This work explores how promoter-proximal mRNA processing signals affect transcription. A promoter-proximal splice donor site is found to shield cryptic pA sites via U1 snRNP binding. The transcription inhibitory effects of exposed pA sites are position-dependent and specific to pA-dependent terminators. The study reveals the importance of gene length for optimal communication between pA-dependent terminators and promoters and has implications for understanding pA-independent mechanisms used by short genes (e.g., histone genes) on a genome-wide scale.
Keywords: coupling between processing and transcription, cryptic pA sites, transcription
Abstract
Gene expression relies on the functional communication between mRNA processing and transcription. We previously described the negative impact of a point-mutated splice donor (SD) site on transcription. Here we demonstrate that this mutation activates an upstream cryptic polyadenylation (CpA) site, which in turn causes reduced transcription. Functional depletion of U1 snRNP in the context of the wild-type SD triggers the same CpA event accompanied by decreased RNA levels. Thus, in accordance with recent findings, U1 snRNP can shield premature pA sites. The negative impact of unshielded pA sites on transcription requires promoter proximity, as demonstrated using artificial constructs and supported by a genome-wide data set. Importantly, transcription down-regulation can be recapitulated in a gene context devoid of splice sites by placing a functional bona fide pA site/transcription terminator within ∼500 base pairs of the promoter. In contrast, promoter-proximal positioning of a pA site-independent histone gene terminator supports high transcription levels. We propose that optimal communication between a pA site-dependent gene terminator and its promoter critically depends on gene length and that short RNA polymerase II-transcribed genes use specialized termination mechanisms to maintain high transcription levels.
The majority of human protein-coding genes produce transcripts that undergo capping, splicing, and polyadenylation (pA) before their nuclear export. These maturation steps are highly regulated and typically cross-talk functionally so that one processing event positively impacts the next (Maniatis and Reed 2002; Moore and Proudfoot 2009; Perales and Bentley 2009). Based on the mainly cotranscriptional nature of mRNA processing, such cross-talk within the same round of transcription extends to the generally stimulatory effect of mRNA processing events onto the transcription reaction as well as the positive impact of transcription back on mRNA processing. A central mediator of these interactions is the C-terminal domain (CTD) of RNA polymerase II (RNAPII). Consisting of 52 heptad repeats with serines (Sers) at positions 2, 5, and 7, the human CTD can attract processing factors in a phosphorylation-dependent manner and position them in close proximity to the emerging pre-mRNA (Cramer et al. 2001; Buratowski 2009). While this orchestrates the timing of mRNA processing, it also provides an efficient platform for molecular signaling between the mRNA processing and transcription machineries.
Evidence for a functional linkage between RNA maturation elements and the efficiency of the subsequent round of transcription of the same gene has also been reported (Furger et al. 2002; Damgaard et al. 2008; Mapendano et al. 2010). In one example, it was demonstrated that pA signals communicate with the transcription initiation machinery, as genes with disrupted 3′ end processing/transcription termination display lowered transcription initiation rates (Mapendano et al. 2010). Although the mechanistic basis for this linkage remains elusive, it was suggested to be caused by defective RNAPII recycling from the gene terminator back to the promoter (Mapendano et al. 2010; Lykke-Andersen et al. 2011). In other cases, the pre-mRNA splicing process and/or splice elements have also been shown to affect transcription. Here, genes with a mutated promoter-proximal splice donor (SD) site exhibited lower transcription initiation rates than their wild-type counterparts (Damgaard et al. 2008). Moreover, a plasmid-borne gene showed decreased transcription when an otherwise promoter-proximal intron was moved to a more distal site (Furger et al. 2002). Since U1 snRNP associates with transcription initiation factors TAF15 (Leichter et al. 2011) and Cyclin H (a subunit of TFIIH) (Kwek et al. 2002) as well as with RNAPII (Tian 2001; Das et al. 2007; Spiluttini et al. 2010), it was suggested that a promoter-proximal U1 snRNP-bound SD might increase the local concentration of general transcription factors (GTFs) around the gene promoter to stimulate initiation of the next round of transcription (Furger et al. 2002; Kwek et al. 2002; Manley 2002; Damgaard et al. 2008). Consistent with such a model, purified U1 snRNP was found to stimulate transcription in vitro (Kwek et al. 2002). However, data demonstrating that U1 snRNP is present at transcription sites independently of splicing (Spiluttini et al. 2010) suggest that other mechanisms accounting for the observed positive correlation between splicing and transcription may also exist.
Here, using a HIV1-derived intron-containing construct, we found that promoter proximity of the SD per se has little bearing on transcription levels. Rather, SD mutation activates a cryptic pA (CpA) site, which in turn negatively impacts transcription. Consistently, inhibiting U1 snRNP function also results in CpA activation and lowered levels of HIV1 RNA. These findings echo the reported property of U1 snRNP to inhibit cleavage at premature pA sites, which are widespread in the human genome (Ashe et al. 1997; Vagner et al. 2000; Kaida et al. 2010; Berg et al. 2012). Importantly, however, our data reveal that, if left unshielded, a promoter-proximal CpA site will negatively impact transcription. This phenomenon is general and can be phenocopied using an intronless gene with a bona fide pA site proximal (<500 base pairs [bp]) to its promoter. However, the effect is specific for a pA site-dependent terminator, since its replacement by a pA site-independent histone gene terminator maintains high transcription levels. In line with this, short genes (<500 bp) rarely use pA site-dependent terminators.
Results
Transcription levels do not vary with increased distance between SD and promoter
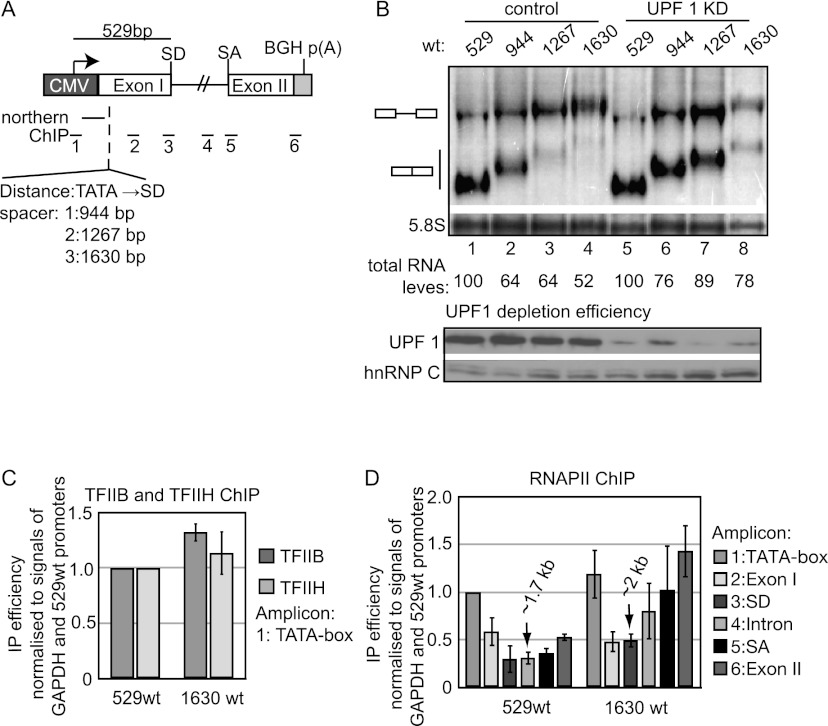
By use of chromatin immunoprecipitation (ChIP) of the GTFs TBP, TFIIB, and TFIIH, together with nuclear run-on (NRO) assays, we previously reported lowered transcription from a CMV promoter upon a (A → T) point mutation of a promoter-proximal SD (denoted as 529m1 in this study) (Supplemental Fig. S1A; Damgaard et al. 2008). These studies did not directly distinguish between SD-assisted tethering of these GTFs to the promoter and the possibility of an inhibitory effect of the SD mutation per se. We therefore examined the impact on transcription activity of increasing the distance between the SD and the promoter, predicting that GTF recruitment would decrease with increased promoter–SD distance, provided the promoter-proximal SD acts in a stimulatory manner. To this end, spacer elements were inserted into exon I of the original HIV1-ENV gene construct (denoted as 529wt in this study), creating the 944wt, 1267wt, and 1630wt constructs, named according to the distance in base pairs between the first T of the TATA box of the CMV promoter and the exon/intron border (Fig. 1A; Supplemental Fig. S1A). These genes were subsequently integrated site-specifically into HEK293 Flp-In T-Rex cells. Upon induction with tetracycline (Tet) for 24 h, all constructs produced unspliced RNA, and levels of spliced RNA were lowered with increasing promoter–SD distance (Fig. 1B, left half of image labeled “control”). Depletion of the essential nonsense-mediated decay (NMD) factor UPF1 (Mendell et al. 2002) led to stabilization of the spacer element-containing mRNA species (Fig. 1B, right half of image labeled “UPF1 KD”). Rather than being caused by lowered transcription, it therefore appears that decreased mRNA levels mainly result from decreases in transcript half-lives, presumably due to the presence of short ORFs in the inserted spacer elements.
