Stem cell changes during aging can affect tissue functions, and the skin is an obvious example of a tissue that undergoes aging-related changes. Doles et al. explore alterations in murine hair follicle stem cells during skin aging and identify an age-associated imbalance in the epidermal Jak–Stat signaling pathway that inhibits the function of these cells. They suggest that aging-related stem cell inhibition may be part of broader tumor-suppressive mechanisms.
Keywords: stem cells, aging, epidermis, Jak–Stat
Abstract
Altered stem cell homeostasis is linked to organismal aging. However, the mechanisms involved remain poorly understood. Here we report novel alterations in hair follicle stem cells during skin aging, including increased numbers, decreased function, and an inability to tolerate stress. Performing high-throughput RNA sequencing on aging stem cells, cytokine arrays, and functional assays, we identify an age-associated imbalance in epidermal Jak–Stat signaling that inhibits stem cell function. Collectively, this study reveals a role for the aging epidermis in the disruption of cytokine and stem cell homeostasis, suggesting that stem cell decline during aging may be part of broader tumor-suppressive mechanisms.
Adult tissue homeostasis requires continual replacement of cells lost due to normal turnover, injury, and disease. However, aging is accompanied by decreased tissue regeneration and homeostasis, both of which are frequently associated with impaired stem cell function. As some stem cell compartments undergo dramatic age-associated changes (including alterations in cell number, decreased regenerative capacity, and fate change), a decline in stem cell function is suggested to contribute to the aging process (Conboy et al. 2003; Rossi et al. 2005; Molofsky et al. 2006). Therefore, characterizing the regulatory mechanisms that mediate normal stem cell aging is critical to understanding age-associated pathologies and disease.
The skin is one of the most obvious tissues to undergo aging-associated phenotypical and functional changes, including decreased hair cycling, epidermal thinning, diminished sebaceous gland function, and an impaired wound response (Balin and Pratt 1989). Although some studies in mice suggest that epidermal stem cells are retained during aging (Stern and Bickenbach 2007; Giangreco et al. 2008), aged human keratinocyte stem cells exhibit decreased colony-forming ability (Barrandon and Green 1987), suggesting that undiscovered stem cell changes may be involved in epidermal aging. Indeed, the skin contains many individual, well-characterized stem cell populations, which not only highlights the complexity of the skin hierarchy, but also suggests that discrete populations may undergo age-associated changes that might ultimately impact tissue function. We set out to investigate this hypothesis using well-established murine hair follicle stem cell models.
Results
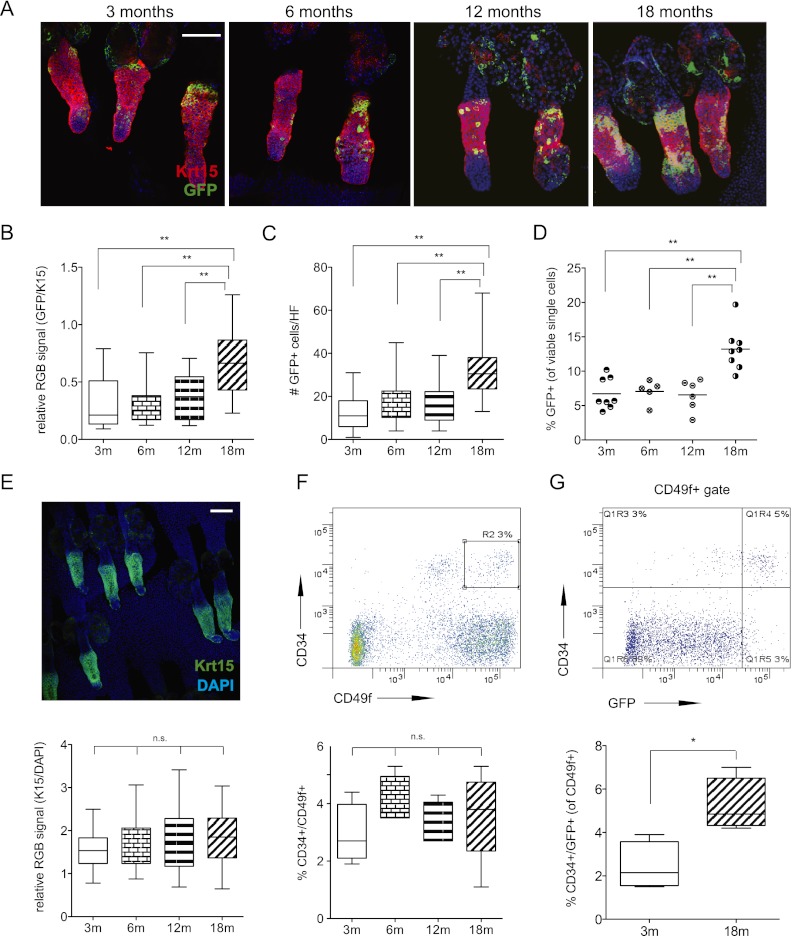
Keratin-15 (Krt-15)-positive hair follicle cells are one of the best-characterized stem cell populations in the skin. Specifically, studies using Krt-15 promoter reporter mouse models have demonstrated that these cells possess stem cell properties, as they are label-retaining cells with multipotent capacity, contributing to hair follicle cycling, sebaceous gland function, and wound repair (Morris et al. 2004; Ito et al. 2005; Petersson et al. 2011). In agreement with published work, whole-mount immunostaining for GFP in the tails of young Krt-15-GFP reporter mice confirmed these cells as a subset of the Krt-15 protein-expressing fraction, with the GFP reporter identifying the most primitive compartment (Fig. 1A; Morris et al. 2004). Although expression is seen in the sebaceous gland and the epidermis, immunohistochemical and FACS analysis supported previous studies demonstrating significant overlap with the CD34+ population of stem cells in the bulge region (Supplemental Fig. 1a–d). However, temporal analyses of Krt-15-GFP expression in the bulge region of the hair follicle revealed a significant increase in GFP with respect to Krt-15 protein in aged mice (Fig. 1A,B) as well as an expansion of the absolute number of GFP+ cells per hair follicle unit (Fig. 1C). Corroborating these observations, we also documented temporal enrichment of GFP+ cells in flow cytometric analyses of aged back skin epidermis (Fig. 1D). As has been previously reported (Giangreco et al. 2008), we did not observe any significant differences in the overall CD34+/CD49f+ or Krt-15 protein-positive populations (Fig. 1E,F, respectively). We did, however, observe that the increasing Krt-15-GFP+ cells coexpressed CD34 (Fig. 1G; Supplemental Fig. 1e), thus highlighting a previously undocumented increase in this hair follicle stem cell population during aging.
