the PAR pathway.
Keywords: PAR, thrombin, GPCR, Hippo, YAP, cancer
Abstract
The Hippo signaling pathway plays a crucial role in tissue growth and tumorigenesis. Core components of the Hippo pathway include the MST1/2 and Lats1/2 kinases. Acting downstream from the Hippo pathway are the YAP/TAZ transcription coactivators, which are inhibited through phosphorylation by Lats. However, upstream signals that regulate the Hippo pathway have not been well delineated. Here we report that stimulation of protease-activated receptors (PARs) activates YAP/TAZ by decreasing phosphorylation and increasing nuclear localization. PAR1 acts through G12/13 and Rho GTPase to inhibit the Lats1/2 kinase. Our observations establish thrombin as a physiological signal for the Hippo pathway and implicate Hippo–YAP as a key downstream signaling branch of PAR activation.
The Hippo signaling pathway, initially discovered by genetic studies in Drosophila as a regulator of organ size, limits cell number via the regulation of cell proliferation and apoptosis (Pan 2010). Core components of the Hippo pathway are highly conserved and also play an important role in mammalian organ size regulation (Zhao et al. 2010a). In the Hippo pathway, a conserved kinase cascade—in which the protein kinase MST1/2 (homolog of the Drosophila Hippo) phosphorylates and activates the Lats1/2 kinases—functions to inhibit the YAP and TAZ transcription coactivators by phosphorylation. Dephosphorylated YAP/TAZ localize in the nucleus and function as transcription coactivators for the TEAD family of transcription factors to induce gene expression, including connective tissue growth factor (CTGF) and Cyr61, thereby promoting cell growth, proliferation, and survival (Lei et al. 2008; Zhao et al. 2008, 2010b).
YAP has been implicated as the candidate oncogene in the human chromosome 11q22 amplicon, and mutations or abnormal expression of Hippo pathway components are associated with human tumorigenesis (Overholtzer et al. 2006; Zeng and Hong 2008). In addition, YAP expression and nuclear localization are elevated in human cancers, and TAZ is up-regulated in high-grade and metastatic breast tumors (Chan et al. 2008; Zhao et al. 2010a; Cordenonsi et al. 2011). Moreover, YAP and TAZ induce an epithelial–mesenchymal transition (EMT), a phenomenon crucial for tumor metastasis (Gupta and Massague 2006). These studies have demonstrated an important role of the Hippo pathway in tumorigenesis.
A large number of proteins have been implicated to act upstream of and regulate the Hippo pathway (Grusche et al. 2010; Zhao et al. 2010a). However, extracellular signals that regulate this pathway are largely unknown. Thrombin acts on protease-activated receptors (PARs), a subfamily of G protein-coupled receptors (GPCR) that participate in a variety of biological process, including chemokine and cytokine release, tissue remodeling, inflammation, proliferation, and angiogenesis (Coughlin 2000; Macfarlane et al. 2001). The PAR1 receptor subtype is activated when the N terminus is proteolytically cleaved by the serine protease thrombin, resulting in an irreversible activation of the receptor. Downstream signaling events are regulated by PAR1 coupling to heterotrimeric GTP-binding proteins, including G12/13, Gq/11, and Gi/o, to activate a variety of signaling cascades (Coughlin 2005; McLaughlin et al. 2005). Several reports have demonstrated that thrombin increases the expression of CTGF and Cyr61 (Chambers et al. 2000; Pendurthi et al. 2002; Walsh et al. 2008). PARs have recently been implicated to play a central role in tumor cell growth, invasion, and cancer metastasis, and up-regulation of receptor expression has been detected in tumor tissues, including melanomas; colon, breast, lung, pancreatic, and prostate cancer; and oral squamous cell carcinoma (Booden et al. 2004; Elste and Petersen 2010; Bar-Shavit et al. 2011; Ramachandran et al. 2012).
In this study, we investigated the functional relationship between thrombin signaling and YAP/TAZ regulation. We show that thrombin and PAR agonist peptides activate YAP/TAZ. Stimulation of PAR by agonist peptides activates YAP/TAZ by inducing dephosphorylation and nuclear localization. PAR1 acts through G12/13, Rho, Lats1/2, and YAP/TAZ to stimulate gene expression, cell migration, and cell invasion. Our study reveals a novel function of thrombin in Hippo pathway regulation and a mechanism linking PAR signaling and the Hippo pathway.