Figure 1.
HIV1 gene transcription activity is unaffected by promoter–SD distance. (A) Schematic representation of exon/intron structure of the assayed HIV1-ENV gene. The insertion site of spacer DNA, together with positions of Northern probes and ChIP amplicons, is shown. (B) Northern blotting analysis of total RNA harvested from the indicated cell lines after 24 h of Tet induction and treated with either control (eGFP) (lanes 1–4) or UPF1 (lanes 5–8) siRNAs. Note the underloading of lane 8. Migration of spliced and unspliced HIV-1 RNA is indicated to the left of the gel. Quantification of total HIV-1 RNA (spliced plus unspliced) normalized to levels of the 5.8S rRNA loading control is shown below the gel. Total RNA levels of 529wt were set to 100% in control and UPF1 KD samples, respectively. Depletion of UPF1 was verified by Western blotting analysis using anti-UPF1 antibody. hnRNP C was used as a loading control. (C) TFIIB and TFIIH CMV promoter ChIP analysis of cells subjected to 24 h of Tet induction. ChIP values were background-subtracted (no antibody control) and normalized to GAPDH promoter signals from the same samples. Signals from the 529wt promoter region were set to 1. Histograms represent averages of three independent biological experiments, each with three qPCR replicates. Standard deviations are shown. (D) RNAPII occupancy throughout the indicated loci measured by RPB1 ChIP using amplicons depicted in A and analyzed and displayed as in C. Numbered arrows denote the distance from the start of the CMV TATA box to the 3′ end of the reverse primer of the respective amplicon.
Hence, to more directly assay for transcriptional activity, we focused on the 529wt and 1630wt constructs and performed ChIP assays of TFIIB and TFIIH (Fig. 1C) as well as the largest subunit of RNAPII, RPB1 (Fig. 1D). Both TFIIB and TFIIH ChIP levels were equal between the two promoters, indicating comparable transcription activities of the 529wt and 1630wt genes. Consistently, RNAPII ChIP levels for the two genes were similar in the promoter and 5′ regions (Fig. 1D, amplicons 1 and 2). We noted an increased RPB1 occupancy in the 3′ part of the 1630wt gene (Fig. 1D, amplicons 4–6). Although the exact basis for this accumulation of RNAPII is not known, it appears to be linked to the distance traveled by RNAPII, since 529wt and 1630wt amplicons positioned at similar distances to the CMV promoter displayed largely equal RBP1 ChIP signals (Fig. 1D, note arrows indicating distances of amplicons to the CMV TATA box). Taken together, the GTF and RNAPII ChIP data imply that transcription initiation rates are equal for the two assayed genes and therefore at odds with SD-directed tethering of GTFs to the gene promoter.
Deficient U1 snRNP recruitment to the SD activates CpA
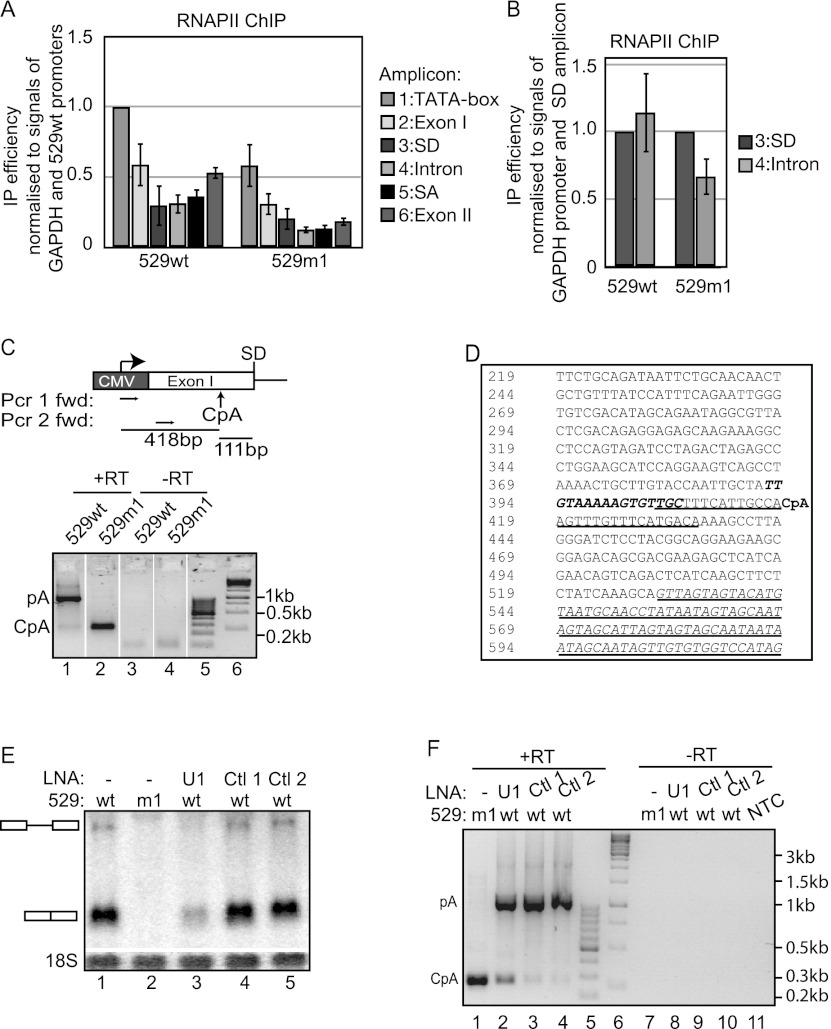
Given the lack of support for a direct transcription stimulatory role of the SD, we next considered the possibility of an inhibitory effect of its mutation. As new constructs containing additional mutations around the SD to further decrease the number of possible hydrogen bonds between the HIV1-ENV SD and U1 snRNA yielded RNA levels and a transcription phenotype comparable with those of the 529m1 gene (data not shown), we surmised that low transcription of 529m1 was not caused by aberrant/residual U1 snRNP binding to the point-mutated m1 site, but rather was due to the complete lack of U1 snRNP recruitment. Moreover, when comparing RNAPII ChIP profiles of the 529m1 and 529wt genes, we noticed that, in addition to a generally lower level of RNAPII over the 529m1 locus (Fig. 2A), the fraction of RNAPII passing the exon/intron border and continuing into the intron of the 529m1 gene was reduced by approximately twofold compared with 529wt (Fig. 2B). This decrease mirrors previous NRO data (Damgaard et al. 2008) and suggested m1-induced premature transcription termination of RNAPII. Given the transcription termination activities of pA sites (Proudfoot 2011) and the reported ability of the U1 snRNP to shield CpA sites (Ashe et al. 1997; Vagner et al. 2000; Kaida et al. 2010; Berg et al. 2012), we used an oligo(dT)-dependent 3′ rapid amplification of cDNA ends (RACE) technique to investigate the production of adenylated RNAs due to the presence of possible premature pA sites in the 529m1 gene. As expected, the method identified full-length spliced 529wt RNA derived from termination at the bovine growth hormone (BGH) pA signal (verified by sequencing) (Fig. 2C, lane 1). However, when assaying 529m1 RNA, 3′ RACE produced a reverse transcriptase-dependent PCR product of a smaller size (Fig. 2C, lane 2). Subsequent sequencing revealed that this product originated from the utilization of a CpA signal triggering RNA cleavage and adenylation at a position located 111 nucleotides (nt) upstream of the exon/intron border (Fig. 2C [top panel], D). The unspliced 529m1 RNA, polyadenylated at the BGH pA site, was not detected due to the unfavorable conditions used for 3′ RACE of this long RNA but could be detected when using 3′ end PCR primers (data not shown).