Figure 1.
Krt-15-GFP hair follicle stem cell number increases with age. (A) Whole-mount coimmunofluorescence of aged tail epidermis stained with antibodies targeting Krt-15 (red) and GFP (green). (B,C) Quantification of absolute GFP cell number per hair follicle (B) and fluorescence intensity relative to Krt-15 protein (n = 32–42 follicles from three mice) (C). (D) Percentage of Krt-15-GFP+ cells in epidermal preparations analyzed by FACS (data points represent independent FACS analyses of one to three mice; n ≥ 7 mice total per time point). (E) Whole-mount immunofluorescence for Krt-15 protein in aged tail epidermis and intensity quantification relative to DAPI. (F) Representative FACS scatterplots and summarized percentages of CD34+/CD49fhi cells in aged epidermis (n = 4–5). (G) FACS analysis of aging GFP+/CD34+/CD49f+ cells (n = 3). Bar, 100 μm. P-values were determined using unpaired, two-tailed Student's t-tests. (*) P < 0.05; (**) P < 0.005; (n.s.) not significant.
To test whether these accumulating aged Krt-15-GFP stem cells retained functional capacity, we FACS-purified GFP-positive and GFP-negative epidermal fractions and plated them in equal numbers to assess clonogenic capacity (Barrandon and Green 1987). In agreement with earlier reports (Morris et al. 2004), young (3-mo) GFP+ cells gave rise to larger and significantly greater numbers of colonies compared with GFP− control cells (Fig. 2A, left panels). Strikingly, we observed a significantly diminished colony-forming ability of aged (18-mo) GFP+ cells cultured under identical conditions (Fig. 2A–C). Similarly, parallel studies using FACS-isolated triple-positive (CD34+/CD49f+/GFP+) stem cells (Fig. 2D) as well as the total CD34+/CD49f+ population (Supplemental Fig. 2a) also revealed an age-associated decline in functional capacity, thus reinforcing the notion that bona fide stem cells are indeed impaired with advanced age. We subsequently tested whether these aged stem cells were functionally impaired in vivo. First, we subjected young and old Krt-15-GFP mice to ionizing radiation (IR) and measured the change in stem cell number in response to exogenous low-level DNA damage (Davies et al. 2008; Liang et al. 2011). Surprisingly, whereas the Krt-15-GFP stem cells in young mice exhibited an approximately twofold increase in response to acute DNA damage, there was no change in old mice (Fig. 2E). Similar results were also seen for the Krt-15-GFP+/CD34+/CD49f+ population (Supplemental Fig. 2b), suggesting that aged stem cells are either unable to respond to the stress or become depleted as a result. To examine this observation in greater detail, we treated shaved, dorsal back skin with 12-O-tetradecanoylphorbol-13-acetate (TPA), an inducer of stem cell activation and epidermal hyperproliferation. Interestingly, at the tissue level, aged skin was not able to tolerate TPA as well as young skin and rapidly developed skin lesions (Supplemental Fig. 2c). In agreement with our earlier data, counting of individual GFP+ stem cells in the hair follicle bulge in untreated young and old dorsal back skin revealed an age-associated increase in absolute cell number with age (Fig. 2F; Supplemental Fig. 2c). However, upon treatment with TPA, whereas young skin exhibited a significant increase in stem cell number in response to stimulus, aged skin displayed the opposite trend, with depletion of both Krt-15-GFP and CD34 immunoreactivity (Fig. 2F,G; Supplemental Fig. 2e). Altogether, this demonstrates an inherent inability of aged stem cells to be maintained following substantial cellular stress.
Figure 2.
Age-associated functional decline in Krt-15-GFP stem cells. (A) Clonogenic colony-forming assays of FACS-sorted GFP+ and GFP− cells from 3- and 18-mo-old epidermis. Pictured are duplicate wells of each condition, representative of n ≥ 3 independent experiments. (B,C) Quantification of colony number (B) and normalized colony size (C) of data depicted in A. (D) Colony assays using FACS-sorted GFP+/CD34+/CD49f+ cells. (E) Aged Krt-15-GFP mice were exposed to 5 Gy of whole-body irradiation and sacrificed 24 h post-IR. Shown is a graph of the FACS-determined fold change in GFP+ cells in IR-treated epidermis relative to non-IR controls at each time point. (F) Absolute GFP+ cell number per hair follicle in shaved back skin epidermis treated with 20 nM TPA every other day for 1 wk. (G) Representative histological images of back skin sections stained with antibodies against Ki67, GFP, and CD34. P-values were determined using unpaired, two-tailed Student's t-tests. (*) P < 0.05; (**) P < 0.001. Error bars for bar graphs represent ±SD.