Results and Discussion
Stimulation of PARs induces YAP dephosphorylation and nuclear localization
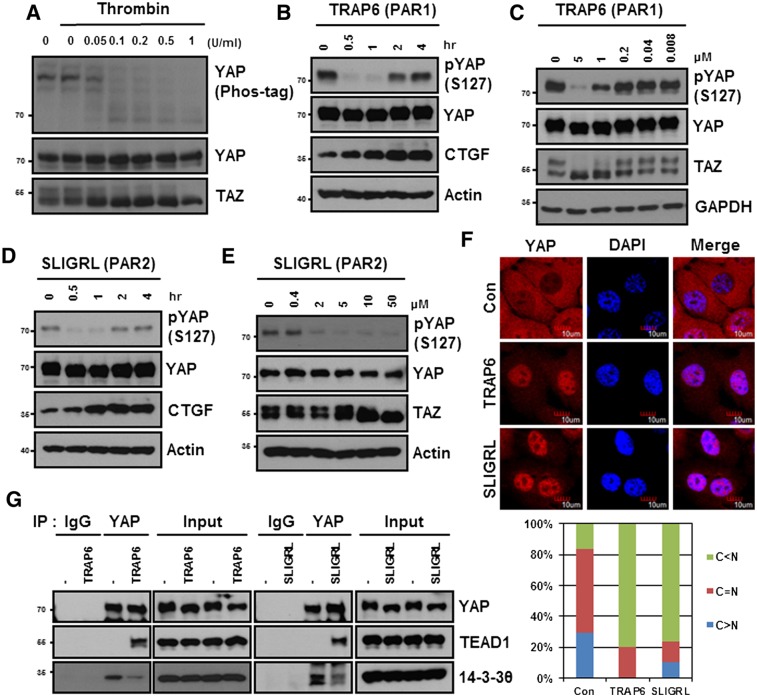
We established that the CTGF and Cyr61 are bona fide YAP target genes (Zhao et al. 2008). Activation of the thrombin receptor (PAR1) has been reported to induce expression of both CTGF and Cyr61 (Chambers et al. 2000; Pendurthi et al. 2000; Walsh et al. 2008). These observations suggested that thrombin may activate YAP to induce CTGF and Cyr61. In serum-starved HEK293A cells, YAP was highly phosphorylated, and addition of thrombin induced robust YAP dephosphorylation as determined by the YAP mobility shift in a phos-tag gel (Fig. 1A). Thrombin also caused a mobility shift of TAZ, suggesting that TAZ was dephosphorylated in response to thrombin. Since thrombin can act on any of three distinct receptors (PAR1, PAR3, and PAR4), we examined the effect of a selective PAR1-activating peptide, TRAP6, on YAP phosphorylation. We found that addition of the PAR1-activating peptide TRAP6 resulted in a rapid YAP dephosphorylation as determined by a phospho-YAP antibody (S127) (Fig. 1B). As a result of YAP activation, CTGF protein expression was also induced by TRAP6 treatment. TRAP6 induced dephosphorylation of YAP in a dose-dependent manner (Fig. 1C). This effect of TRAP6 on YAP phosphorylation was transient, as the YAP phosphorylation was partially recovered 2 h after TRAP6 treatment (Fig. 1B; Supplemental Fig. S1A). However, we examined multiple cell lines and found that TRAP6 but not SLIGRL decreased YAP phosphorylation in HeLa cells (Supplemental Fig. S1C). Activation of PAR2 by a SLIGRL peptide potently induced YAP dephosphorylation and CTGF accumulation in both a time- and a dose-dependent manner (Fig. 1D,E; Supplemental Fig. S1B). SLIGRL also induced YAP dephosphorylation in human umbilical vein endothelial cells (HUVECs) (Supplemental Fig. S1D). Both TRAP6 and SLIGRL potently repressed the phosphorylation of YAP in MDA-MB-231 cells (Supplemental Fig. S1E). The differential response of PAR-activating peptides in various cell lines could be due to the relative expression of different PAR receptors in these cell lines. The above data demonstrate a physiological regulation of YAP/TAZ activation by thrombin receptor activation.
Figure 1.
Agonists of PARs induce dephosphorylation and activation of YAP and TAZ. (A) Thrombin induces YAP dephosphorylation. HEK293A cells were serum-starved for 24 h and then stimulated with thrombin at various doses (units per milliliter) for 30 min. YAP phosphorylation was determined by phos-tag. Thrombin also induced TAZ mobility shift. The numbers on the left of each panel denote molecular weight markers. (B,D) HEK293A cells were serum-starved for 12 h and then stimulated with TRAP6 or SLIGRL for the indicated durations. Cells were lysed and subjected to immunoblotting with the indicated antibodies. (C,E). HEK293A cells were stimulated with different concentrations of TRAP6 or SLIGRL for 1 h. (F) YAP subcellular localization was determined by immunofluorescence staining for endogenous YAP (red); DAPI (blue) was used for cell nuclei. Cells were stimulated with TRAP6 and SLIGRL for 1 h. The images shown represent typical staining pattern of the majority cells. Quantifications are shown in the bottom panel. (G) TRAP6 and SLIGRL stimulate YAP interaction with TEAD1. Cell lysates were subjected to immunoprecipitation with control IgG or YAP antibody. The coimmunoprecipitated TEAD1 and 14-3-3 were detected by immunoblotting.
We next determined the effect of PAR1 on YAP nuclear localization. Phosphorylation of YAP S127 by Lats1/2 causes cytoplasmic sequestration of YAP due to 14-3-3 binding. The dephosphorylated YAP resides in the cell nucleus and induces gene expression by interacting with its cognate transcription factors, TEAD1–4 (Zhao et al. 2007, 2008). As expected, TRAP6 caused significant nuclear accumulation of YAP (Fig. 1F; Supplemental Fig. S1F). Moreover, TRAP6 increased the interaction between YAP and TEAD1 and decreased the interaction between YAP and 14-3-3 (Fig. 1G). SLIGRL similarly increased YAP nuclear localization and interaction with TEAD1 (Fig. 1F,G). Based on the above data, we conclude that stimulation of PAR1 and PAR2 activates YAP by inducing dephosphorylation and increasing nuclear localization.