Figure 2.
U1 snRNP/SD interaction suppresses a CpA site upstream of the HIV-1 ENV exon/intron border. (A) RNAPII ChIP analysis of 529wt and 529m1 cells subjected to 24 h of Tet induction using ChIP amplicons depicted in Figure 1A and analyzed and displayed as in Figure 1D. (B) RNAPII SD and Intron amplicon signals from A plotted with SD signal set to 1 for both 529wt and 529m1 genes. (C, top) Schematic drawing of the positions of 3′ RACE forward primers and the mapped CpA site in relation to the CMV promoter and the exon/intron border. (Bottom) 3′ RACE seminested RT–PCR of total RNA harvested from 18-h Tet-induced 529wt or 529m1 cells as indicated. “+RT” and “−RT” denote the addition or omission of reverse transcriptase, respectively. The sizes of DNA markers run in lanes 5 and 6 are indicated at the right. (D) Sequence of the DNA region surrounding the mapped CpA site (denoted by bold “CpA”). The intron sequence is underlined, bold italic letters denote a deleted upstream region in 529wt/m1Δ−27– −12, and a plain underlined sequence around the CpA site denotes the deleted sequence of 529wt/m1ΔCpA. Numbering annotates the distance in base pairs to the first T of the TATA box. (E) Northern blotting analysis of HIV-1 RNA harvested from 12-h Tet-induced 529wt or 529m1 cells treated with either “anti-U1” or control LNA/DNA hybrids. The Ctl1 and Ctl2 sequences were directed toward the antisense strand of eGFP and the yeast SSA4 RNA, respectively. Migration of spliced and unspliced HIV1 RNA is indicated at the left of the gel. 18S rRNA was used as a loading control. (F) 3′ RACE seminested RT–PCR on RNA samples from E analyzed and displayed as in C. (NTC) No template control.
To gain further support for the indicated role of U1 snRNP in suppressing cryptic cleavage/pA in the 529wt context, we functionally depleted U1 snRNP in 529wt cells by transfecting an LNA/DNA hybrid oligo (anti-U1) complementary to the 5′ arm of U1 snRNA, which is necessary for interaction with the SD site. Northern blotting analysis revealed reduced steady-state levels of both spliced and unspliced 529wt RNA upon anti-U1 transfection, which was not observed after the transfection of control LNA/DNA hybrids (Fig. 2E). Moreover, 3′ RACE analysis demonstrated that U1 snRNP inhibition triggered some utilization of the same CpA site that was activated in the 529m1 context (Fig. 2F). Taken together, we conclude that U1 snRNP is preventing the usage of an upstream CpA site in the 529wt context, which can be activated by disrupting the interaction between the nascent HIV1 RNA and the U1 snRNP. We note that this is a newly discovered HIV1 CpA site, not to be confused with the promoter-proximal pA site residing in the 5′ long terminal repeat (LTR) of a full-length HIV-1 construct (Ashe et al. 1995).
CpA site usage depends on conventional 3′ end processing factors and causes lower transcriptional activity
To obtain more direct evidence linking the newly found CpA site to transcription down-regulation, we sought to eliminate its usage. Since no clear consensus motif expected to promote cleavage/pA (Beaudoing et al. 2000; Ozsolak et al. 2010) is present in the vicinity of the mapped CpA site, we initially constructed a 529m1 gene variant lacking the −27- to −12-bp segment upstream of the CpA cleavage site (529m1Δ−27– −12) (Fig. 2D; Supplemental Figs. S1A, S2A), usually important at bona fide pA sites. However, this deletion did not alter steady-state RNA levels (Supplemental Fig. S2B) and gave rise to several 3′ RACE products, including that of the CpA (see Supplemental Fig. S2C, lane 4). Thus, while the upstream region has some influence on CpA site usage, its deletion appears to simply activate new ones. As a next attempt, we deleted 30 bp encompassing the CpA site itself (529m1 ΔCpA) (Supplemental Figs. S1A, S2D), but again, steady-state RNA levels from this cell line were similar to its parental 529m1 counterpart (Supplemental Fig. S2E). 3′ RACE and sequencing analyses of RNA derived from the 529m1 ΔCpA construct revealed a fast-migrating PCR product (Supplemental Fig. S2F), corresponding to a cleavage event 157 nt upstream of the SD (Supplemental Fig. S2G), which was also detectable from the 529m1Δ−27– −12 gene construct (Supplemental Fig. S2C, arrow). We conclude that this particular region is generally susceptible to CpA in the absence of U1 snRNP binding.
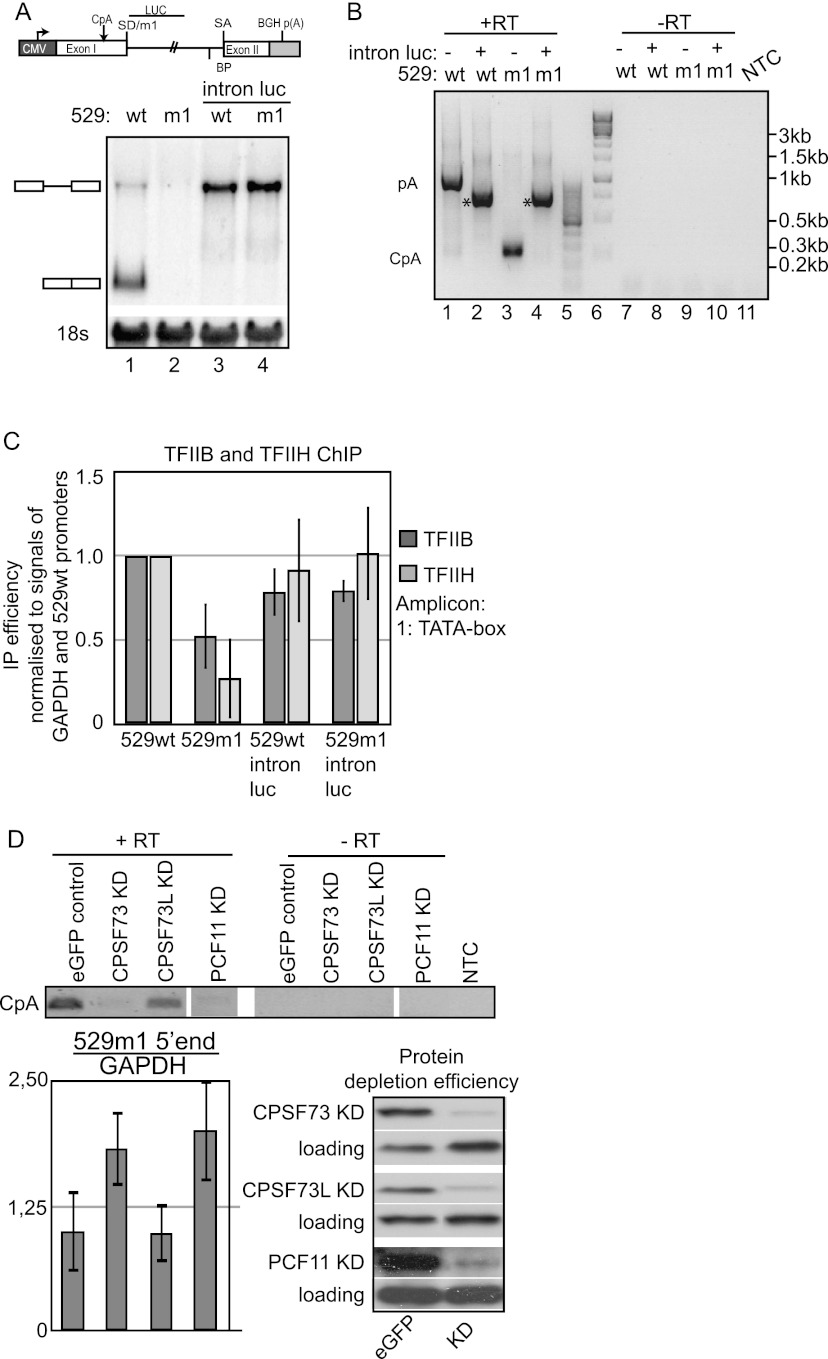
Being unable to eliminate the CpA by deletions in the proximity of the CpA site, we next focused on downstream sequence information and constructed cell lines harboring 529m1 or 529wt genes with the first half of their introns exchanged with a fragment from the Luciferase gene (yielding 529m1-intronLuc and 529wt-intronLuc, respectively) but leaving the original m1 and wild-type SD sequences intact (Fig. 3A, top; Supplemental Fig. S1A). For both constructs, Northern blotting analysis of steady-state RNA revealed robust and similar levels of solely unspliced transcripts, which were polyadenylated at the BGH pA site (verified by sequencing) (Fig. 3A, bottom). Consistently, 3′ RACE analysis did not detect spliced RNA from either of the constructs and did not provide any evidence for CpA site usage (Fig. 3B). Thus, the 5′ part of the HIV-ENV intron contains sequences necessary for CpA site utilization. Moreover, since the 529wt-intronLuc pre-mRNA does not splice, the 5′ part of the HIV-ENV intron also harbors elements required for splicing. More importantly, however, when measuring transcription activity by TFIIB and TFIIH ChIP analysis, we observed that both 529wt-intronLuc and 529m1-intronLuc constructs display TFIIB and TFIIH promoter occupancies comparable with the 529wt promoter (Fig. 3C). Thus, direct abolishment of CpA site utilization restores transcriptional activity in the 529m1 gene context.