To gain deeper insight into the molecular mechanisms underlying these age-related changes, we performed high-throughput RNA sequencing (RNA-seq) on 3- and 18-mo Krt-15-GFP cells freshly isolated from the skin (data provided in Supplemental Table 1). Importantly, expression (fragments per kilobase of exon per million of fragments mapped [FPKM]) values generated by sequencing and further selectively validated by quantitative RT–PCR (qRT–PCR) demonstrated that with age, the GFP+ stem cell population retains, and possibly increases, the relative expression of a core stem cell signature (Supplemental Figs. 3, 4; Tumbar et al. 2004; Lien et al. 2011). Interestingly, while the core signature of these cells increased, we observed little change or even possible decreases in the alternate fate signatures (namely, interfollicular epidermis and sebaceous gland) (Tumbar et al. 2004; Lien et al. 2011), suggesting that there may be fate changes within this population with age (Supplemental Fig. 3).
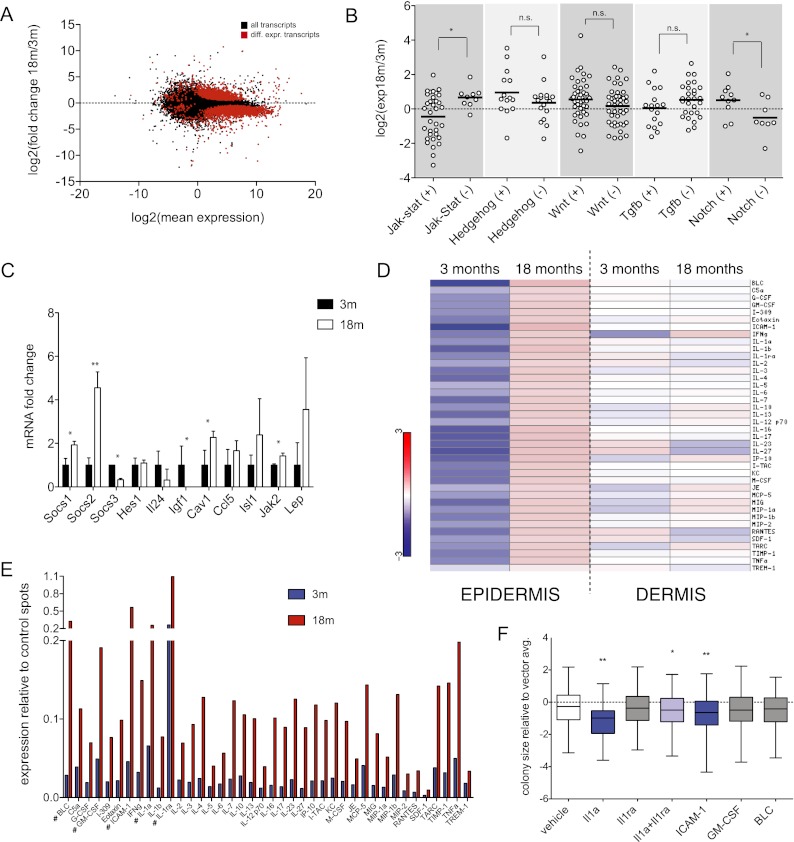
Unbiased, global analyses of transcript expression in highly purified Krt-15-GFP cells revealed substantial changes in many genes and biological processes (Fig. 3A; Supplemental Fig. 5). Given that stem cell functional decline has been linked with changes in key signaling pathways (Silva-Vargas et al. 2005; Brack et al. 2007), we initially focused on these for subsequent analyses. On the basis of gene ontology (GO) annotations, we manually extracted transcripts associated with positive and negative regulation of Jak–Stat, Wnt, Hedgehog, Tgf-β, and Notch signaling to survey canonical signal transduction activity. Interestingly, two pathways (Jak–Stat and Notch) stood out as being significantly altered with age, as evidenced by a robust inverse relationship between positive and negative signaling regulators (Fig. 3B). We also validated many of these gene expression changes by qRT–PCR in independent biological replicates (Fig. 3C). Collectively, our deep-sequencing results demonstrate that while Krt-15-GFP cells maintain a stem-like signature during aging, they also exhibit marked alterations in critical signal transduction cascades.
Figure 3.
Dynamic changes in cytokine signaling networks in aging epidermis. (A) A balloon graph depicting RNA-seq-determined individual transcript changes of aged, Krt-15-GFP stem cells. Red data points represent significantly up-regulated or down-regulated transcripts. (B) Differential expression values of transcripts associated with positive (+) or negative (−) regulation of selected signaling pathways. (C) qRT–PCR validation of selected Jak–Stat signaling transcripts. Data represent n = 3 independently FACS-sorted RNA pools (mice) for each time point. (D) Heat maps summarizing normalized fold change values for individual cytokines/chemokines detected in murine epidermis (left) and dermis (right). (E) Raw expression values for assayed cytokines/chemokines in aged (3-mo vs. 18-mo) epidermis. (F) A summary of clonogenic colony size of young (3-mo) epidermal cells exposed to selected recombinant cytokines. P-values were determined using unpaired, two-tailed Student's t-tests. (*) P < 0.05; (**) P < 0.005; (n.s.) not significant; (#) cytokines selected for further detailed analyses. Error bars for bar graphs represent ±SD.