TRAP6 acts through GPCRs and G12/13 to induce YAP/TAZ dephosphorylation
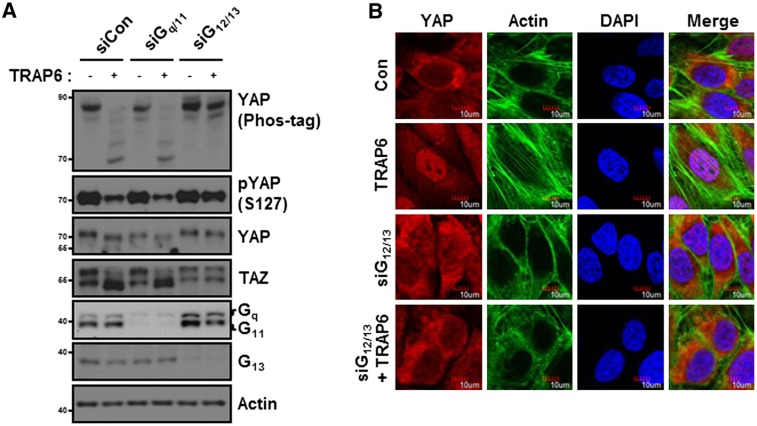
PAR1 activates several trimeric Gα proteins (G12/13, Gq/11, or Gi/o) to initiate intracellular signaling pathways (Offermanns et al. 1994; McLaughlin et al. 2005). To determine which Gα proteins are involved in YAP regulation, we tested the effect of constitutively active Gα mutants on YAP phosphorylation. Expression of active G12/13 and G11 strongly decreased the basal phosphorylation of YAP and TAZ, whereas expression of Gi had a minor effect on YAP dephosphorylation (Supplemental Fig. S2). We next tested the function of endogenous Gα proteins for PAR to regulate YAP/TAZ phosphorylation by RNAi (Fig. 2A). Knockdown of Gq and G11 had little effect on TRAP6-induced dephosphorylation of YAP and TAZ. However, knockdown of G12/13 strongly blocked the dephosphorylation of YAP by TRAP6 (Fig. 2A). These data show that G12/13 are required for PAR1 to induce dephosphorylation of YAP and TAZ. To further confirm the role of G12/13 in YAP regulation by PAR1, we examined intracellular localization of YAP. In control siRNA-treated cells, TRAP6 induced YAP nuclear localization (Fig. 2B). Consistent with the YAP phosphorylation data, TRAP6 was unable to induce YAP nuclear accumulation in cells treated with G12/13 siRNA. Based on the above observations, we conclude that activation of PAR1 by TRAP6 signals through G12/13 to induce YAP/TAZ dephosphorylation and activation.
Figure 2.
TRAP6 activates YAP/TAZ via G12/13. (A) HEK293A cells were transfected with the indicated siRNAs. YAP phosphorylation was determined by both phos-tag, which detects phosphorylation by enhancing mobility shift, and phospho-YAP (S127) antibody. TAZ phosphorylation can be visualized by mobility shift. The knockdown efficiency of Gq/11 and G12/13 is indicated by immunoblotting. (B) MCF10A cells were transfected with siG12/13 as indicated. Cells were treated with 2 μM TRAP6 for 1 h before fixation. YAP subcellular localization was determined by immunofluorescence staining for endogenous YAP (red) and F-actin (green); DAPI (blue) was used for cell nuclei.
TRAP6 modulates YAP/TAZ dephosphorylation via a Rho and actin cytoskeleton, but Rho kinase (ROCK)-independent, pathway
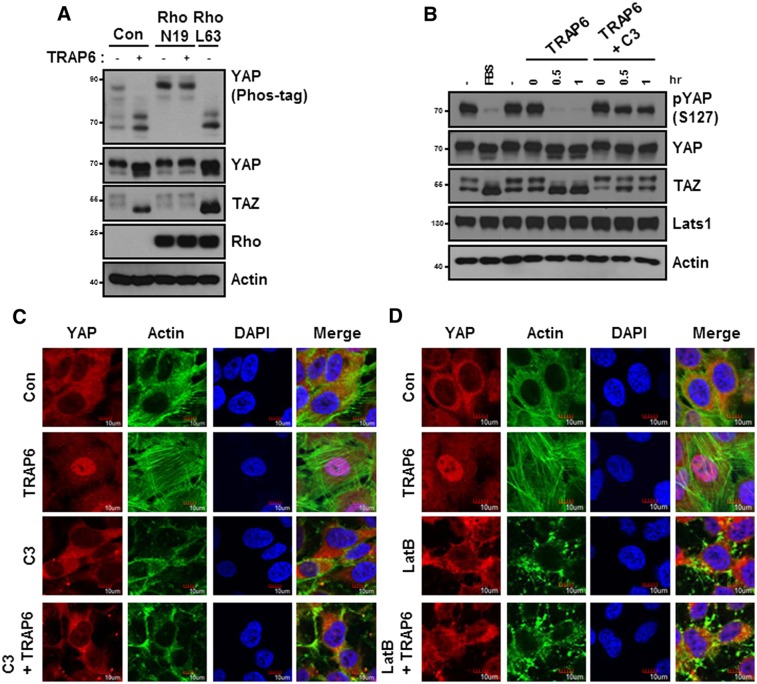
The Rho GTPases represent the major downstream signaling module of G12/13, which binds to and activates the Rho guanine nucleotide exchange factor (GEF) (Martin et al. 2001). Consistent with a role of G12/13 in PAR signaling to YAP, Rho GTPase has been recently reported to induce YAP dephosphorylation (Dupont et al. 2011; Zhao et al. 2012). We tested the function of Rho in TRAP6-induced YAP dephosphorylation by using dominant-negative Rho-N19. Notably, expression of Rho-N19 blocked TRAP6-induced YAP dephosphorylation (Fig. 3A). Conversely, the constitutively active Rho-L63 mutant induced dephosphorylation of YAP. To further determine the function of endogenous Rho in YAP regulation, we tested the effect of botulinum toxin C3, a specific inhibitor of Rho GTPase, on YAP phosphorylation. C3 treatment strongly suppressed YAP/TAZ dephosphorylation in response to TRAP6 treatment (Fig. 3B). Consistent with the changes in YAP phosphorylation, C3 treatment blocked the TRAP6-induced YAP nuclear localization, indicating that Rho activation is required for TRAP6 to modulate the Hippo pathway activity (Fig. 3C).