Figure 3.
Inhibition of CpA usage causes transcription derepression. (A, top) Schematic drawing of HIV1-ENV constructs, indicating the part of the intron exchanged by luciferase sequence to create 529wt-intronLuc and 529m1-intronLuc. SD/m1, splice acceptor (SA), and branch point (BP) sequences were all left intact. (Bottom) Northern blotting analysis of total RNA harvested from 529wt-intronLuc and 529m1-intronLuc cells after 24 h of Tet induction. The loading control and gel annotation are as in Figure 2E. (B) 3′ RACE seminested RT–PCR on RNA samples from A analyzed and displayed as in Figure 2C. Asterisks indicate an RT–PCR product arising from internal dT priming of a short stretch of As in the luciferase insertion. (C) TFIIB and TFIIH CMV promoter ChIP analyses of cells from A analyzed and displayed as in Figure 1C. (D) Semiquantitative 3′ RACE of 529m1 RNA harvested from cells treated with siRNA against eGFP, CPSF73, CPSF73L, or PCF11 as indicated. First and second PCRs contained 15 and 35 cycles, respectively. RT-qPCR levels of the extreme 5′ end of 529m1 RNA are shown relative to GAPDH RT-qPCR levels of the same samples. Standard deviations were calculated from three independent qPCR reactions. Protein depletions were verified by Western blotting analysis using the indicated antibodies. Loading was controlled by the display of cross-reacting protein species.
Our inability to define the cis requirements for CpA site usage prompted us to inquire whether this site uses conventional 3′ end processing factors for its recognition at all. To this end, we depleted cells expressing the 529m1 construct for the pre-mRNA 3′ end endonuclease CPSF73 and the 3′ end processing/termination factor PCF11, respectively, and conducted semiquantitative 3′ RACE to monitor CpA site usage (Fig. 3D). In both depletion contexts, CpA usage was robustly decreased, whereas control depletion of CPSF73L, the catalytic subunit of the Integrator complex involved in snRNA 3′ end processing (Baillat et al. 2005), had no effect. Lack of CpA site usage upon CPSF73 and PCF11 depletion was not simply due to lower transcript levels, as RT-qPCR analysis of the extreme 5′ end of 529m1 RNA revealed an approximately twofold increase in both experimental situations (Fig. 3D, “5′ end”). These results suggest that 3′ end processing at the CpA site relies on bona fide cleavage/pA factors.
The HIV1 CpA site down-regulates transcription only when placed in a promoter-proximal position
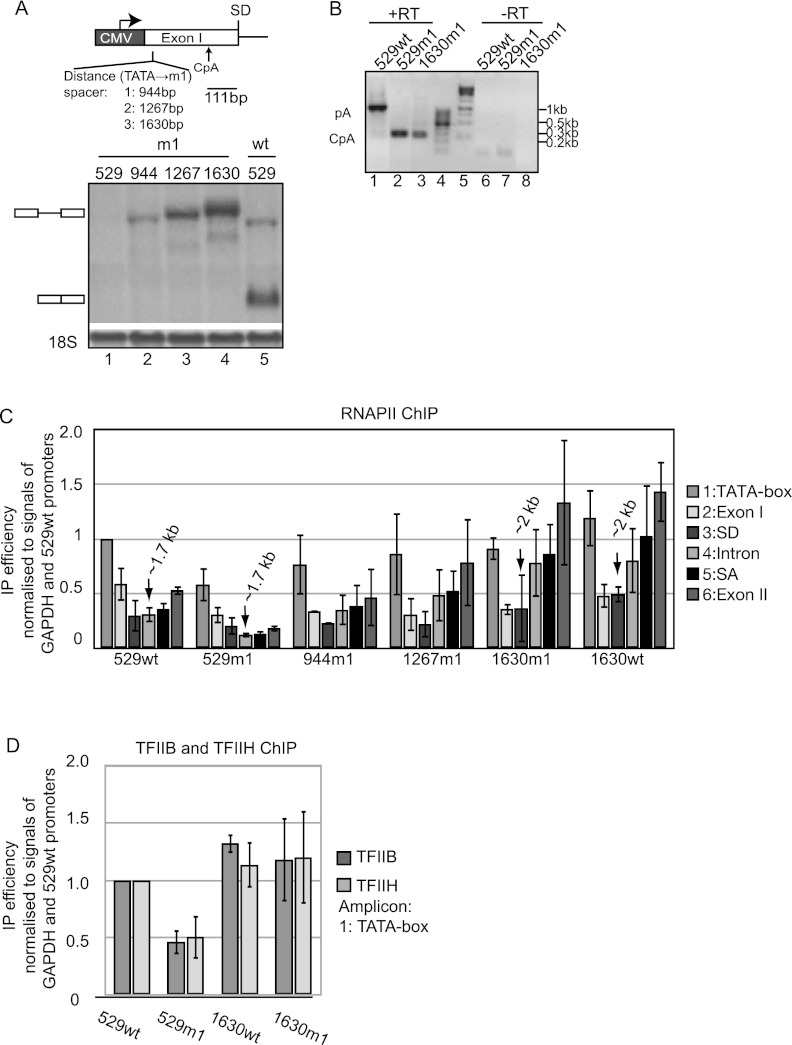
Typically, CpA sites have been detected in introns or 3′ untranslated regions (UTRs), but negative effects on transcription have not been reported (Kaida et al. 2010; Berg et al. 2012). In contrast, the CpA site discovered in this study is positioned in the first exon and gives rise to an ∼430-nt intronless transcript. To elucidate whether the relatively short distance between the promoter and the CpA site caused the observed impact on transcription, we gradually increased this length by inserting spacer elements into exon I of the 529m1 construct, creating the new reporters 944m1, 1267m1, and 1630m1 (Fig. 4A, top; Supplemental Fig. S1A). Northern blotting analysis of steady-state RNA showed increased levels of unspliced RNA when increasing the promoter/m1 (CpA) distance (Fig. 4A, bottom). The dramatic difference between 529m1 and 1630m1 RNA levels occurred despite the fact that the SDm1-activated CpA site was being used in both cases (Fig. 4B). Moreover, increased RNA levels were not observed when inserting an equally long spacer into either the intron or exon II downstream from the CpA site, demonstrating that rescue of RNA levels was not simply due to extension of the transcription unit (data not shown). Measuring transcription activity by RNAPII ChIP (Fig. 4C, cf. amplicons denoted with arrows) demonstrated that at least part of the increase in RNA levels was based on increased transcription activity. The virtually identical RNAPII ChIP profiles of the 1630wt and 1630m1 loci revealed that altering the position of the CpA to a promoter-distal site also suppressed the predominant premature transcription termination phenotype of 529m1 (Fig. 4C). This occurred with a concomitant reversal of the suppressed transcription initiation rate, as assessed by TFIIB and TFIIH ChIP (Fig. 4D). We conclude that the CpA site needs to be promoter-proximal in order to lower transcription.