The transcriptional change in regulators of Jak–Stat signaling was of particular interest to us for several reasons: First, it represented the signaling pathway most prominently deregulated in our analysis. Second, the increase in Jak–Stat negative regulators, including SOCS1 and SOCS2, is consistent with activation of a negative feedback loop (Krebs and Hilton 2001; Kimura et al. 2005) and suggests that aged Krt15-GFP-positive cells may be engaging feedback mechanisms in response to a proinflammatory environment. To investigate this, we performed protein cytokine arrays that simultaneously analyze the relative abundance of 40 cytokines and chemokines on freshly prepared epidermal and dermal cell lysates collected from 3- and 18-mo-old mice (Fig. 3D; Supplemental Fig. 6). Interestingly, while significant changes in cytokine levels were not detected in aged dermis (Supplemental Figs. 6, 7), we observed a striking age-dependent increase of nearly every tested cytokine in epidermal samples (Fig. 3E; Supplemental Fig. 6), including several factors—e.g., BLC/CXCL13, GM-CSF, ICAM-1/CD54, Il-1α, and Il-1ra—that reached particularly high expression levels in 18-mo-old epidermis.
As alterations in cytokine abundance are generally reflected at the transcript level (Freund et al. 2010), we examined our sequencing expression data for these same 40 cytokines within the purified aging stem cells and found no corresponding trend, suggesting a stem cell-extrinsic, epidermis-derived origin of the proinflammatory signal (Supplemental Fig. 8). Given that specific cytokine signaling has recently been suggested to inhibit stem cell function in other settings (Villeda et al. 2011), we suspected that the enhanced cytokine networks might also be contributing to decreased stem cell function in the skin. To investigate this, we endeavored to directly test whether any of these deregulated and highly expressed cytokines could affect the clonogenic capacity of young, healthy keratinocytes in vitro. Of the six recombinant proteins assayed (Il-1α, Il-1ra, Il-6, GM-CSF, ICAM-1, and BLC), two—Il-1α and ICAM-1—were able to individually suppress growth of 3-mo-old clonogenic keratinocyte cultures (Fig. 3F; Supplemental Fig. 9). Interestingly, cotreatment of Il-1α cultures with the receptor antagonist Il-1ra ameliorated much of the growth inhibitory effect, suggesting that the stoichiometric balance of Il-1 signaling components is particularly critical in regulating stem cell function (Fig. 3F; Supplemental Fig. 9). Importantly, while we did not observe growth-interfering effects with every tested cytokine, we cannot rule out important contributions of these (and other) factors on stem cell function that require precise dosing or combinatorial coactivation to realize their potential. These data, however, demonstrate that individual cytokines that increase during tissue aging can contribute to functional stem cell decline.
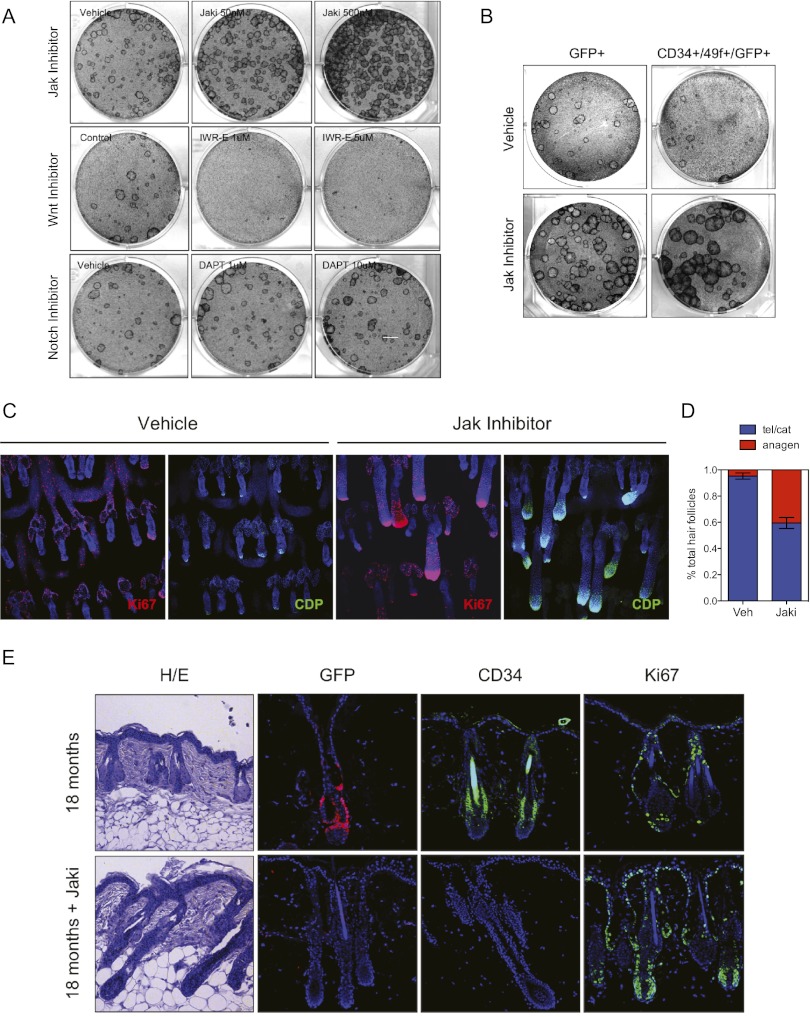
A striking feature of the aging epidermal arrays was the expression increase across such a diverse set of cytokines. Taking into account our observation that positive regulation of Il-1α signaling had an adverse effect on keratinocyte stem cell function, we hypothesized that global repression of cytokine signaling may have potentially growth-promoting effects in aged epidermis. To address this, we generated clonogenic cultures from aged (18-mo) wild-type mice and treated these with the protein kinase inhibitor pyridone-6 (Jak Inhibitor I, Jaki), a global inhibitor of the Jak–Stat pathway. Strikingly, this resulted in a robust rescue of aged stem cell clonogenic activity, as evidenced by more abundant and significantly faster-growing colonies compared with vehicle-treated control cultures that maintained expression of prototypical epidermal markers, including p63, Krt-14, and CD49f (Fig. 4A; Supplemental Fig. 10a). This rescue effect was not seen when we tested inhibitors of other pathways that showed little change