Figure 3.
TRAP6 activates YAP/TAZ through Rho and cytoskeletons. (A) HEK293A cells were transiently cotransfected with the indicated plasmids and were treated with 2 μM TRAP6 for 1 h. The cells were then lysed and subjected to immunoblotting analysis with the indicated antibodies. (B) HEK293A cells were pretreated with 2 μg/mL C3 for 4 h and then stimulated with TRAP6 for the indicated durations. The presence of FBS is indicated. (C) MCF10A cells were pretreated with 2 μg/mL C3 for 4 h and then incubated with TRAP6 for 1 h. After fixation, YAP subcellular localization was determined by immunofluorescence staining for endogenous YAP (red), F-actin (green); DAPI (blue) was used for cell nuclei. (D) MCF10A cells were pretreated with Latrunculin B for 10 min and then stimulated with TRAP6 for 1 h. Immunofluorescence staining is similar to that shown in C.
One of the direct downstream effectors of Rho is the ROCK kinase, which binds to and is stimulated by active Rho. We examined the function of ROCK. Inhibition of ROCK by H1152 effectively inhibited phosphorylation of its known downstream target, MYPT, but had a marginal effect on TRAP6-induced YAP dephosphorylation and nuclear localization. These data suggest that Rho regulates YAP/TAZ phosphorylation by a mechanism independent of ROCK (Supplemental Fig. S3A,B). The Rho GTPase potently modulates actin cytoskeletal organization, particularly stress fiber formation (Vouret-Craviari et al. 1998). Interestingly, cytoskeletal rearrangement has been shown to regulate the Hippo pathway and YAP phosphorylation, although the precise biochemical mechanism is currently unknown (Dupont et al. 2011; Fernandez et al. 2011; Rauskolb et al. 2011; Sansores-Garcia et al. 2011; Zhao et al. 2012). To test the involvement of cytoskeletal reorganization in YAP regulation by TRAP6, we used cytoskeleton-disrupting reagents. Latrunculin B, which disrupts F-actin, blocked TRAP6-induced YAP dephosphorylation. Similar to Latrunculin B, Cytochalasin D also suppressed the effect of TRAP6, although less efficiently (Supplemental Fig. S4). Consistent with changes in YAP phosphorylation, Latrunculin B treatment prevented YAP nuclear localization in response to TRAP6 (Fig. 3D). There is an apparent correlation between actin stress fiber and YAP nuclear localization. TRAP6 induces actin stress fiber and YAP nuclear accumulation. Knockdown of G12/13 or treatment with the Rho-inactivating toxin C3 disrupted actin stress fiber and YAP nuclear localization (Figs. 2B, 3C,D). These data suggest that the actin cytoskeleton plays an important role in the regulation of YAP phosphorylation and subcellular localization, and the actin cytoskeleton may act downstream from Rho GTPase to affect YAP phosphorylation.
TRAP6 inhibits Lats1/2 but not MST1/2 kinase activity
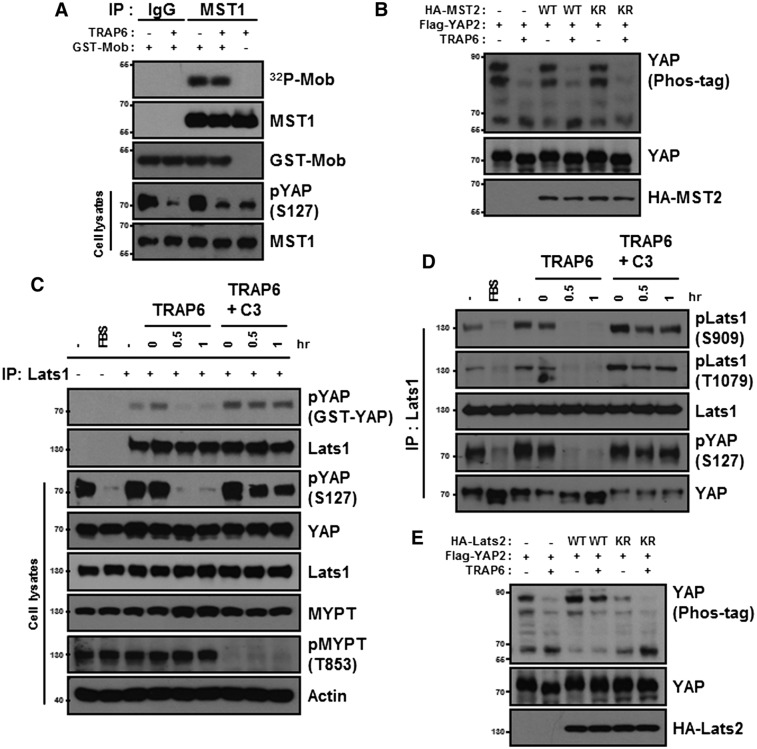
MST1/2 and Lats1/2 form a kinase cascade to regulate YAP/TAZ phosphorylation (Chan et al. 2005). To determine whether MST1/2 are involved, we measured endogenous MST1 kinase activity and found that TRAP6 stimulation had no effect on MST1 kinase activity as indicated by Mob phosphorylation (Fig. 4A). Under the same conditions, endogenous YAP phosphorylation was reduced by TRAP6. Similarly, TRAP6 had no effect on the activity of the ectopically expressed MST2 (Supplemental Fig. S5). Ectopic expression of either MST2-WT or the inactive MST2-KR mutant had little effect on YAP phosphorylation in either the presence or absence of TRAP6 (Fig. 4B). These data suggest that MST1/2 kinase activity is not regulated by TRAP6 and is unlikely to be involved in TRAP6-induced YAP regulation.