Figure 4.
Promoter-proximal CpA utilization down-regulates transcription. (A, top) Schematic drawing showing the spacer insertion site relative to the CpA position. (Bottom) Northern blotting analysis of total RNA harvested from indicated cells Tet-induced for 24 h. The loading control and gel annotation are as in Figure 2E. (B) 3′ RACE seminested RT–PCR on total RNA samples from the 529wt, 529m1, and 1630m1 cell lines from A analyzed and displayed as in Figure 2C. Sizes estimated from DNA markers in lanes 4 and 5 are indicated at the right. (C) RNAPII ChIP analysis of the indicated loci using conditions and amplicons as in Figure 1D. Data were analyzed and displayed as in Figure 1D. (D) TFIIB and TFIIH CMV promoter ChIP analysis of indicated loci analyzed and displayed as in Figure 1C.
Bona fide pA sites lower transcription when placed promoter-proximally
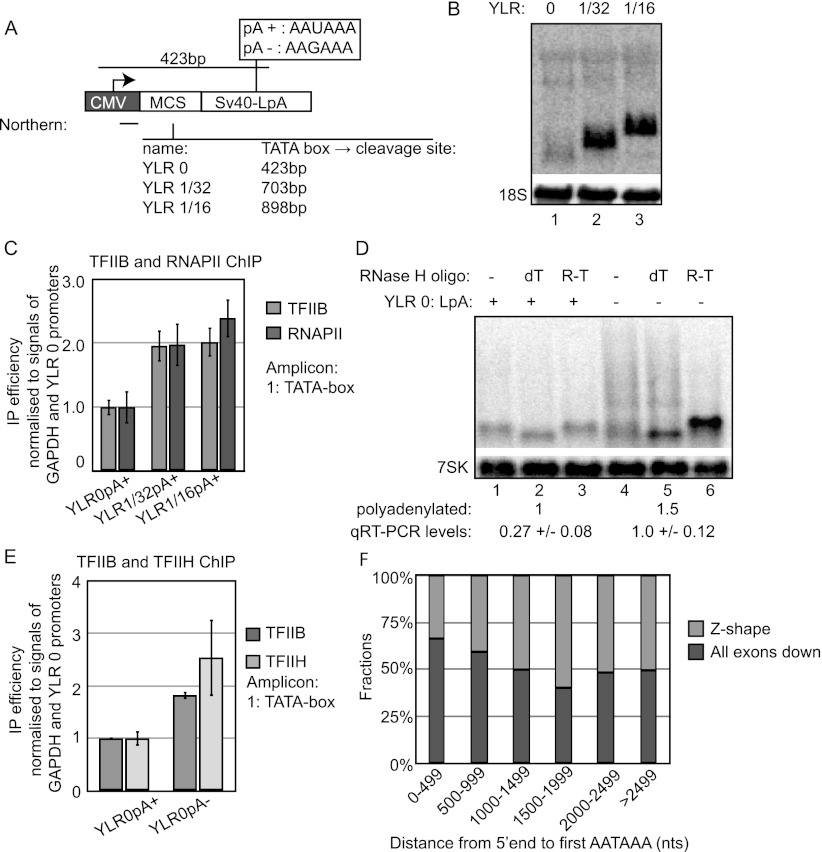
We speculated whether the observed transcriptional decrease by a promoter-proximal CpA site was a specific HIV1-ENV example. Moreover, lacking a full sequence characterization of the HIV1 CpA, we also wondered whether a bona fide pA site would exert a similar activity. To examine these questions, we measured steady-state RNA and transcription levels from newly constructed and stably integrated loci harboring a “normal” SV40-late pA (LpA) site situated at varying distances from the CMV promoter. To circumvent any possible interference from processing signals, we selected splicing-inert DNA derived from the pcDNA5 multiple cloning site as well as from the YLR454W Saccharomyces cerevisiae gene as spacers between the promoter and pA site (Fig. 5A; Supplemental Fig. S1B). The YLR0pA+ gene, producing a mature RNA with an estimated length comparable with the 529m1 CpA locus, yielded low steady-state RNA levels, as revealed by Northern blotting analysis (Fig. 5B). In contrast, longer YLR gene variants with distances from the TATA box to the pA cleavage site of 703 bp and 898 bp, respectively, yielded considerably higher levels. TFIIB and RNAPII promoter occupancies, as measured by ChIP, demonstrated that part of this difference was based on lowered transcription of the YLR0pA+ gene (Fig. 5C). Thus, promoter-proximal positioning of a bona fide pA site also challenges transcription.
Figure 5.
A functional promoter-proximal SV40-LpA site negatively affects transcription. (A) Schematic overview of the “YLR constructs.” YLR0 contains a multiple cloning site (MCS) sequence, while the fractions in YLR1/32 and YLR1/16 indicate which portions (from the 5′ end) of the YLR454W gene were inserted. All constructs harbor either a wild-type (wt) (pA+) or a point-mutated (pA−) SV40-LpA site as indicated in the box. The horizontal black line below “CMV” denotes the position of the radiolabeled probe used for Northern blotting analysis. Distances from the CMV TATA box to the SV40-LpA cleavage site of the various constructs are indicated. (B) Northern blotting analysis of total RNA harvested from the indicated cells Tet-induced for 24 h. The loading control is as in Figure 2E. (C) TFIIB and RNAPII CMV promoter ChIP analysis of the indicated loci analyzed and displayed as in Figure 1C. (D) RNase H/Northern blotting analysis of RNA harvested from the indicated cells Tet-induced for 24 h. Total RNA was treated with RNase H and DNA oligonucleotides directed against either the pA tail (“dT”) or the readthough region (“R-T”) immediately downstream from the SV40-LpA site. Numbers below the gel show quantification of transcripts polyadenylated at the SV40-LpA site as well as total transcript levels as measured by qRT–PCR using an mRNA-specific reverse transcription reaction followed by PCR of the YLR 5′ end and normalized to endogenous GAPDH levels. Standard deviations are based on qPCR triplicates. (E) TFIIB and TFIIH CMV promoter ChIP analysis of the indicated loci analyzed and displayed as in Figure 1C. (F) Fractions of the “all exons down” (dark gray) and “Z-shaped” (light gray) distributions (as previously defined by Berg et al. 2012) as a function of the distance between the 5′ end and the first AAUAAA hexamer in transcripts.