in our transcriptome analysis (Wnt, Hedgehog, and Notch) or the Wnt agonist BIO (Fig. 4A; Supplemental Fig. 10b), highlighting the functional significance of Jak–Stat transcript changes derived from our sequencing data. Rescue of aged colony-forming ability was also seen with two different inhibitors of the Jak–Stat pathway, highlighting the specificity of these results (Supplemental Fig. 11a). This effect of Jak kinase inhibition was also evident when we treated aged GFP+ or GFP+/CD34+/CD49f+ cultures, indicating that Jak inhibition has a proproliferative effect directly on aged stem cells (Fig. 4B), including on cells derived from mice exposed to γ-irradiation in vivo (Supplemental Fig. 12b). Ultimately, however, the proliferative advantage with Jak inhibition seemed temporary, as serial passaging of the aged colonies treated with Jak inhibitor restored an aged phenotype to these cells (Supplemental Fig. 11b). Interestingly, nonbulge stem cell populations (GFP+/CD34−) also exhibited an age-associated decrease in proliferation that was rescued with the inhibitor, suggesting that Jak kinase suppression may have effects on nonstem cells or other stem cell populations within the culture (Supplemental Fig. 12a).
Figure 4.
Jak kinase inhibition promotes aged epidermal stem cell function in vitro and in vivo. (A) Clonogenic colony formation assays of aged (18-mo) keratinocytes cultured in the presence of Jak, Wnt, or Notch pathway inhibitors. (B) Colony assays of FACS-sorted GFP+ or GFP+/CD34+/CD49f+ cells treated with 500 nM Jak inhibitor. (C) Representative whole-mount immunofluorescence images of Jak inhibitor-treated, aged tail epidermis stained with antibodies targeting Ki67 (left, red) and CDP (right, green). Bar, 200 μm. (D) Quantification of anagen hair follicles in Jak inhibitor-treated tail hair follicles compared with vehicle controls. Data are representative of three independent in vivo experiments (n = 5 mice). (E) Representative histological images of Jak inhibitor-treated back skin epidermis stained using antibodies targeting GFP, CD34, or Ki67. P-values were determined using unpaired, two-tailed Student's t-tests. (*) P < 0.05; (**) P < 0.0001. Error bars for bar graphs represent ±SD.
Having identified inhibition of cytokine signaling as a potent means of restoring aspects of stem cell function in vitro, we tested its potential biological relevance in vivo. Consistent with our clonogenicity results, a 1-wk topical Jak inhibitor regimen on 18-mo tail epidermis resulted in a substantial increase in the number of active hair follicles. By whole-mount immunohistochemistry, we observed an increase in Ki67 and CCAAT displacement protein (CDP) expression, both of which are associated with actively cycling hair follicles (Fig. 4C). Quantitatively, we observed <5% anagen (active) hair follicles in vehicle-treated epidermis. However, this increased to >35% when exposed to the inhibitor (Fig. 4D; Supplemental Fig. 13). This effect seemed to be independent of the stage of the mature hair follicle when treated, as follicles were largely in telogen (resting) at both time points examined using anagen hair follicle counting and analysis of relevant signaling pathways (Supplemental Fig. 14). Furthermore, histological examination of shaved back skin treated with vehicle or Jak inhibitor also revealed significant hair follicle reactivation (Fig. 4E). However, upon further investigation, we observed a marked depletion in both Krt-15-GFP and CD34 immunoreactivity in Jak inhibitor-treated samples, suggesting that while aged hair follicles are capable of being stimulated into an active state, enhanced proliferative/activating signals may ultimately drive the stem cells to exhaustion and depletion (Fig. 4E). Taken together, these data uncover a complex relationship between epidermal stem cell proliferation and self-renewal in aged skin and show that aspects of this relationship can be partially restored through inhibition of the Jak–Stat pathway.