Figure 4.
TRAP6 inhibits Lats kinase activity. (A) HEK293A cells were treated with 2 μM TRAP6 for 1 h. The activity of immunoprecipitated MST1 was determined by in vitro kinase assay using GST-Mob as a substrate. P32 incoporation into GST-Mob was detected by autoradiograph. (B) HEK293A cells were cotransfected with the indicated plasmids. After transfection, cells were treated with 2 μM TRAP6 for 1 h. YAP phosphorylation was determined. (C) HEK293A cells were pretreated with 2 μg/mL C3 for 4 h and then incubated with TRAP6 for the indicated times. The presence of FBS is indicated. Lats1 immunoprecipitated from the cell lysates was subjected to in vitro kinase assays using GST-YAP as a substrate. YAP phosphorylation was detected by phospho-YAP (S127) antibody. (D) Stimulation conditions were similar to those in C. Lats1 was immunoprecipitated with anti-Lats1 antibody from the cell lysates and then subjected to immunoblotting as indicated. (E) Flag-YAP was cotransfected with wild-type or a kinase-dead (KR) mutant of HA-Lats2. After transfection, cells were treated with 2 μM TRAP6 for 1 h. (B,E) Phosphorylation of Flag-YAP was determined by phos-tag gel.
To determine whether Lats1/2 kinase is involved in YAP regulation by TRAP6, we measured Lats1 kinase activity in vitro (Fig. 4C). Interestingly, Lats1 was potently inhibited by TRAP6 treatment. Moreover, there is a strong correlation between Lats1 inactivation, as determined by the in vitro kinase assays, and endogenous YAP dephosphorylation, as determined by phospho-YAP (S127) immunoblotting (Fig. 4C). Consistently, inhibition of Rho by C3 blocked the inhibitory effect of TRAP6 on Lats1 kinase activity (Fig. 4D), indicating that Rho activation is required for Lats1 inhibition by TRAP6. Lats1/2 are activated by phosphorylation of the activation loop and the hydrophobic motif (Chan et al. 2005). Using two phospho-specific antibodies, we detected a significant decrease in phosphorylation in both the activation loop and the hydrophobic motif of endogenous Lats1 upon TRAP6 treatment (Fig. 4D). Inhibition of Rho by C3 increased basal phosphorylation of Lats1, and more importantly, C3 effectively blocked the inhibitory effect of TRAP6 on Lats1 phosphorylation (Fig. 4D). These data further support a model in which TRAP6 inhibits Lats1 via Rho activation. Finally, we examined the effect of Lats2 overexpression on YAP phosphorylation in response to TRAP6. Coexpression of wild-type Lats2 effectively blocked TRAP6-induced dephosphorylation of YAP (Fig. 4E). Complementarily, expression of the inactive Lats2-KR mutant decreased YAP2 basal phosphorylation, presumably through a dominant-negative effect on endogenous Lats function. Together, these data demonstrate a direct role of Lats kinase in the regulation of YAP by PAR signaling.
YAP and TAZ are required for PAR1 to stimulate cell migration and cell invasion
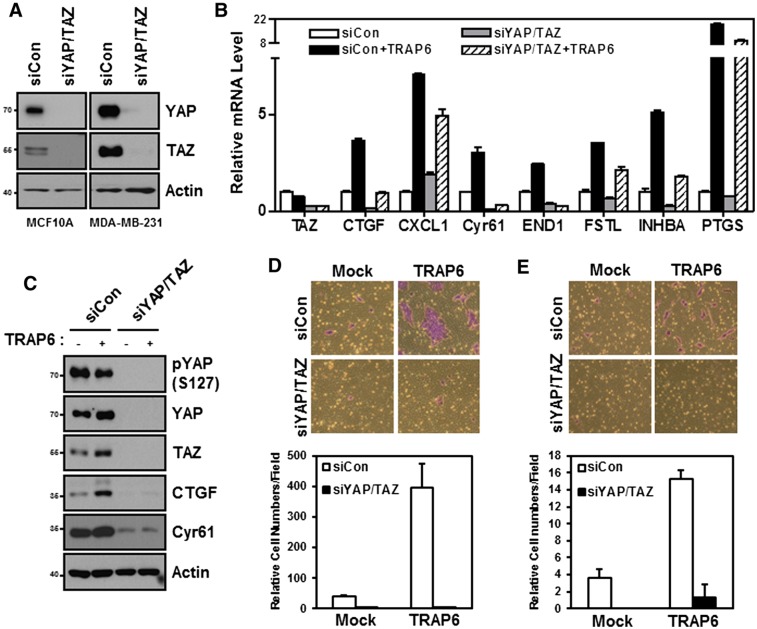
PAR1 stimulation induces the expression of CTGF and Cyr61 (Chambers et al. 2000; Pendurthi et al. 2000; Walsh et al. 2008), which are also the best-characterized YAP target genes. To determine the function of YAP/TAZ in TRAP6-induced gene expression, YAP/TAZ were knocked down by siRNA. The knockdown efficiency was confirmed by immunoblotting (Fig. 5A). Several of the TRAP6-inducible genes, including CTGF, Cyr61, END1, FSTL, INHBA, PTGS, and CXCL1, were examined (Fig. 5B). Knockdown of YAP/TAZ blocked the mRNA induction of CTFG, Cyr61, and END1 in response to TRAP6. The reduced expression of CTGF and Cyr61 was confirmed by Western blotting (Fig. 5C). In addition, YAP/TAZ knockdown partially suppressed the induction of FSTL and PTGS. In contrast, induction of CXCL1 by TRAP6 was largely insensitive to YAP/TAZ knockdown. These results support an important role of YAP/TAZ in TRAP6-dependent gene expression. Not surprisingly, expression of some TRAP6-inducible genes, such as CXCL1, did not depend on YAP/TAZ. This is likely due to the ability of PAR to activate other downstream transcription factors parallel to YAP/TAZ (Bar-Shavit et al. 2011).