To cement this interpretation, we constructed the YLR0pA− gene, harboring a point-mutated pA site (AAUAAA → AAGAAA), which dramatically lowers the efficiency of pA site recognition with a consequent decrease in transcription termination efficiency (Supplemental Fig. S1B; Mapendano et al. 2010). As expected, Northern blotting analysis demonstrated that stable cells harboring the YLR0pA− gene produced elevated levels of readthrough RNA as compared with YLR0pA+ gene-containing cells (Fig. 5D). DNA oligo-triggered RNase H cleavage of added pA tails (Fig. 5D, “dT,” lanes 2,5) or readthrough RNA (Fig. 5D, “R-T,” lanes 3,6) revealed that, although the mutated pA to a minor extent was used, the clear majority of transcription events ignored this sequence. Thus, effectively, the mutated pA site resulted in a longer distance traveled by the majority of RNAPII complexes, hence mimicking a longer transcription unit. Moreover, quantification of YLR0pA+ and YLR0pA− transcripts polyadenylated at the SV40-LpA site—and thus presumed to have similar half-lives—revealed a 1.5-fold increase of YLR0pA− species (Fig. 5D, “polyadenylated”). In addition, more readthrough RNA was produced in YLR0pA− cells, amounting to an approximately fourfold increase when both polyadenylated and readthrough RNA were quantified by quantitative RT–PCR (qRT-PCR) using an amplicon close to the transcription start site (Fig. 5D, “qRT–PCR levels”). Thus, the YLR0pA− gene produces more transcript than YLR0pA+. Accordingly, TFIIB and TFIIH ChIP levels were elevated in the promoter of the YLR0pA− compared with the YLR0pA+ gene (Fig. 5E). To ascertain that pA site mutation would only cause transcription derepression of genes harboring a promoter-proximal pA site, we also altered the pA sites of the YLR1/32pA+ and YLR1/16pA+ constructs, creating YLR1/32pA− and YLR1/16pA− (Supplemental Fig. S1B). Although pA site usage was virtually abolished on these genes (Supplemental Fig. S3A, lanes 9,11), their steady-state level RNA output did not differ considerably from that of their pA+ counterparts (Supplemental Fig. S3A, cf. lanes 4 and 10, and lanes 6 and 12). Moreover, TFIIB and TFIIH occupancies of the YLR1/16pA+ and YLR1/16pA− promoters were similar (Supplemental Fig. S3B). We conclude that repression of transcription is triggered by a promoter-proximal bona fide pA site, provided it is efficiently recognized.
To investigate the generality of this observation—i.e., whether a similar phenomenon can be observed for endogenous human genes—we analyzed data from a recent study by Dreyfuss and colleagues (Berg et al. 2012). Here, a U1 antisense morpholino was transfected into HeLa cells for 8 h to inhibit U1 snRNP function, and differential RNA expression was assayed with a massive parallel sequencing method called HIDE-seq. The investigators defined 10 different response categories. The so-called “Z-shaped” response refers to transcripts where the 5′ part is up-regulated upon U1 snRNP inhibition, whereas the 3′ part is down-regulated. This is indicative of premature 3′ end formation at the transition between the 5′ and 3′ parts, which was confirmed for a subset of transcripts (Berg et al. 2012). Another response category, “all exons down,” where the entire transcript is down upon U1 snRNP inhibition, could expectedly hold transcripts subjected to premature 3′ end formation and ensuing transcriptional down-regulation. We therefore selected these two categories and compared the distance between transcript 5′ ends and their first AAUAAA hexamer. A statistical significant tendency was revealed toward promoter proximity of AAUAAA sites in the “all exons down” group (Fig. 5F, P-value < 0.05). This is in accordance with the hypothesis that early pA sites are general triggers of transcriptional down-regulation.
Short genes use pA site-independent terminators
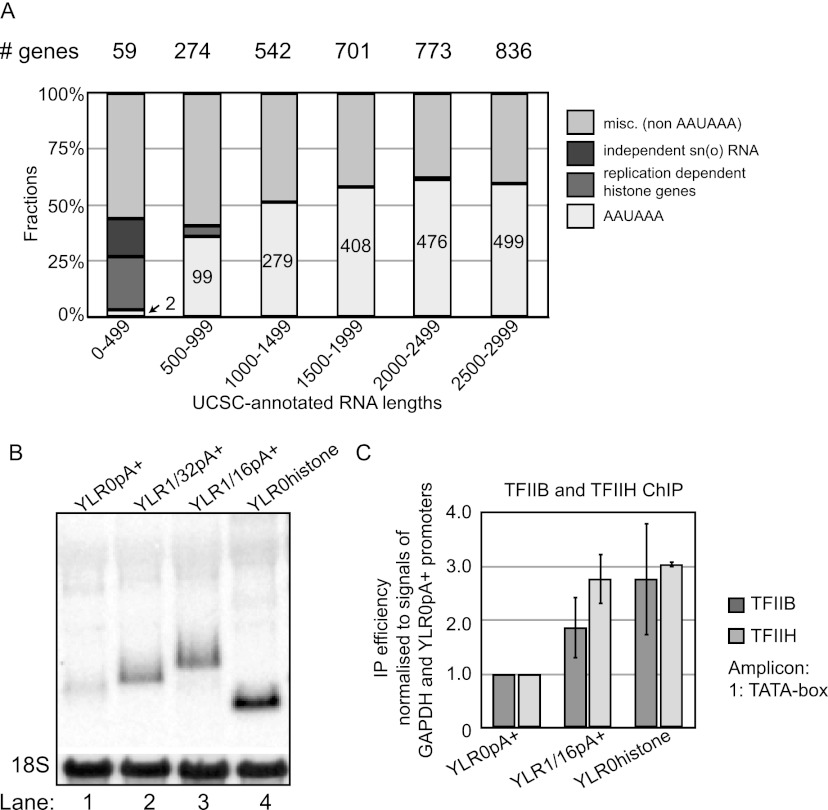
Results obtained so far beg the question: How many human genes that produce RNAs below the ∼500-nt cutoff demonstrated here to be nonsupportive of optimal transcription use a pA site-dependent transcription terminator? To address this question, we calculated the fraction of genes annotated in the University of California at Santa Cruz (UCSC) Genome Browser and harboring a consensus AAUAAA hexameric pA site motif in their 3′ UTRs as a function of their annotated RNA length. Of 59 genes predicted to produce RNAs <500 nt with annotated ORFs as well as 3′ and 5′ UTRs, together with snRNAs and independent snoRNAs, only two (3.3%) harbor a conserved AAUAAA motif in the 3′ UTR (Fig. 6A). This fraction rises to 99 of 274 (36.1%) when considering AAUAAA-containing genes producing RNAs of 500–999 nt in length and stays high with increasing RNA lengths. Thus, human RNAPII transcribed genes yielding products <500 nt in length and using a consensus pA site-dependent terminator are extremely rare. Instead, 14 of the 59 RNAs <500 nt are made from replication-dependent histone genes, which use a stem–loop-dependent 3′ end processing system to elicit transcription termination (Fig. 6A; Marzluff et al. 2002). Other major RNAPII transcribed genes producing transcripts <500 nt are snRNA loci (U1, U2, U4, U5, U11, U12, and U4atac) (Montzka and Steitz 1988; Jawdekar and Henry 2008) and loci producing the independently transcribed and capped snoRNAs (U3, U8, and U13). These genes also have specialized 3′ end processing/transcription termination systems (Kim et al. 2006; Egloff et al. 2008).
Figure 6.
Short human genes acquire pA site-independent terminators for their optimal expression. (A) Fraction of human genes harboring different transcription terminators plotted as a function of the predicted lengths of their produced RNAs. Annotated genes from the UCSC Genome Browser (http://genome.ucsc.edu) hg18 assembly were analyzed as follows: Genes with no ORF and without either a 5′ UTR or a 3′ UTR were discarded. Remaining examples were divided based on the estimated length of their produced RNA as indicated in the histogram. “AAUAAA” denotes RNAs with this consensus sequence in the 3′ UTR, and the actual number of genes with AAUAAA in the 3′ UTR is annotated in the respective bar. “Replication dependent histone genes” were taken from Marzluff et al. (2002). Furthermore, independently transcribed and capped sn(o)RNAs (U1, U2, U3, U4, U5, U8, U13, U11, U12, and U4atac) were included. “Misc. (non-AAUAAA)”denotes annotated genes that did not fall into the above-mentioned groups. The number of genes in each size category is indicated above the histogram, and the total fraction is set to 100%. (B) Northern blotting analysis of total RNA harvested from the indicated cells Tet-induced for 24 h. The loading control is as in Figure 2E. (C) TFIIB and TFIIH occupancies at the promoters of the indicated gene constructs analyzed and displayed as in Figure 1C.