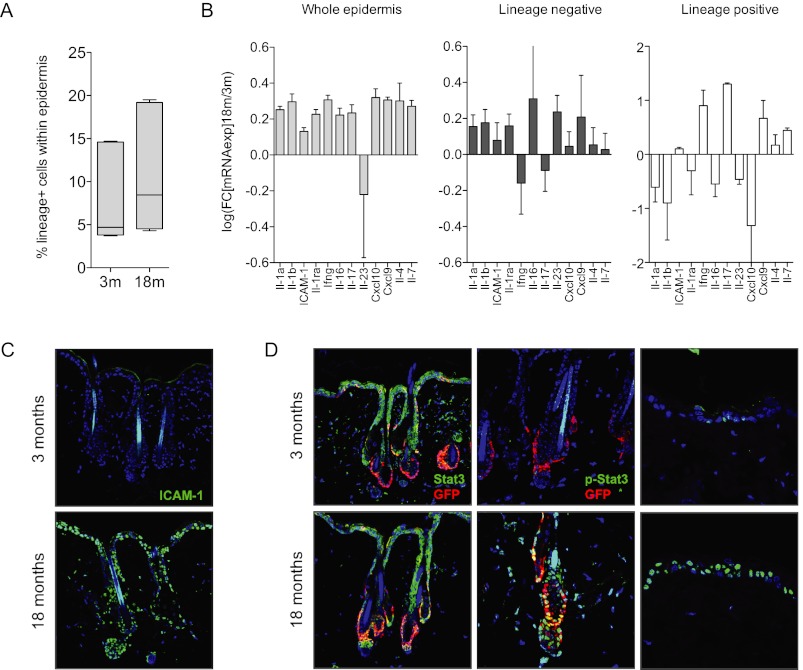
Given the robust ability of cytokine signaling to modulate stem cell function and hair follicle status in aged mice, we sought to elucidate the cellular sources of these increased signals. Using antibodies against immune cell subtypes, we performed FACS analyses on whole epidermal preparations generated from young (3-mo) and aged (18-mo) mice. We did not observe a substantial increase in the percentage of cells labeled using lineage antibodies against an array of mature immune subtypes, including lymphocytes, monocytes/macrophages, NK cells, erythrocytes, and granulocytes (Fig. 5A; Supplemental Fig. 15). We then separated the bulk lineage-positive and lineage-negative fractions at each time point and analyzed them by qRT–PCR in order to ascertain whether cytokine expression levels were changing over time. Surprisingly, we observed a significant increase in cytokine transcript expression emanating from the lineage-negative fraction, suggesting that the age-associated increase in certain cytokine levels derived from the epidermal compartment itself and not solely from bona fide immune cells (Fig. 5B). These data were further supported and refined in additional FACS/qPCR experiments, which specifically implicated the CD34−/CD49fmed, CD49fhi subpopulations as likely sources of several cytokines, including those involved in Il-1 signaling (Supplemental Fig. 15). In addition, immunofluorescent staining of back skin tissue sections using an ICAM-1 antibody revealed an epidermal-specific staining pattern increasing with age throughout the interfollicular epidermis, the hair follicles, and associated sebaceous glands (Fig. 5C). Interestingly, and in accordance with the overall up-regulation of cytokine signaling, we also observed a robust increase in epidermal phospho-Stat3 staining throughout the aged tissue, including in the interfollicular epidermis and within the Krt-15-GFP stem cell population (Fig. 5D), a pattern that was not observed with various stages of the hair follicle cycle in young mice (Supplemental Fig. 16). Thus, our data show that the epidermis contributes to the age-associated cytokine imbalance, with activity and expression detectable throughout the tissue.
Figure 5.
Epidermal-derived cytokines promote Stat3 hyperactivation in aged skin. (A) Box plot depicting the percentage of lineage-positive cells present in aging epidermal preps as determined by FACS. (B) qPCR of selected cytokine transcripts in preparations of whole epidermis (left) or FACS-sorted lineage-negative (middle) or lineage-positive (right) cell fractions. (C,D) Immunofluorescent images of aged back skin stained with antibodies targeting ICAM-1 (C) or Stat3, phospho-Stat3, or GFP (D). Error bars represent ±SD.
Discussion
Collectively, our data identify for the first time that hair follicle stem cells are susceptible to age-associated changes that may contribute to aging phenotypes. As has been described for other stem cell populations (Rossi et al. 2005), these cells increase significantly in number but decrease in functional capacity with normal aging, suggesting that the mechanisms we identify here may also be general mediators of stem cell aging in other tissues. That these cells retain stem-like properties while simultaneously being repressed by the external environment implies a degree of reversibility, which we show is partially feasible with chemical compounds. Whether this is possible in other stem cell populations remains to be seen, but studies suggest that this is likely (Conboy et al. 2005; Boyle et al. 2007; Pan et al. 2007; Villeda et al. 2011).
Conceptually, the biological function behind such an inhibition of stem cell function remains intriguing. Given that age is a primary determinant of tumor initiation and that this Krt-15 stem cell population was recently identified as a target cell of origin in squamous cell carcinoma (Keyes et al. 2011; Lapouge et al. 2011; White et al. 2011), the expansion of this population during aging could significantly increase the chance of malignant transformation—through either up-regulation of stem cell-maintaining proto-oncogene signaling, as demonstrated here, or simply increased opportunities for mutation. Certainly, as inflammation has long been linked to tumor progression, this suggests that stem cell impairment and, consequently, tissue aging may ultimately be the result of tumor-suppressive mechanisms. Our observation of increased phospho-Stat3 signaling throughout the aging epidermis and within the stem cell population, coupled with evidence that hyperactivated Stat3 can drive tumor progression (Bromberg et al. 1999; Demaria et al. 2012), provides a rationale for the engagement of this suppressive phenotype, particularly within vulnerable stem cell populations. Further detailed analyses of this aging stem cell population are needed to assess whether these cells might be primed for tumor initiation by age-associated inflammation as well as to uncover cell-intrinsic mechanisms likely contributing to the aging process.
Materials and methods
Animal use
Aged Krt-15-GFP (Morris et al. 2004) or wild-type C57Bl6/J mice were housed in accordance with the CEEA (Ethical Committee for Animal Experimentation) of the Government of Catalonia. For chemical epidermal activation assays, shaved back or tail skin was treated either once or three times over 1 wk with 20 nM TPA (Sigma-Aldrich). Skin samples were harvested the day following the final treatment and processed as described below. IR experiments were performed by subjecting mice to 5 Gy of whole-body irradiation followed by flow cytometric analysis of single-cell epidermal preparations at the indicated time points. For in vivo pyridone-6 (Jak inhibitor I; Calbiochem) assays, a 1 mM inhibitor preparation (in DMSO/acetone) was applied to tail skin and harvested as described for TPA experiments.