Figure 5.
YAP/TAZ mediate physiological functions of PARs in gene induction, cell migration, and invasion. (A) YAP/TAZ knockdown by siRNAs in MCF10A cells (left) and MDA-MB-231 cells (right). Cells were transfected with the indicated siRNAs. The protein levels were determined by immunoblotting. (B,C) MDA-MB-231 cells were transfected with the indicated siRNAs and serum-starved for 12 h. (B) After treating with 2 μM TRAP6 for 4 h, mRNA levels of CTGF, Cyr61, CXCL1, END1, FSTL, INHBA, and PTGS were measured by quantitative PCR. (C) Protein levels of YAP/TAZ, CTGF, and Cyr61 were determined by immunoblotting. (D) MCF10A cells were transiently transfected with the indicated siRNAs. Cell migration was determined by transwell cell migration assay. Cells were stained with crystal violet (top panel) and quantified (bottom panel). (E) MDA-MB-231 cells were transiently transfected with the indicated siRNAs. Cell invasion was determined by Matrigel invasion assay.
The role of YAP/TAZ in modulating cell migration and cell invasion is well established (Chan et al. 2008; Zhang et al. 2008). PAR is known to stimulate cell migration and has been implicated in tumor metastasis (Even-Ram et al. 1998; Tellez and Bar-Eli 2003; Bar-Shavit et al. 2011). We therefore examined the function of YAP/TAZ in mediating the cellular function of PAR. Compared with control cells, TRAP6 stimulated MCF10A cell migration. Knockdown of YAP/TAZ efficiently blocked the cell migration induced by TRAP6 (Fig. 5D; Supplemental Fig. S7). Furthermore, TRAP6-induced cell invasion was suppressed in YAP/TAZ knockdown cells (Fig. 5E). Taken together, these data suggest an important role of YAP and TAZ in mediating the physiological functions of PAR1 on gene induction, cell migration, and invasion.
In this study, we demonstrate that the PAR regulates the Hippo pathway. In addition, we showed recently that the Hippo–YAP pathway is regulated by other GPCR signaling cascades (Yu et al. 2012). Our observations provide new insight into the role of extracellular ligands and GPCRs in the Hippo pathway. We propose that activation of PAR1 and other GPCRs that couple to G12/13 triggers the activation of Rho GTPase, which works through the actin cytoskeleton to inhibit Lats1/2 kinase, leading to eventual YAP/TAZ dephosphorylation, nuclear localization, and gene expression. The precise mechanism by which Rho inhibits Lats requires further investigation. Several recent studies have provided evidence that the actin cytoskeleton modulates the Hippo pathway (Dupont et al. 2011; Fernandez et al. 2011; Rauskolb et al. 2011; Sansores-Garcia et al. 2011; Zhao et al. 2012). Another report suggested that cell morphology and stress fiber/F-actin regulate YAP in a phosphorylation-dependent manner (Wada et al. 2011). We speculate that ligands that signal from outside the cell to Rho GTPase may inhibit Lats1/2 kinases via modulating the actin cytoskeleton.
The connection between PAR1 and the Hippo pathway revealed by this study has several important implications. PAR1 is widely expressed in multiple cell types and has been implicated in a broad spectrum of physiological regulations. Altered PAR signaling is also implicated in pathological conditions, including tumor cell invasion and diseases in the cardiovascular, gastrointestinal, respiratory, and nervous systems (Ramachandran et al. 2012). Activation of PAR1 and other GPCRs that couple to G12/13 and Rho signaling pathways can lead to aberrant cell growth, and ligands for these receptors are increased under pathophysiological conditions. For example, thrombin and PAR1 expression are up-regulated in a tumor microenvironment and malignant or invasive cancer cells (Bar-Shavit et al. 2011; Ramachandran et al. 2012). On the other hand, thrombin is generated at sites of tissue injury to promote wound healing. We speculate that activation of YAP/TAZ by thrombin and PAR1 may play prominent roles in both pathophysiological conditions, such as cancer, and normal physiology, such as wound healing.