If “short” RNAPII transcribed genes generally avoid the undesirable consequences of having promoter-proximal pA sites by switching to a different termination system optimized for short loci, one should be able to relieve transcription repression of the YLR0pA+ gene by replacing its SV40-LpA site with a pA site-independent terminator. To test this idea, we inserted the 3′ end of the replication-dependent histone gene HISTH2AA3 in place of the SV40-LpA site to construct the YLR0Histone gene (Supplemental Fig. S1B), designed to produce a RNA with a length comparable with that of the YLR0pA+ transcript. Northern blotting analysis revealed a YLR0Histone RNA species appearing slightly shorter than the YLR0pA+ RNA, presumably due to its lack of a pA tail (Fig. 6B, cf. lanes 1 and 4). This transcript was present in higher levels than YLR0pA+ RNA. Consistent with an underlying derepression of transcription, we found that the YLR0Histone promoter was occupied by approximately threefold more TFIIB and TFIIH than that of the YLR0pA+ promoter (Fig. 6C). Thus, the negative effect on transcription caused by a promoter-proximal termination site is not due to the promoter proximity itself but depends on the nature of the terminator.
Discussion
With the original aim to dissect the mechanism governing transcription down-regulation associated with a point-mutated HIV1-ENV SD site (Damgaard et al. 2008), we discovered a negative impact of promoter-proximal pA sites on transcription. The insertion of spacer elements in between the CMV promoter and the wild-type or mutated HIV1-ENV SD sites established that while the wild-type SD exhibited no distance-dependent effect on transcription, the mutated SD exercised a strong and distance-dependent negative impact. Thus, in the studied context, rather than the promoter-proximal SD site being transcriptionally stimulatory, as previously suggested (Damgaard et al. 2008), the lowered HIV1-ENV transcription phenotype is better explained by a negative influence of SD mutation. We go on to show that such negative effect is mediated through the activation of premature cleavage and pA (PCPA) at a newly identified CpA site upstream of the HIV1-ENV SD. While we were unable to determine a consensus sequence element that constitutes the CpA signal, we used RNAi-mediated depletion to show that it requires bona fide cleavage and pA factors for its usage.
As introduction of LNA/DNA anti-U1 also resulted in PCPA at the 529wt locus, we conclude that utilization of the HIV1-ENV CpA site under normal circumstances is suppressed through a U1 snRNP-dependent process. Deletion of the HIV1-ENV splice acceptor did not elicit transcription down-regulation (data not shown). Thus, consistent with the lack of CpA activation by disruption of U2 snRNP-dependent processes (Kaida et al. 2010), we suggest that U1 snRNP binding per se, rather than active splicing, leads to CpA site shielding. The notion that U1 snRNP can protect transcripts against PCPA has been made earlier (Ashe et al. 1997; Vagner et al. 2000). Although these studies did not report direct effects of PCPA back on transcription, it is interesting to note that a significant fraction of genes analyzed by Dreyfuss and colleagues (Kaida et al. 2010; Berg et al. 2012) displayed reduced RNA levels throughout the transcription unit when U1 snRNP was inhibited. We propose that some of these undergo PCPA within the first few hundred promoter-proximal nucleotides of the transcript, eliciting transcription down-regulation. Consistently, a larger fraction of genes in the “all exon down” subgroup defined by Berg et al. (2012) harbors an AAUAAA hexamer within 500 nt of their annotated 5′ ends as compared with the “Z- shaped” subgroup. Transcriptional down-regulation in combination with the apparent reduced stability of short transcripts (Figs. 2E, 5B) likely explains the low RNA levels in this subgroup. It is also interesting to note that while Berg et al. (2012) mainly found PCPA events occurring in introns (downstream from SD sites), the two reported cases of promoter-proximal CpA shielding (Ashe et al. 1997; this study) disclose shielded CpA sites upstream of SD sequences. Thus, it is apparent that U1 snRNP can shield both upstream and downstream pA sites. The ability of U1 snRNP to suppress CpA usage may be less kinetically constrained in a promoter-proximal context where other features (e.g., RNAPII CTD status) contributing to pA site usage are suboptimal. Conversely, pA sites placed in their functional context of gene 3′ ends are likely in a more ideal position to fend off U1 snRNP suppression. However, further dissection of the requirements for CpA site utility is needed to completely understand its kinetic and structural relationship to transcription.
When moving the mutated HIV1-ENV SD site away from the CMV promoter, PCPA still occurred but without affecting transcription rates. Therefore, promoter-proximal PCPA is not simply a consequence of low gene activity, but rather the cause of the transcription phenotype. To study this phenomenon in a gene context devoid of splicing signals, we produced a set of constructs of variable lengths terminated by the bona fide SV40-LpA signal and found that a gene with a distance between the TATA box and the pA cleavage site of 424 bp is transcriptionally challenged. Remarkably, extending this gene to produce an RNA just 279 nt longer triggered “maximal” transcriptional activity. This leads us to propose that (1) cross-talk between mRNA processing and transcription can, at least in some cases, be simplified to the functional communication between pA site utility and transcription reinitiation and (2) the mechanism accounting for pA site-induced down-regulation of transcription functions within a relatively narrow window of ∼500 nt from the promoter. Consistently, Guo et al. (2011) observed a similar gene shortening-dependent decrease in mRNA levels with a largely similar cutoff between genes, giving rise to high and low transcript levels. We suggest that these short genes were also subject to promoter-proximal pA site-dependent transcriptional down-regulation.
Our bioinformatics analysis implies that “short” RNAPII transcribed genes circumvent the undesirable consequences of having promoter-proximal pA sites by switching to a different transcription termination system optimized for such short loci. Consistent with this notion, replacing the transcription inhibitory pA site of YLR0pA+ with the terminator of the replication-dependent HISTH2AA3 histone gene fully restored both steady-state RNA and transcription levels. Thus, short gene length does not, per se, elicit a negative effect on transcription, which rather appears to be a specific feature of pA site-mediated termination. Why are short genes with pA site-dependent terminators transcriptionally challenged? We previously demonstrated that, for longer genes, optimal 3′ end processing positively impacts transcription initiation, possibly through the enhanced recycling of initiation-prone RNAPII from the gene terminator back to the promoter (Mapendano et al. 2010; Lykke-Andersen et al. 2011). In line with this notion, pA site-containing genes below a certain length may inherently not support RNAPII recycling. Perhaps a too high CTD-Ser5/7P to Ser2P ratio in the early phases of transcription is refractory to efficient pA site-dependent termination and its coupling to reinitiation and preparation for a new transcription event. In addition, potential physical contacts between promoter and terminator regions may be suboptimal for genes below a certain length due to spatial constraints. Highly expressed short RNAPII transcribed genes, like U snRNA and replication-dependent histone genes, have presumably instead evolved an initiation/termination system that circumvents the apparent inefficiency of recycling RNAPII. In fact, the U1 snRNA gene uses promoter and termination sequences that are codependent (de Vegvar et al. 1986; Hernandez and Weiner 1986; Ramamurthy et al. 1996; Egloff et al. 2008), and proper 3′ end formation of U1 snRNA is only optimal when the total length of the transcript is <280 nt (Ramamurthy et al. 1996). Exploring which features allow such robust expression of a short gene may help delineate the seemingly different mechanisms in gene expression strategies used by pA site-dependent and -independent terminators.
Why do CpA sites reside in so many viral and human genes (Furth et al. 1994; Ashe et al. 1995; Kaida et al. 2010; Berg et al. 2012)? Although most CpA sites have been identified through the artificial disruption of the interaction between U1 snRNP and its target RNA, recent findings show that U1 snRNA shortage in activated neurons triggers the isoform switching of two genes due to altered pA site usage (Berg et al. 2012). It remains to be seen how many genes use U1 snRNA-mediated shielding of CpA sites as a regulatory mechanism. However, it is also feasible that, in some cases, constraints on the regional sequence information, such as certain essential codons or, e.g., a splicing signal, will fortuitously create a CpA site. Efficient shielding by the highly abundant U1 snRNP may ensure that there is no evolutionary pressure to rid genomes of such signals. Finally, the present study raises the possibility that unshielding of promoter-proximal pA sites may also serve as a powerful mechanism for the cell to shut down gene expression.