Primary mouse keratinocyte clonogenicity assays
Primary mouse keratinocytes were harvested from dorsal back skin of aged mice as described previously (Jensen et al. 2010). Chemical and cytokine treatments were initiated ∼8 h following establishment of the coculture in order to allow for equal initial seeding numbers. All cytokines were purchased from Peprotech and used as follows: 0.4–4 ng/mL Il-1α, 5–50 ng/mL Il-1ra, 1–10 ng/mL GM-CSF, 4–40 ng/mL ICAM-1, and 1–10 ng/mL BLC. Chemical activators/inhibitors were used as follows: 50–500 nM pyridone-6 (Calbiochem), 1–10 μM DAPT (Sigma), 500 nM–1 μM BIO (Cayman Chemical), 1–5 μM IWR-1 endo and exo (Cayman Chemical), 1–5 μM cyclopamine (Calbiochem), 80–400 nM tofacitinib (Selleck Chemicals), and 80–400 nM ruxolitinib (Selleck Chemicals). To visualize clonogenic keratinocyte expansion, cocultures were fixed in 10% formalin, stained with crystal violet/methanol, and imaged using conventional scanning and imaging tools.
Whole-mount immunofluorescence
Preparation of tail skin and whole-mount stainings was performed as previously described (Braun et al. 2003). Primary antibodies were incubated overnight at room temperature and used at the following concentrations: anti-GFP (1:200; Invitrogen); anti-Ki67 (1:200; Abcam); anti-Krt-15 (1:200; NeoMarkers); and anti-CDP (1:100; Santa Cruz Biotechnology). Alexa-conjugated secondary antibodies (1:500; Molecular Probes) were used for 2 h at room temperature. Nuclei were then stained with DAPI (1:10,000; Roche), and epidermal sheets were mounted in Mowiol. Pictures were acquired with a Leica TCS SP5 confocal microscope and, when applicable, quantified using ImageJ software.
Immunohistochemistry
Back and/or tail skin was fixed in 10% NBF (Sigma-Aldrich) overnight, washed in PBS, and then embedded in paraffin. Deparaffinized sections were boiled for 10 min in 0.01 M citric acid for antigen retrieval. Primary antibodies were incubated overnight at 4°C, and secondary antibodies were incubated for 2 h at room temperature in 1% serum/PBS. Nuclei were stained with DAPI (1:10,000; Roche), and the slides were mounted in VectaShield (Vector Laboratories). Primary antibodies were used at the following dilutions: anti-Krt-15 (1:200; MS-1068-P1, NeoMarkers); anti-Ki67 (1:200; ab15580, Abcam); anti-GFP (1:200; A11122, Invitrogen); anti-CD34 (1:50; 560233, BD Biosciences); anti-ICAM-1 (1:50; 4915, Cell Signaling); anti-Stat3 (1:50; 9132, Cell Signaling); and anti-p-Stat3 (Tyr705) (1:50; 9145, Cell Signaling). Alexa-conjugated secondary antibodies (Molecular Probes) were all used at a dilution of 1:2000. Pictures were acquired with a Leica DMI 6000B or a Leica TCS SP5 confocal microscope.
Flow cytometry
Epidermal cells from back skin of Krt-15-GFP or wild-type C57Bl6/J mice were isolated as described above. Cell suspensions were incubated for 30 min on ice with the following antibodies at the given dilutions: APC-conjugated anti-CD34 (1:200; clone RAM34, BD Pharmingen) and FITC-conjugated anti-α6-integrin (1:200; CD49f clone NKI-GoH3, Serotec). Keratinocytes were sorted on the basis of single cellularity, viability (DAPI), GFP positivity (Krt-15+), and/or CD34/CD49f status. FACS purification was performed on a FACS Aria system equipped with FACS DiVa software (BD Bioscience). Sorted cells were collected in keratinocyte medium supplemented to 50% FBS and either plated for clonogenic assays or pelleted and stored at −80°C for RNA isolation. FACS analysis was performed using LSRII FACS Analysers (BD Biosciences) and analyzed using Flowjo software.
RNA-seq and analysis
Total RNA was isolated from pooled, FACS-sorted GFP+ cells using the RNeasy microkit (Qiagen). RNA-seq libraries were prepared using an Illumina RNA prep kit and sequenced using the Illumina HiSeq2000 platform. A total of 233 million 46-base-pair (bp) paired-end reads were mapped to the Mus musculus genome (NCBIM37.57) by using the TopHat aligner (version 1.3.0) (Trapnell et al. 2009), a RNA-seq mapper specifically designed for detecting splice junctions between exons and based on the fast NGS mapper Bowtie (Langmead et al. 2009). TopHat parameters were set to default with a mate pair distance of 150. Final transcript levels of all mouse Ensembl known genes (v57) were calculated in units of FPKM by counting the number of mapped and spliced reads to exons, normalized by the length of the exons and averaged over all used exons for each transcript. On average, ∼81% of the total reads (equivalent to 188 million sequences) were mapped to the genome or the splice junctions. Of these, 85.6% mapped uniquely, 8.88% mapped twice, 2.8% mapped three times, and 2.9% mapped from four to 20 times. Cuffdiff tool from the Cufflinks package (version 1.0.3) (Roberts et al. 2011) was then used to detect the transcripts whose expression statistically changed between 3- and 18-mo Krt-15-GFP+ cells.