Materials and methods
Plasmids
The pCMV-Flag-YAP2, pcDNA3-HA-Lats2 (wild-type and KR), pcDNA3-HA-MST2 (wild-type and KR), Flag-MST2, HA-Sav, GST-Mob, and pRK5-MYC-Rho (wild-type, L63, and N19) constructs were described before (Zhao et al. 2007). G protein plasmids were purchased from the Missouri S&T cDNA Resource Center.
Cell culture and transfection
HEK293A and MDA-MB-231 cells were cultured in DMEM with 10% FBS. MCF10A cells were cultured in DMEM/F12 supplemented with 5% horse serum, 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, 10 μg/mL insulin, 100 ng/mL cholera toxin, and 50 μg/mL penicillin/streptomycin. For serum starvation, cells were incubated in DMEM or DMEM/F12 without other supplements. Cells were transfected with plasmid DNA using PolyJet DNA in vitro tranfection reagent (SinaGen Laboratories).
Immunoprecipitation
Cells were lysed with mild lysis buffer (50 mM HEPES at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF, 1.5 mM Na3VO4, 1 mM PMSF, 1 mM DTT, protease inhibitor cocktail). The supernatants obtained by centrifugation were incubated with the appropriate antibodies for 2 h at 4°C, and protein G- or protein A-conjugated beads were added in for 1 h. Immunoprecipitates were washed four times with lysis buffer, and proteins were eluted with SDS-PAGE sample buffer.
Kinase assay
Lats and MST kinase assays were performed as described (Zhao et al. 2007). The immunoprecipitated MST was subjected to a kinase assay in the presence of GST-Mob as substrate. Lats kinase assays were performed similarly using GST-YAP as a substrate.
siRNA
YAP, TAZ, Gq, G11, G12, G13, and control siRNAs were from Dharmacon SMARTpool and were introduced into cells by transient transfection with RNAi MAX (Invitrogen) in accordance with the manufacturer's instructions.
Immunoblotting
Immunoblotting was performed as per standard protocol, and antibodies are indicated in the Supplemental Material.
Immunofluorescence staining
Immunofluorescence staining was performed as described (Zhao et al. 2007) and using MCF10A cells. Antibodies are described in the Supplemental Material.
RNA isolation and real-time PCR
Cells were washed with cold PBS, and total RNA was extracted using an RNesay kit (Qiagen) treated with RNA-free DNase. One microgram of RNA was used for reverse transcription with iScript reverse transcriptase (Bio-Rad). cDNA was then diluted and used for real-time PCR with gene-specific primers using KAPA SYBR Fast qPCR master mix (Kapa Biosystems) and the 7300 real-time PCR system (Applied Biosystems). The relative abundance of mRNA was calculated by normalization to β-actin mRNA. Primers are described in the Supplemental Material.
Cell migration assay
The cell migration assay was performed using BD Falcon cell culture inserts for 24-well plates with 8.0-μm pores, and the inserts were precoated with 20 μg/mL fibronectin. MCF10A cells transfected with control siRNA or YAP/TAZ siRNA were serum-starved for 24 h and then seeded into the upper chamber of the insert (2 × 105 cells per well) in DMEM/F12 without other supplements, and the lower chamber was filled with DMEM/F12 with or without 2 μM TRAP6. After 24 h, cells were fixed using 4% paraformaldehyde and stained using 0.05% crystal violet. Nonmigrating cells were removed from the upper membrane by scrubbing with a cotton swab. Cells in the upper chamber were carefully removed, and cells migrated through the filter were assessed by photography. Cells were counted from photographs of the membrane for quantification.
Cell invasion assay
The cell invasion assay was performed using BioCoat Matrigel invasion chambers (BD Biosciences) containing inserts with a Matrigel and an underlying membrane with 8.0-μm pores. The inserts were hydrated with warm DMEM without FBS in 5% CO2 for 2 h at 37°C. MDA-MB-231 cells transfected with control siRNA or YAP/TAZ siRNA were serum-starved for 24 h and then seeded into the upper chamber of the insert (2 × 105 cells per well) in DMEM with no FBS, and the lower chamber contained DMEM with or without 2 μM TRAP6. TRAP6 was replenished every 4 h. After 24 h, the invading cells were fixed using 4% paraformaldehyde and stained using 0.05% crystal violet. Noninvading cells were removed from the upper membrane by scrubbing with a cotton swab. The invading cells were visualized under an inverted light microscope. Cells were counted from photographs of the membrane. The experiments were repeated in duplicate, and cell counts were based on three different fields per experiment.
Acknowledgment
We thank Dr. Rick Neubig, JoAnn Trejo, and Wolfram Ruf for advice, and Karen Tumaneng and Dr. Hyun Woo Park for critical reading of the manuscript. This work is supported by grants from NIH (to K.L.G. and J.H.B.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.197582.112.