Materials and methods
Cell lines, plasmids, and cell culture
529wt and 529m1 cell lines were used previously (Damgaard et al. 2008) under the names 5′SS wild type and 5′SS-m1, respectively (for details, see the Supplemental Material). Construction of plasmids 944wt/m1, 1267wt/m1, 1630wt/m1, 529wt/m1ΔCpA, 529wt/m1Δ−27– −12, and 550wt/m1intron-Luc as well as YLR0pA+/−, YLR1/32pA+/−, YLR1/16pA+/−, and YLR0Histone was done by conventional methods (see the Supplemental Material). Production of corresponding stable cell lines was done as previously described (Damgaard et al. 2008). To induce transcription from the CMV promoter, cells were treated with 250 ng/mL Tet for 24 h unless otherwise stated.
siRNA and LNA/DNA transfections
Cells (6 × 105) were seeded in a 6-cm dish in growth medium without antibiotics. Twenty-four hours later, cells were transfected with siRNA at a final concentration of 22.5 nM using SilentFect (Bio-Rad), according to the manufacturer's guidelines. Growth medium was changed 4 h after transfection. Forty-eight hours after the first transfection, cells were retransfected with a final concentration of 22.5 nM siRNA using Lipofectamine 2000 (Invitrogen) as per the manufacturer's protocol. Again, growth medium was changed after 4 h, and, 20 h later, transcription was induced by Tet addition.
U1 LNA/DNA was transfected into 50% confluent cells at a final concentration of 100 nM using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Six hours after transfection, growth medium was changed, and Tet was added for 12 h.
RNA analysis
RNA purification, DNA oligonucleotide-directed RNase H cleavage, Northern blotting analysis, and reverse transcription reactions were done as previously described (Mapendano et al. 2010) using the oligonucleotides listed in Supplemental Table S1. 3′ RACE assays were performed using SuperScript II (Invitrogen) according to the manufacturer's guidelines. Unless otherwise stated, the 3′ RACE dT primer contained an adapter sequence used for subsequent PCR. 3′ RACE seminested PCR fragments were cloned for sequencing using the pCR4 TOPO Vector (Invitrogen) according to the manufacturer's protocol.
ChIP analysis
ChIP experiments were carried out largely as described in Mapendano et al. (2010). Details are described in the Supplemental Material.
qPCR analysis
qPCR reactions were performed with SYBR Green mix (Invitrogen) and processed on a Stratagene Mx3000 or Mx3005 PCR instrument.
Bioinformatics analyses
For the analysis presented in Figure 5F, a list of gene names and their corresponding U1 morpholino response categories were kindly provided by Michael G. Berg and Gideon Dreyfuss (Berg et al. 2012). Transcripts from the response categories “all exons down” (n = 862, median = 5402 nt) and “Z-shaped” (n = 1023, median = 5764 nt) were selected and assigned the most conservative (i.e., the longest possible) distance between the 5′ end of the transcript and the first AAUAAA hexamer. Distance measures from the two response categories were then compared with the Wilcoxon rank-sum test to assess statistical significance. The null hypothesis that there is a location shift of 0 between the two distributions was rejected with a P-value of 0.03847. Furthermore, the proportion of genes with distances <500 nt is significantly larger for the “all exons down” category (25 of 862) compared with the “Z-shaped” category (15 of 1023; P = 0.001548). The bioinformatics approach used for Figure 6A is described in the corresponding legend.
Acknowledgments
Christian K. Damgaard, Christophe K. Mapendano, Manfred Schmid, Marta Lloret Llinares, Evgenia Ntini, Dominico Libri, and Edouard Bertrand are thanked for critical reading of the manuscript. Karina Jurgensen and Dorthe Riishøj are acknowledged for technical assistance. Jesper Bertram Bramsen and Edouard Bertrand are thanked for gifts of reagents. Thomas Birkballe Hansen, Michael G. Berg, and Gideon Dreyfuss are thanked for help with bioinformatics. This work was supported by the Danish National Research Foundation (T.H.J.), the Danish National Research Council (T.H.J.), the Novo Nordisk Foundation (T.H.J.), and the Lundbeck Foundation (S.L.A.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.189126.112.
References
- Ashe MP, Griffin P, James W, Proudfoot NJ 1995. Poly(A) site selection in the HIV-1 provirus: Inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev 9: 3008–3025 [DOI] [PubMed] [Google Scholar]
- Ashe MP, Pearson LH, Proudfoot NJ 1997. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J 16: 5752–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repreat of RNA polymerase II. Cell 123: 265–276 [DOI] [PubMed] [Google Scholar]
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res 10: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zang Z, Cho S, Sherrill-Mix M, Wan L, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD 2001. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science 292: 1863–1876 [DOI] [PubMed] [Google Scholar]
- Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol Cell 29: 271–278 [DOI] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R 2007. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell 26: 867–881 [DOI] [PubMed] [Google Scholar]
- de Vegvar HE, Lund E, Dahlberg JE 1986. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell 47: 259–266 [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Murphy S 2008. Expression of human snRNA genes from beginning to end. Biochem Soc Trans 36: 590–594 [DOI] [PubMed] [Google Scholar]
- Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ 2002. Promoter proximal splice sites enhance transcription. Genes Dev 16: 2792–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth PA, Choe WT, Rex JH, Byrne JC, Baker CC 1994. Sequences homologoues to 5′splice site are required for the inhibitory activity of papillomavirus late 3′ untranslated region. Mol Cell Biol 14: 5278–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Garrett M, Micklem G, Brogna S 2011. Poly(A) signals located near the 5′ end of genes are silenced by a general mechanism that prevents premature 3′-end processing. Mol Cell Biol 31: 639–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N, Weiner AM 1986. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell 47: 249–258 [DOI] [PubMed] [Google Scholar]
- Jawdekar GW, Henry RW 2008. Transcriptional regulation of hman small nuclear RNA genes. Biochim Biophys Acta 1779: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S 2006. Distinct pathways for snoRNA and mRNA termination. Mol Cell 24: 723–734 [DOI] [PubMed] [Google Scholar]
- Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, Proudfoot NJ, Akoulitchev A 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol 9: 800–805 [DOI] [PubMed] [Google Scholar]
- Leichter M, Marko M, Ganou V, Patrinou-Georgoula M, Tora L, Guialis A 2011. A fraction of the transcription factor TAF15 participates in interactions with a subset of the spliceosomal U1 snRNP complex. Biochim Biophys Acta 1814: 1812–1824 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Mapendano CK, Jensen TH 2011. An ending is a new beginning: Transcription termination supports re-initiation. Cell Cycle 10: 863–865 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Manley JL 2002. Nuclear coupling: RNA processing reaches back to transcription. Nat Struct Biol 9: 790–791 [DOI] [PubMed] [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH 2010. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 40: 410–422 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ 2002. The human and mouse replication-dependent histone genes. Genomics 80: 487–498 [PubMed] [Google Scholar]
- Mendell JT, ap Rhys CM, Dietz HC 2002. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science 298: 419–422 [DOI] [PubMed] [Google Scholar]
- Montzka KA, Steitz JA 1988. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc Natl Acad Sci 85: 8885–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D 2009. ‘Cotranscriptionality’: The transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ 2011. Ending the message: Poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy L, Ingledue TC, Pilch DR, Kay BK, Marzluff WF 1996. Increasing the distance between the snRNA promoter and the 3′ box decreases the efficiency of snRNA 3′-end formation. Nucleic Acids Res 24: 4525–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiluttini B, Gu B, Belagal P, Smirnova AS, Nguyen VT, Hebert C, Schmidt U, Bertrand E, Darzacq X, Bensaude O 2010. Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J Cell Sci 123: 2085–2093 [DOI] [PubMed] [Google Scholar]
- Tian H 2001. RNA ligands generated against complex nuclear targets indicate a role for U1 snRNP in coordinating transcription and RNA splicing. FEBS Lett 509: 282–286 [DOI] [PubMed] [Google Scholar]
- Vagner S, Ruegsegger U, Gunderson SI, Keller W, Mattaj IW 2000. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA 6: 178–188 [DOI] [PMC free article] [PubMed] [Google Scholar]