RT-qPCR
Total RNA from cultured or FACS-sorted cells was purified using the RNeasy microkit or minikit (Qiagen). Equal amounts of RNA were reverse-transcribed using random hexamer priming and SuperScript III (Invitrogen). RT-qPCR was performed with SYBR Green master mix (Roche) and gene-specific primers (sequences provided in Supplemental Table 2) using a LightCycler 480 instrument (Roche). Relative levels of expression were determined using the ΔΔCt method relative to the housekeeping gene actin.
Acknowledgments
We are grateful to Salvador Aznar-Benitah, Michael Hemann, Victoria Aranda, and Luke Gilbert for helpful discussion and critical reading of the manuscript, and to Heinz Himmelbauer and the CRG core facilities for technical assistance. J.D. is supported by an EMBO long-term fellowship, and M.A.S. is funded by a La Caixa bank studentship. This work was funded in part by a Plan Nacional grant to W.M.K. from the Spanish Ministry for Science and Innovation (SAF2010-18829).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.192294.112.
References
- Balin AK, Pratt LA 1989. Physiological consequences of human skin aging. Cutis 43: 431–436 [PubMed] [Google Scholar]
- Barrandon Y, Green H 1987. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci 84: 2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle M, Wong C, Rocha M, Jones DL 2007. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell 1: 470–478 [DOI] [PubMed] [Google Scholar]
- Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA 2007. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317: 807–810 [DOI] [PubMed] [Google Scholar]
- Braun KM, Niemann C, Jensen UB, Sundberg JP, Silva-Vargas V, Watt FM 2003. Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in whole mounts of mouse epidermis. Development 130: 5241–5255 [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr 1999. Stat3 as an oncogene. Cell 98: 295–303 [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, Rando TA 2003. Notch-mediated restoration of regenerative potential to aged muscle. Science 302: 1575–1577 [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA 2005. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433: 760–764 [DOI] [PubMed] [Google Scholar]
- Davies PS, Dismuke AD, Powell AE, Carroll KH, Wong MH 2008. Wnt-reporter expression pattern in the mouse intestine during homeostasis. BMC Gastroenterol 8: 57 doi: 10.1186/1471-230X-8-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria M, Misale S, Giorgi C, Miano V, Camporeale A, Campisi J, Pinton P, Poli V 2012. STAT3 can serve as a hit in the process of malignant transformation of primary cells. Cell Death Differ 19: 1390–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A, Orjalo AV, Desprez PY, Campisi J 2010. Inflammatory networks during cellular senescence: Causes and consequences. Trends Mol Med 16: 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangreco A, Qin M, Pintar JE, Watt FM 2008. Epidermal stem cells are retained in vivo throughout skin aging. Aging Cell 7: 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G 2005. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med 11: 1351–1354 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Driskell RR, Watt FM 2010. Assaying proliferation and differentiation capacity of stem cells using disaggregated adult mouse epidermis. Nat Protoc 5: 898–911 [DOI] [PubMed] [Google Scholar]
- Keyes WM, Pecoraro M, Aranda V, Vernersson-Lindahl E, Li W, Vogel H, Guo X, Garcia EL, Michurina TV, Enikolopov G, et al. 2011. ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell 8: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Naka T, Muta T, Takeuchi O, Akira S, Kawase I, Kishimoto T 2005. Suppressor of cytokine signaling-1 selectively inhibits LPS-induced IL-6 production by regulating JAK–STAT. Proc Natl Acad Sci 102: 17089–17094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs DL, Hilton DJ 2001. SOCS proteins: Negative regulators of cytokine signaling. Stem Cells 19: 378–387 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge G, Youssef KK, Vokaer B, Achouri Y, Michaux C, Sotiropoulou PA, Blanpain C 2011. Identifying the cellular origin of squamous skin tumors. Proc Natl Acad Sci 108: 7431–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, So YH, Cui J, Ma K, Xu X, Zhao Y, Cai L, Li W 2011. The low-dose ionizing radiation stimulates cell proliferation via activation of the MAPK/ERK pathway in rat cultured mesenchymal stem cells. J Radiat Res (Tokyo) 52: 380–386 [DOI] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E 2011. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 9: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ 2006. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G 2004. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol 22: 411–417 [DOI] [PubMed] [Google Scholar]
- Pan L, Chen S, Weng C, Call G, Zhu D, Tang H, Zhang N, Xie T 2007. Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell 1: 458–469 [DOI] [PubMed] [Google Scholar]
- Petersson M, Brylka H, Kraus A, John S, Rappl G, Schettina P, Niemann C 2011. TCF/Lef1 activity controls establishment of diverse stem and progenitor cell compartments in mouse epidermis. EMBO J 30: 3004–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L 2011. Identification of novel transcripts in annotated genomes using RNA-seq. Bioinformatics 27: 2325–2329 [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, Weissman IL 2005. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci 102: 9194–9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Vargas V, Lo Celso C, Giangreco A, Ofstad T, Prowse DM, Braun KM, Watt FM 2005. β-Catenin and Hedgehog signal strength can specify number and location of hair follicles in adult epidermis without recruitment of bulge stem cells. Dev Cell 9: 121–131 [DOI] [PubMed] [Google Scholar]
- Stern MM, Bickenbach JR 2007. Epidermal stem cells are resistant to cellular aging. Aging Cell 6: 439–452 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E 2004. Defining the epithelial stem cell niche in skin. Science 303: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, Stan TM, Fainberg N, Ding Z, Eggel A, et al. 2011. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AC, Tran K, Khuu J, Dang C, Cui Y, Binder SW, Lowry WE 2011. Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc Natl Acad Sci 108: 7425–7430 [DOI] [PMC free article] [PubMed] [Google Scholar]