References
- Bar-Shavit R, Turm H, Salah Z, Maoz M, Cohen I, Weiss E, Uziely B, Grisaru-Granovsky S 2011. PAR1 plays a role in epithelial malignancies: Transcriptional regulation and novel signaling pathway. IUBMB Life 63: 397–402 [DOI] [PubMed] [Google Scholar]
- Booden MA, Eckert LB, Der CJ, Trejo J 2004. Persistent signaling by dysregulated thrombin receptor trafficking promotes breast carcinoma cell invasion. Mol Cell Biol 24: 1990–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers RC, Leoni P, Blanc-Brude OP, Wembridge DE, Laurent GJ 2000. Thrombin is a potent inducer of connective tissue growth factor production via proteolytic activation of protease-activated receptor-1. J Biol Chem 275: 35584–35591 [DOI] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24: 2076–2086 [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W 2008. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res 68: 2592–2598 [DOI] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al. 2011. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147: 759–772 [DOI] [PubMed] [Google Scholar]
- Coughlin SR 2000. Thrombin signalling and protease-activated receptors. Nature 407: 258–264 [DOI] [PubMed] [Google Scholar]
- Coughlin SR 2005. Protease-activated receptors in hemostasis, thrombosis and vascular biology. J Thromb Haemost 3: 1800–1814 [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183 [DOI] [PubMed] [Google Scholar]
- Elste AP, Petersen I 2010. Expression of proteinase-activated receptor 1–4 (PAR 1–4) in human cancer. J Mol Histol 41: 89–99 [DOI] [PubMed] [Google Scholar]
- Even-Ram S, Uziely B, Cohen P, Grisaru-Granovsky S, Maoz M, Ginzburg Y, Reich R, Vlodavsky I, Bar-Shavit R 1998. Thrombin receptor overexpression in malignant and physiological invasion processes. Nat Med 4: 909–914 [DOI] [PubMed] [Google Scholar]
- Fernandez BG, Gaspar P, Bras-Pereira C, Jezowska B, Rebelo SR, Janody F 2011. Actin-capping protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138: 2337–2346 [DOI] [PubMed] [Google Scholar]
- Grusche FA, Richardson HE, Harvey KF 2010. Upstream regulation of the hippo size control pathway. Curr Biol 20: R574–R582 doi: 10.1016/j.cub.2010.05.023 [DOI] [PubMed] [Google Scholar]
- Gupta GP, Massague J 2006. Cancer metastasis: Building a framework. Cell 127: 679–695 [DOI] [PubMed] [Google Scholar]
- Lei QY, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL 2008. TAZ promotes cell proliferation and epithelial–mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol 28: 2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane SR, Seatter MJ, Kanke T, Hunter GD, Plevin R 2001. Proteinase-activated receptors. Pharmacol Rev 53: 245–282 [PubMed] [Google Scholar]
- Martin CB, Mahon GM, Klinger MB, Kay RJ, Symons M, Der CJ, Whitehead IP 2001. The thrombin receptor, PAR-1, causes transformation by activation of Rho-mediated signaling pathways. Oncogene 20: 1953–1963 [DOI] [PubMed] [Google Scholar]
- McLaughlin JN, Shen L, Holinstat M, Brooks JD, Dibenedetto E, Hamm HE 2005. Functional selectivity of G protein signaling by agonist peptides and thrombin for the protease-activated receptor-1. J Biol Chem 280: 25048–25059 [DOI] [PubMed] [Google Scholar]
- Offermanns S, Jones SV, Bombien E, Schultz G 1994. Stimulation of mitogen-activated protein kinase activity by different secretory stimuli in rat basophilic leukemia cells. J Immunol 152: 250–261 [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA 2006. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci 103: 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D 2010. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendurthi UR, Allen KE, Ezban M, Rao LV 2000. Factor VIIa and thrombin induce the expression of Cyr61 and connective tissue growth factor, extracellular matrix signaling proteins that could act as possible downstream mediators in factor VIIa·tissue factor-induced signal transduction. J Biol Chem 275: 14632–14641 [DOI] [PubMed] [Google Scholar]
- Pendurthi UR, Ngyuen M, Andrade-Gordon P, Petersen LC, Rao LV 2002. Plasmin induces Cyr61 gene expression in fibroblasts via protease-activated receptor-1 and p44/42 mitogen-activated protein kinase-dependent signaling pathway. Arterioscler Thromb Vasc Biol 22: 1421–1426 [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Noorbakhsh F, Defea K, Hollenberg MD 2012. Targeting proteinase-activated receptors: Therapeutic potential and challenges. Nat Rev Drug Discov 11: 69–86 [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Pan G, Reddy BV, Oh H, Irvine KD 2011. Zyxin links fat signaling to the hippo pathway. PLoS Biol 9: e1000624 doi: 10.1371/journal.pbio.1000624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, Halder G 2011. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J 30: 2325–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez C, Bar-Eli M 2003. Role and regulation of the thrombin receptor (PAR-1) in human melanoma. Oncogene 22: 3130–3137 [DOI] [PubMed] [Google Scholar]
- Vouret-Craviari V, Boquet P, Pouyssegur J, Van Obberghen-Schilling E 1998. Regulation of the actin cytoskeleton by thrombin in human endothelial cells: Role of Rho proteins in endothelial barrier function. Mol Biol Cell 9: 2639–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Itoga K, Okano T, Yonemura S, Sasaki H 2011. Hippo pathway regulation by cell morphology and stress fibers. Development 138: 3907–3914 [DOI] [PubMed] [Google Scholar]
- Walsh CT, Radeff-Huang J, Matteo R, Hsiao A, Subramaniam S, Stupack D, Brown JH 2008. Thrombin receptor and RhoA mediate cell proliferation through integrins and cysteine-rich protein 61. FASEB J 22: 4011–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. 2012. Regulation of the Hippo–YAP pathway by G-protein-coupled receptor signaling. Cell 150: 780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Hong W 2008. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell 13: 188–192 [DOI] [PubMed] [Google Scholar]
- Zhang J, Smolen GA, Haber DA 2008. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res 68: 2789–2794 [DOI] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL 2010a. The Hippo–YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev 24: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL 2010b. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev 24: 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL 2012. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev 26: 54–68 [DOI] [PMC free article] [PubMed] [Google Scholar]